Abstract
Photobacterium damsela
α2,6-sialyltransferase was cloned as N- and C- His-tagged fusion proteins with different lengths (16–497 aa or 113–497 aa). Expression and activity assays indicated that the N-terminal 112 amino acid residues of the protein were not required for its α2,6-sialyltransferase activity. Among four truncated forms tested, N-His-tagged Δ15Pd2,6ST(N) containing 16–497 amino acid residues had the highest expression level. Similar to the Δ15Pd2,6ST(N), the shorter Δ112Pd2,6ST(N) was active in a wide pH range of 7.5–10.0. A divalent metal ion was not required for the sialyltransferase activity, and the addition of EDTA and dithiothreitol did not affect the activity significantly.
Keywords: Carbohydrate, Expression, Photobacterium damsela, Sialic acid Sialyltransferase
Introduction
Sialyltransferases (EC 2.4.99.-) are key enzymes in the biosynthesis of sialoglycoconjugates (sialic acid-containing glycoconjugates) (Harduin-Lepers et al. 1995). They catalyze the reaction that transfers a sialic acid residue from its activated sugar nucleotide donor cytidine 5′-monophosphate sialic acid (CMP-sialic acid) to an acceptor, usually a structure with a galactose, N-acetylgalactosamine (GalNAc) or another sialic acid at the non-reducing terminus.
Photobacterium damsela α2,6-sialyltransferase (Pd2,6ST or sialyltransferase 0160) (GenBank accession number: BAA25316) was the first bacterial α 2,6-sialyltransferase that has ever been cloned and expressed in E. coli (Yamamoto et al. 1998). The flexible donor and acceptor substrate specificities of the enzyme were described and the enzyme was applied in the synthesis of α2,6-linked sialosides and glycopeptides (Yamamoto et al. 1998a; Yu et al. 2006; Yamamoto et al. 1998b; Kajihara et al. 1996; Teo et al. 2005; Yamamoto et al. 1996; Endo et al. 2001). Pd2,6ST shares amino acid sequence homology with a multifunctional Pasteurella multocida sialyltransferase PmST1 encoded by gene Pm0188 (GenBank accession numbers: AAY89061, AAK02272) (Yu et al. 2005), a Haemophilus ducreyi α2,3-sialyltransferase encoded by gene Hd0053 (GenBank accession number: AAP95068) (Li et al. 2007), a Photobacterium phosphoreum α2,3-sialyltransferase encoded by gene pst3–467 (GenBank accession number: BAF63530) (Tsukamoto et al. 2007), and a Photobacterium leiognathi α2,6-sialyltransferase (Yamamoto et al. 2007). These proteins do not share sequence homology with any other reported bacterial or mammalian glycosyltransferases. They have been classified into glycosyltransferase family GT80 on the Carbohydrate-Active enZyme database (CAZy – http://www.cazy.org/index.html) (Coutinho et al. 2003; Campbell et al. 1997).
Unlike other sialyltransferases in the GT80 family, Pd2,6ST has an extra C-terminal domain (D498 to D675) showing high homology to the bacterial phosphate transport system regulatory protein PhoU. This PhoU domain has been proven to be unrelated to the sialyltransferase activity of Pd2,6ST (Yamamoto et al. 1998a; Yu et al. 2006). The lipobox-containing N-terminal 15 amino acids deleted in the purified native Pd2,6ST enzyme (Tsukamoto et al. 2007) may function as a signal sequence. This is similar to the Photobacterium phosphoreum α2,3-sialyltransferase, for which a lipobox signal sequence was also identified (Tsukamoto et al.2007).
Compared to other sialyltransferses in the GT80 family, Pd2,6ST has an extra N-terminal domain (C16 to P112). By cloning and characterizing four constructs of Pd2,6ST fusion proteins differing in terms of the length of the protein and the position of the Histidine tag, we show here that the N-terminal 112 amino acid residues are not required for the sialyltransferase activity of Pd2,6ST. This finding will help the structural characterization of this important enzyme by NMR and crystal structure studies.
Material and methods
Chemicals and reagents
Restriction enzymes and T4 DNA ligase were purchased from Promega and New England Biolabs. E. coli competent cell DH5α and BL21(DE3) were purchased from Invitrogen. Pasteurella damsela (ATCC #33539) α2,6-sialyltransferase gene in a TOPO vector was a kind gift from Dr. Ajit Varki at the University of California – San Diego.
