Abstract
We report a preliminary demonstration of the accurate mass and time (AMT) tag approach for lipidomics. Initial data-dependent LC-MS/MS analyses of human plasma, erythrocyte, and lymphocyte lipids were performed in order to identify lipid molecular species in conjunction with complementary accurate mass and isotopic distribution information. Identified lipids were used to populate initial lipid AMT tag databases containing 250 and 45 entries for those species detected in positive and negative electrospray ionization (ESI) modes, respectively. The positive ESI database was then utilized to identify human plasma, erythrocyte, and lymphocyte lipids in high-throughput LC-MS analyses based on the AMT tag approach. We were able to define the lipid profiles of human plasma, erythrocytes, and lymphocytes based on qualitative and quantitative differences in lipid abundance.
Keywords: lipidomics, AMT tag approach, capillary liquid chromatography, mass spectrometry
INTRODUCTION
Lipids traditionally have been thought to play no other role in biological systems other than as components of cell membranes. However, research over the last two decades has proven that lipids can perform functions typically assigned to nucleic acids, proteins, and metabolites. It is now established that lipids play direct roles in exocytosis [1], ion-channel regulation [2–6], membrane domain formation [7,8], and cell signaling [9–12]. They are also known to display more systemic functions. For example, prostaglandins are enzymatic oxidation products of arachidonic acid that promote inflammation [13]. Alternatively, isoprostanes are non-enzymatic oxidation products of arachidonic acid that have been traditionally used as indicators of oxidative stress [14] but are now being shown to have a number of biological activities [15]. Finally, reactive products of lipid peroxidation are known to accumulate on tissue proteins and negatively impact their structures and functions [16,17].
The combination of the “-omics” era and the emerging roles of lipids in biological processes has logically resulted in the field of lipidomics, which can be defined as the quantitative measurement of time-related or stimuli-dependent perturbations in the lipid complement of an integrated biological system, cell, or cell types. The application of lipidomics in the study of comparative samples (e.g. for biomarker discovery) results in a paradox, i.e. how does one obtain comprehensive yet quantitative coverage of the lipidome in a high-throughput, cost-effective manner? Typical lipidomics experiments involve mass spectrometry (MS)-based analyses of lipid extracts either with or without online separation of individual species by liquid chromatography (LC) [18–22]. Tandem mass spectrometry (MS/MS) methods such as precursor-ion, product-ion, and neutral-loss scans are often used to both identify and quantify lipid species, but it is difficult to perform all three scan types simultaneously. Therefore, MS data acquisition methods that stagger the scan types or multiple MS experiments each consisting of unique scan types are required in order to obtain comprehensive and quantitative coverage of the lipidome. Such approaches are time consuming, particularly when coupled with online LC separations that provide increased coverage of the lipidome.
We have developed the accurate mass and time (AMT) tag approach [23] to address an analogous situation in LC-MS-based proteomics studies. In this approach, initial LC-MS/MS analyses are performed on pre-fractionated peptide samples in order to provide peptide sequence identifications. These experiments are relatively low throughput because the peptide pre-fractionation can be quite extensive and require separate LC-MS/MS analyses for each fraction. However, the identified peptide sequences are cataloged in an AMT tag database that includes, among other parameters, their calculated monoisotopic masses (based on their identified sequences) and observed normalized LC elution times (NETs). Subsequent quantitative LC-MS analyses of the same or similar samples are performed utilizing a time-of-flight (TOF) or Fourier transform ion cyclotron resonance (FTICR) MS, and peptide features (characterized by observed monoisotopic mass and elution time) detected during these experiments are then compared to entries in the AMT tag database in order to identify individual peptides. The high mass measurement accuracy (MMA) obtained during LC-TOF or LC-FTICR MS experiments is exploited together with chromatographic alignment in order to provide confident peptide matches. These are high-throughput analyses relative to the initial LC-MS/ MS experiments because the TOF or FTICR MS provides higher dynamic range and sensitivity that allows the analysis of unfractionated samples without the undersampling issues associated with MS/MS analyses [23].
In this study, we report a preliminary demonstration of the AMT tag approach for lipidomics. Initial data-dependent LC-MS/MS analyses of human blood lipids were performed in order to identify as many lipid species as possible for entry into a lipid AMT tag database. High-throughput LC-MS analyses were then performed to provide relative quantitation data for those species matching the database. This preliminary work has resulted in initial lipid AMT tag databases containing 272 total entries for species detected in both positive and negative electrospray ionization (ESI) and corresponding to several lipid classes. The positive ESI database was then utilized to identify human plasma, erythrocyte, and lymphocyte lipids in high-throughput quantitative LC-MS analyses based on the AMT tag approach.