Cloning
Using the plasmid of Photobacterium damsela (ATCC # 33539) α2,6-sialyltransferase gene in a TOPO vector as the template for polymerase chain reactions (PCR), four different constructs in pET15b or pET22b(+) vector were obtained for expressing N- or C- His6-tagged fusion proteins. Forward primer 5′ GATCCATATGTGTAATAGTGACAATACCAGC 3′ (NdeI restriction site is underlined) was used to clone 15Pd2,6ST(N) in pET15b vector and 15Pd2,6ST(C) in pET22b(+) vector. Forward primer 5′ GATCCATATGACGCTAGAAGTTTACATCGATC 3′ (NdeI restriction site is underlined) was used to clone Δ112Pd2,6ST(N) in pET15b vector and Δ112Pd2,6ST(C) in pET22b(+) vector. Reverse primer 5′ CGCGGATCCTTAAGCCCAGAACAGAACATC 3′ (BamHI restriction site is underlined) was used to clone both Δ15Pd2,6ST(N) and Δ112Pd2,6ST(N) in pET15b vector, and reverse primer 5′CGCGGATCCTTAGTGATGATGATGATGATGAGCCC AGAACAGAACATCTTTTTC 3′ (BamHI restriction site is underlined, codons encoding hexahistidine are in italics) was used to clone both Δ15Pd2,6ST(C) and Δ112Pd2,6ST(C) in pET22b(+) vector. PCR reactions were performed in a 50 μl reaction mixture using Herculase enhanced DNA polymerase (Stratagene) according to manufacturer’s instruction. PCR product was purified and digested with NdeI and BamHI restriction enzymes. The purified and digested PCR product was ligated with predigested pET15b or pET22b(+) vector and transformed into E. coli DH5α electrocompetent cells. Selected clones were grown for minipreps and characterization by restriction mapping. DNA sequencing was performed by Davis Sequencing Facility at the University of California-Davis.
Expression
Positive plasmids were selected and transformed into BL21 (DE3) chemical competent cells. Plasmid-bearing E. coli strain was cultured in LB rich medium (10 g l−1 tryptone, 5 g l−1 yeast extract, and 10 g l−1 NaCl) supplemented with ampicillin (100 μg ml−1). Overexpression of target proteins was achieved by inducing the E. coli culture with 0.1 mM of isopropyl-1-thio-β-D-galactopyranoside (IPTG) when the OD600 nm of the culture reached 0.8–1.0, followed by incubating the culture at 20 °C for 24 h with vigorous shaking at 250 rpm in a C25KC incubator shaker (New Brunswick Scientific, Edison, NJ).
Purification of Pd2,6ST by affinity chromatography
His6-tagged target proteins were purified from the cell lysate using ÄKTA FPLC system (GE Healthcare Life Sciences). To obtain the cell lysate, cells were harvested by centrifugation at 4000 × g for 2 hrs. Cell pellets were then re-suspended in 20 ml of lysis buffer (Tris-HCl, pH 8.0, 100 mM, 0.1% Triton X-100) containing lysozyme (100 μg ml−1) and DNaseI (3 μg ml−1). After incubating the mixture at 37 °C for 50 min with vigorous shaking, the cell lysate was collected by centrifugation at 12,000 × g for 30 min. The supernatant (lysate) was applied to a HisTrap™ FF 5 ml column (GE Healthcare). The column was then washed with 10 volumes of binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl, pH 7.5), 15 volumes of washing buffer (30 – 50 mM imidazole, 0.5 NaCl, 20 mM Tris-HCl, pH 7.5), followed by 8 volumes of elute buffer (200 mM imidazole, 0.5 NaCl, 20 mM Tris-HCl, pH 7.5). The fractions containing the purified enzyme were collected, dialyzed against Tris-HCl buffer (20 mM, pH 7.5) containing 10% glycerol, and stored at 4 °C.
Quantification of purified protein
Protein concentration was determined in a 96-well plate using Bicinchoninic acid (BCA) Protein Assay Kit (Pierce Biotechnology, Rockford, IL) with bovine serum albumin as a protein standard. The absorbance of each sample was measured at 562 nm using a Bio-Tek EL311SX plate reader.
pH profile by HPLC
Typical enzymatic assays were performed in a 20 μl reaction mixture containing a buffer (200 mM) with a pH in the range of 5.0–11.0, CMP-Neu5Ac (1 mM), 4-methylumbelliferyl β-D-lactoside (LacMU) (1 mM), and Δ15Pd2,6ST(N) (240 ng) or Δ112Pd2,6ST(N) (285 ng). Reactions were allowed to proceed for 30 min at 37 °C and were quenched by adding ice-cold 12% acetonitrile to make 60-fold dilutions. The samples were then kept on ice until an aliquot of 8 μl was injected and analyzed by a Shimadzu LC-2010A system equipped with a membrane online degasser, a temperature control unit and a fluorescence detector. A reverse phase Premier C18 column (250 × 4.6 mm I.D., 5 μm particle size, Shimadzu) protected with a C18 guard column cartridge was used. The mobile phase was 12% acetonitrile. The fluorescent compounds LacMU and Neu5Acα2,6LacMU were detected by excitation at 325 nm and emission at 372 nm (Kajihara et al. 1999; Kajihara et al. 2004).
Results and discussion
Cloning, expression and purification of four constructs of Pd2,6ST
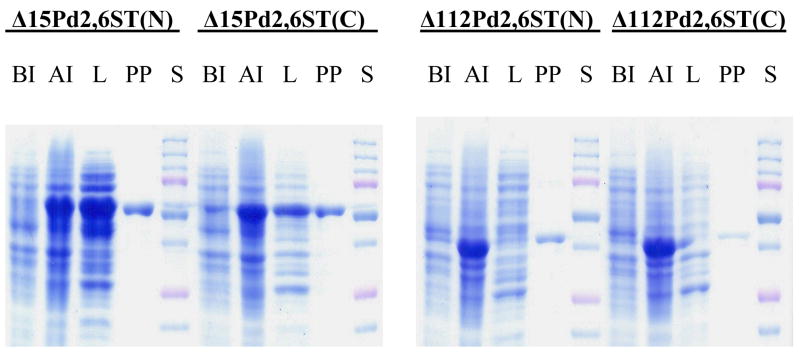
The N-terminal 15 amino acids and the C-terminal domain containing amino acid residues D498-D675 of Pd2,6ST have been shown to be unrelated to its α2,6-sialyltransferase activity (Yu et al. 2006). Amino acid sequence alignments of the five reported sialyltransferase members in the GT80 family indicate that compared to the multifunctional Pasteurella multocida sialyltransferase encoded by Pm0188 (Yu et al. 2005), the Haemophilus ducreyi α2,3-sialyltransferase encoded by gene Hd0053 (Li et al. 2007), and the Photobacterium phosphoreum α2,3-sialyltransferase encoded by gene pst3–467 (Tsukamoto et al. 2007), Pd2,6ST has an extra domain containing amino acid residues C16-P112. A similar domain is also found in the Photobacterium leiognathi α2,6-sialyltransferase (Yamamoto et al. 2007). In order to determine whether this extra domain in Pd2,6ST is related to its sialyltransferase activity, four different Pd2,6ST constructs including Δ15Pd2,6ST(N), Δ15Pd2,6ST(C), Δ112Pd2,6ST(N), and Δ112Pd2,6ST(C) were cloned. They differ from each other in terms of the length of the protein and the position of the Histidine tag (at N- or C-terminus). As shown in Fig. 1, all four Pd2,6ST proteins could be expressed in large quantity (> 60% of the whole cell extract) in E. coli after IPTG induction (lanes AI). While both Δ15Pd2,6ST(N) and Δ15Pd2,6ST(C) having an N-terminal 15 amino acid residues deletion were quite soluble, only minimal amounts of the targeted proteins were present in the soluble portion of the cell extracts (cell lysate, lanes L in Fig. 1) of Δ112Pd2,6ST(N) and Δ112Pd2,6ST(C) having an N-terminal 112 amino acid residues deletion. A single protein band in the purified fractions (lanes PP in Fig. 1) indicated that onestep affinity column purification using a HisTrap™ FF 5 ml column in ÄKTA FPLC system was efficient for all four constructs.
Fig. 1.
Sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analyses for four different Pd2,6ST constructs. Lanes: S, Bio-Rad Precision Plus Protein Standards (10–250 kDa); BI, whole cell extract before induction; AI, whole cell extract after induction; L, lysate; PP, HisTrap™ FF 5 ml column purified protein. SDS-PAGE was performed in 10% Tris–glycine gels using Bio-Rad Mini-protein III cell gel electrophoresis unit (Bio-Rad, Hercules, CA) at DC = 150 V. Gels were stained with Coomassie Blue R-250.
As a result, up to 36 mg of purified Δ15Pd2,6ST(N) could be routinely obtained from one liter of E. coli culture (Table 1). The expression level (24 mg per liter culture) of soluble Δ15Pd2,6ST(C) was 33% less than that of the Δ15Pd2,6ST(N). The amounts of soluble Δ112Pd2,6ST(N) and Δ112Pd2,6ST(C) that could be purified from one liter E. coli cell culture were about 20 fold less than the ones missing only the first 15 amino acid residues. These data indicated that the amino acid residues C16-P112 of Pd2,6ST were important for the solubility of the recombinant proteins. The position of the Histidine-tag also affected, to some extent, the solubility of the resulted fusion protein.
Table 1.
Expression levels of purified recombinant Pd2,6STs from E. coli
| Enzymes | Δ15Pd2,6ST(N) | Δ15Pd2,6ST(C) | Δ112Pd2,6ST(N) | Δ112Pd2,6ST(C) |
|---|---|---|---|---|
| Amino acid residues | 16 – 497 aa | 16 – 497 aa | 113 – 497 aa | 113 – 497 aa |
| His-tag position | N-His tag | C-His tag | N-His tag | C-His tag |
| Expression level (mg l−1) | 36.0 | 24.4 | 1.6 | 1.1 |
Kinetics of Δ15Pd2,6ST(N) and Δ112Pd2,6ST(N)
In order to determine whether deleting the amino acid residues C16-P112 of Pd2,6ST affects the kinetic parameters of the recombinant proteins, kinetic studies were carried out for Δ15Pd2,6ST(N) and Δ112Pd2,6ST(N). The results are liste in Table 2. The apparent KM values of both donor CMP-Neu5Ac and acceptor LacMU for Δ Δ112Pd2,6ST(N) are similar to those of Δ15Pd2,6ST(N). The KM values of CMP-Neu5Ac for Δ15Pd2,6ST(N) and Δ112Pd2,6ST(N) (both are 0.9 mM) are in the same magnitude as the KM of Pd2,6ST 0160 purified from Photobacterium damsela (0.32 mM) (Yamamoto et al. 1996). They are about 100-fold lower than the KM of rat liver α2,6-sialyltransferase (7.59 μM) (Angata et al. 1998). The KM values of LacMU for Δ15Pd2,6ST(N) (0.8 mM) and Δ112Pd2,6ST(N) (0.6 mM) are about one magnitude less than the KM of lactose (6.82 mM) and N-acetyllactosaminide (8.95 mM) for Pd2,6ST 0160 (Yamamoto et al. 1996). The kcat values of both CMP-Neu5Ac and LacMU for Δ15Pd2,6ST(N) are about 2-fold of those for Δ112Pd2,6ST(N), leading to about 2-fold increase in kcat/KM values for Δ15Pd2,6ST(N). Taken together, these results indicate that the amino acid residues C16-P112 of Pd2,6ST do not affect the binding affinity of the protein to the substrates, but slightly affect the turnover rate of the enzyme.
Table 2.
Apparent kinetic parameters of Pd2,6STsa
| Enzymes | Δ15Pd2,6ST(N) | Δ112Pd2,6ST(N) | ||
|---|---|---|---|---|
| Substrates | CMP-Neu5Ac | LacMU | CMP-Neu5Ac | LacMU |
| KM (mM) | 0.9 ± 0.2 | 0.8 ± 0.1 | 0.9 ± 0.3 | 0.6 ± 0.1 |
| Vmax (mMs−1) | (4.6 ± 0.4) × 10−4 | (4.0 ± 0.2) × 10−4 | (4.2 ± 0.3) × 10−4 | (3.4 ± 0.2) × 10−4 |
| kcat (s−1) | 2.7 ± 0.2 | 2.3 ± 0.1 | 1.4 ± 0.1 | 1.1 ± 0.1 |
| kcat/KM (s−1 mM−1) | 2.9 | 3.0 | 1.6 | 1.9 |
Assays were performed in duplicate in a reaction mixture of 20 μl containing Tris-HCl buffer (100 mM, pH 8.5) using varied concentrations (0.1, 0.25, 0.4, 1, 2, and 4 mM) of LacMU and a fixed concentration (1 mM) of CMP-Neu5Ac, or a fixed concentration (1 mM) of LacMU and varied concentrations (0.1, 0.25, 0.4, 1, 2, and 4 mM) of CMP-Neu5Ac. The amount of Δ15Pd2,6ST(N) used was 190 ng, and the amount of Δ112Pd2,6ST(N) used was 285 ng. Reactions were allowed to proceed for 30 min at 37 °C. Apparent kinetic parameters were obtained by fitting the data (the average values of duplicate assay results) into the Michaelis-Menten equation using Grafit 5.0.
The native function of the amino acid sequence C16-P112 in marine bacterium Photobacterium damsela is currently unknown. Pd2,6ST itself may be involved in the biosynthesis of α2,6-linked sialosides on the surface of the bacterium. Indeed, glycans containing α2,6-linked sialic acid have been detected on the surface of Photobacterium leiognathi JT-SHIZ-145 using SSA lectin staining (Yamamoto et al. 2007). More detailed glycobiology studies are yet to be carried out in order to fully understand the biological functions of these bacterial sialyltransferases.
pH profile of Δ15Pd2,6ST(N) and Δ112Pd2,6ST(N)
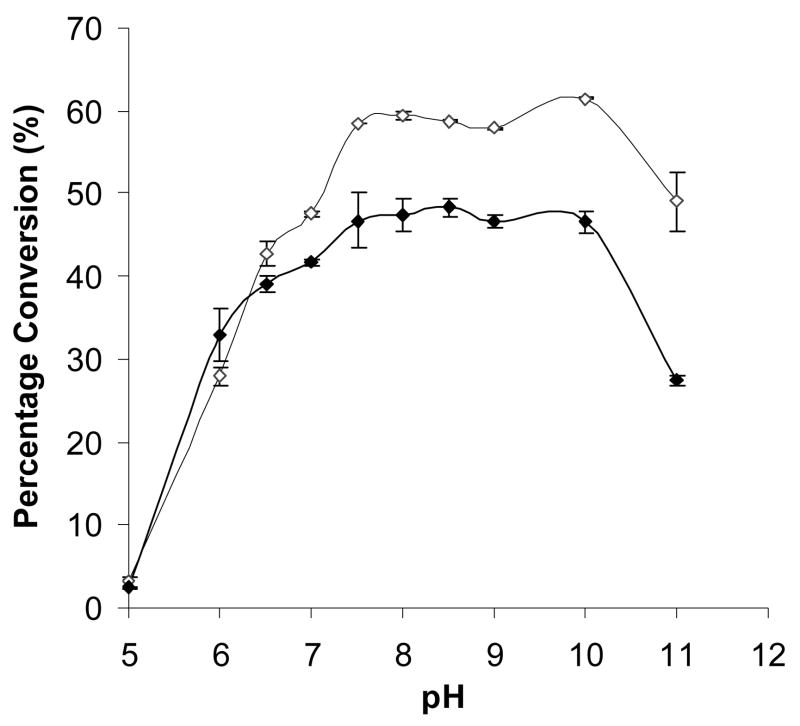
As shown in Fig. 2, the pH profiles of Δ15Pd2,6ST(N) and Δ112Pd2,6ST(N) are very similar. Both enzymes are active in a wide pH range varying from 7.5 to 10.0 with about 5% variation in the production of Neu5Acα2,6LacMU. The activities of both enzymes decline quickly when pH goes below 6.5. The activities drop to about 5% when the pH is at 5.0 (Fig. 2). This property is different from previous reports which found optimal pH values of 5.0 and 6.0 for Pd2,6ST 0160 (Yamamoto et al. 1996) and a truncated recombinant enzyme (Teo et al. 2005), respectively.
Fig. 2.
The pH profile of Δ15Pd2,6ST(N) and Δ112Pd2,6ST(N) (100% conversion was defined as the formation of 1 mM Neu5Acα2,6LacMU). Activity was measured in duplicate at indicated pH at 37 °C for 30 min. Buffers (200 mM) used were: MES, pH 5.0–6.0; Hepes, pH 7.0; Tris–HCl, pH 7.5–9.0; and CAPS, pH 10.0–11.0. Symbols and amounts of enzymes used: Δ15Pd2,6ST(N) (⋄, unfilled diamond), 240 ng; Δ112Pd2,6ST(N) (◆, filled diamond), 285 ng.
Effects of metal ions, reducing agent DTT, and chelating agent EDTA
A Tris-HCl buffer of pH 8.5 was chosen for the assays to compare the effects of metal ions Mg2+ and Mn2+, reducing agent DTT, and chelating agent EDTA on the activity of Δ15Pd2,6ST(N) and Δ112Pd2,6ST(N). and Δ112Pd2,6ST(N). As shown in Table 3, the presence of various concentrations of metal ion Mg2+ did not affect the α2,6-sialyltransferase activity of either Δ15Pd2,6ST(N) or Δ112Pd2,6ST(N). The addition of 5 mM chelating agent EDTA did not affect the activity significantly, indicating a metal ion was not required for the activity of the enzymes. However, addition of 10 mM of Mn2+ ion decreased the activity of Δ15Pd2,6ST(N) to 60% but did not affect the activity of Δ112Pd2,6ST(N). Increasing the concentration of Mn2+ to 20 mM caused the decrease of the activity of both Δ15Pd2,6ST(N) and Δ112Pd2,6ST(N). The detrimental effect of Mn2+ ion was similar to that reported for PmST1 (Yu et al. 2005).
Table 3.
Effects of metal ions, EDTA and DTT on the α2,6-sialyltransferase activity of Δ15Pd2,6ST(N) and Δ112Pd2,6ST(N)a
| Reactions | Δ15Pd2,6ST(N) | Δ112Pd2,6ST(N) |
|---|---|---|
| Control | 50% | 43% |
| MgCl2 | 48% | 43–44% |
| 5 mM MnCl2 | 44% | 39% |
| 10 mM MnCl2 | 28% | 38% |
| 20 mM MnCl2 | 15% | 21% |
| DTT | 51–52% | 45–46% |
| 5 mM EDTA | 43% | 35% |
The numbers in the table represent the percentage conversions (Standard Deviation ≤ 1.4%). The activity was determined in duplicate at 37 °C for 30 min (100% conversion was defined as the formation of 1 mM Neu5Acα2,6LacMU). Different concentrations (5 mM, 10 mM, and 20 mM) of MgCl2 or MnCl2, 5 mM EDTA, and various concentrations (0.2 mM, 1 mM, and 5 mM) of DTT were used in a reaction mixture of 20μl containing Tris-HCl buffer (pH 8.5, 100 mM), CMP-Neu5Ac (1 mM), LacMU (1 mM), and Δ15Pd2,6ST(N) (190 ng) or Δ112Pd2,6ST(N) (285ng). Reaction in the absence of DTT, EDTA, or metal ion was used as a control.
There are three cysteine residues (C16, C60, and C89) in the amino acid sequence C16-P112 of Pd2,6ST. The addition of DTT up to 5 mM did not affect the sialyltransferase activity of both Δ15Pd2,6ST(N) and Δ112Pd2,6ST(N), indicating that disulfide bond formation in Δ15Pd2,6ST(N) was unlikely.
In conclusion, we show here that the N-terminal 112 amino acid residues in Pd2,6ST are not required for the α2,6-sialyltransferase activity, but affect the solubility of the recombinant protein expressed in E. coli.
Acknowledgments
This work was supported by NIH R01GM076360 and the start-up funds from the Regents of the University of California.
References
- Angata T, Matsuda T, Kitajima K. Synthesis of neoglycoconjugates containing deaminated neuraminic acid (KDN) using rat liver α2,6-sialyltransferase. Glycobiology. 1998;8:277–284. doi: 10.1093/glycob/8.3.277. [DOI] [PubMed] [Google Scholar]
- Campbell JA, Davies GJ, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326:929–939. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- Endo T, Koizumi S, Tabata K, Kakita S, Ozaki A. Large-scale production of the carbohydrate portion of the sialyl-Tn epitope, alpha-Neup5Ac-(2→6)-D-GalpNAc, through bacterial coupling. Carbohydr Res. 2001;330:439–443. doi: 10.1016/s0008-6215(01)00007-6. [DOI] [PubMed] [Google Scholar]
- Harduin-Lepers A, Recchi MA, Delannoy P. 1994, the year of sialyltransferases. Glycobiology. 1995;5:741–758. doi: 10.1093/glycob/5.8.741. [DOI] [PubMed] [Google Scholar]
- Kajihara Y, Yamamoto T, Nagae H, Nakashizuka M, Sakakibara T, Terada I. A novel alpha-2,6-sialyltransferase: transfer of sialic acid to fucosyl and sialyl trisaccharides. J Org Chem. 1996;61:8632–8635. [Google Scholar]
- Kajihara Y, Akai S, Nakagawa T, Sato R, Ebata T, Kodama H, Sato K. Enzymatic synthesis of Kdn oligosaccharides by a bacterial alpha-(2→6)-sialyltransferase. Carbohydr Res. 1999;315:137–141. doi: 10.1016/s0008-6215(98)00331-0. [DOI] [PubMed] [Google Scholar]
- Kajihara Y, Kamiyama D, Yamamoto N, Sakakibara T, Izumi M, Hashimoto H. Synthesis of 2-[(2-pyridyl)amino]ethyl beta-D-lactosaminide and evaluation of its acceptor ability for sialyltransferase: a comparison with 4-methylumbelliferyl and dansyl beta-D-lactosaminide. Carbohydr Res. 2004;339:1545–1550. doi: 10.1016/j.carres.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Li Y, Sun M, Huang S, Yu H, Chokhawala HA, Thon V, Chen X. The Hd0053 gene of Haemophilus ducreyi encodes an alpha2,3-sialyltransferase. Biochem Biophys Res Commun. 2007;361:555–560. doi: 10.1016/j.bbrc.2007.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo C, Hwang T, Chen P, Hung C, Gao H, Chang L, Lin C. Synthesis of sialyl TN glycopeptides - enzymatic sialylation by α2,6-sialyltransferase from Photobacterium damsela. Adv Synth Catal. 2005;347:967–972. [Google Scholar]
- Tsukamoto H, Takakura Y, Yamamoto T. Purification, cloning, and expression of an α/β-galactoside α-2,3-sialyltransferase from a luminous marine bacterium, Photobacterium phosphoreum. J Biol Chem. 2007;282:29794–29802. doi: 10.1074/jbc.M701907200. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Hamada Y, Ichikawa M, Kajiwara H, Mine T, Tsukamoto H, Takakura Y. A β-galactoside α2,6-sialyltransferase produced by a marine bacterium, Photobacterium leiognathi JT-SHIZ-145, is active at pH 8. Glycobiology. 2007 doi: 10.1093/glycob/cwm086. (Advance Access published on August 17, 2007) [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Nagae H, Kajihara Y, Terada I. Mass production of bacterial alpha 2,6-sialyltransferase and enzymatic syntheses of sialyloligosaccharides. Biosci Biotechnol Biochem. 1998b;62:210–214. doi: 10.1271/bbb.62.210. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Nakashizuka M, Kodama H, Kajihara Y, Terada I. Purification and characterization of a marine bacterial beta-galactoside alpha 2,6-sialyltransferase from Photobacterium damsela JT0160. J Biochem (Tokyo) 1996;120:104–110. doi: 10.1093/oxfordjournals.jbchem.a021370. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Nakashizuka M, Terada I. Cloning and expression of a marine bacterial beta-galactoside alpha2,6-sialyltransferase gene from Photobacterium damsela JT0160. J Biochem (Tokyo) 1998a;123:94–100. doi: 10.1093/oxfordjournals.jbchem.a021921. [DOI] [PubMed] [Google Scholar]
- Yu H, Chokhawala H, Karpel R, Wu B, Zhang J, Zhang Y, Jia Q, Chen X. A multifunctional Pasteurella multocida sialyltransferase: a powerful tool for the synthesis of sialoside libraries. J Am Chem Soc. 2005;127:17618–17619. doi: 10.1021/ja0561690. [DOI] [PubMed] [Google Scholar]
- Yu H, Huang S, Chokhawala H, Sun M, Zheng H, Chen X. Highly efficient chemoenzymatic synthesis of naturally occurring and non-natural alpha-2,6-linked sialosides: a P. damsela alpha-2,6-sialyltransferase with extremely flexible donor-substrate specificity. Angew Chem Int Ed Engl. 2006;45:3938–3944. doi: 10.1002/anie.200600572. [DOI] [PMC free article] [PubMed] [Google Scholar]