EXPERIMENTAL
Reagents and materials
HPLC-grade (Optima) water, acetonitrile, and methanol were purchased from Fisher Scientific (Fair Lawn, NJ). MS-grade ammonium acetate and triethylamine (TEA) were obtained from Fluka (St. Louis, MO). Human citrated plasma samples were received frozen on dry ice from Dr. Lawrence S. Phillips of Emory University via the National Institutes of Health (NIH, Bethesda, MD) Sample Repository. Human erythrocyte and lymphocyte cells suspended in 0.9% saline solution containing 10% dimethylsulfoxide were also received from Dr. Phillips via the NIDDK Central Repository. Plasma, erythrocytes, and lymphocytes were isolated from the same individuals.
Human plasma lipid extraction
This work was approved by the Institutional Review Board of the Pacific Northwest National Laboratory. For LC optimization and lipid identification (LC-MS/MS) experiments, 30 µL of plasma from 10 healthy individuals was pooled. Plasma lipids were then extracted via the addition of 120 µL cold (−20°C) chloroform/methanol (2:1, v/v) [24] to 30 µL plasma, and the mixture was vortexed for 10 s (chloroform is a carcinogen, therefore necessary precautions should be taken to limit human exposure). The sample was then allowed to stand at room temperature for 5 min, followed by vortexing for 10 s. Protein was separated from the two liquid phases by centrifugation at 2,000 rpm for 10 min. The lower lipid phase was removed by pipetting and placed into a sterile, siliconized eppendorf tube, while the protein interlayer and upper aqueous phase were discarded. The lipid phase was then dried in vacuo and stored at −80°C until analysis. Prior to analysis, dried lipid extracts were reconstituted in 125 µL methanol and centrifuged at 12,000 rpm for 10 min to remove any particulates.
For lipid profiling experiments of individual samples, 30 µL of plasma was extracted in triplicate with chloroform/methanol as described above. The lower lipid phase was removed by pipetting, dried in vacuo, and stored at −80°C until analysis. Prior to analysis, dried lipid extracts were reconstituted in 125 µL methanol and centrifuged at 12,000 rpm for 10 min to remove any particulates.
Human erythrocyte and lymphocyte lipids
For human erythrocyte lipid extraction, 100 µL of erythrocyte cell suspension from 10 healthy individuals was pooled to create a uniform sample for use in LC optimization and lipid identification (LC-MS/MS) experiments. Erythrocytes were then lysed over 2 min by sonicating water bath, and lipids were extracted via the addition of 400 µL cold (−20°C) chloroform/methanol (2:1 v/v) to 100 µL of pooled erythrocyte cell suspension. The mixture was vortexed for 10 s, allowed to stand at room temperature for 5 min, and again vortexed for 10 s. Protein was separated from the two liquid phases by centrifugation at 2,000 rpm for 10 min. The lower lipid phase was removed by pipetting and placed into sterile, siliconized eppendorf tubes, while the protein interlayer and upper aqueous phase were discarded. The lipid phase was then dried in vacuo and stored at −80°C until analysis. Prior to analysis, dried lipid extracts were reconstituted in 125 µL methanol and centrifuged at 12,000 rpm for 10 min to remove any particulates.
For lipid profiling experiments of individual samples, 100 µL of erythrocyte cell suspension was extracted in triplicate with chloroform/methanol as described above. The lower lipid phase was removed by pipetting, dried in vacuo, and stored at −80°C until analysis. Prior to analysis, dried lipid extracts were reconstituted in 125 µL methanol and centrifuged at 12,000 rpm for 10 min to remove any particulates.
For human lymphocyte lipid extraction, 30 µL of lymphocyte cell suspension from 10 healthy individuals was pooled to create a uniform sample for use in LC optimization and lipid identification (LC-MS/MS) experiments. Lymphocytes were then lysed over 2 min by sonicating water bath, and lipids were extracted via the addition of 120 µL cold (−20°C) chloroform/methanol (2:1 v/v) to 30 µL of pooled lymphocyte cell suspension. The mixture was vortexed for 10 s, allowed to stand at room temperature for 5 min, and again vortexed for 10 s. Protein was separated from the two liquid phases by centrifugation at 2,000 rpm for 10 min. The lower lipid phase was removed by pipetting and placed into sterile, siliconized eppendorf tubes, while the protein interlayer and upper aqueous phase were discarded. The lipid phase was then dried in vacuo and stored at −80°C until analysis. Prior to analysis, dried lipid extracts were reconstituted in 125 µL methanol and centrifuged at 12,000 rpm for 10 min to remove any particulates.
For lipid profiling experiments of individual samples, 30 µL of lymphocyte cell suspension was extracted in triplicate with chloroform/methanol as described above. The lower lipid phase was removed by pipetting, dried in vacuo, and stored at −80°C until analysis. Prior to analysis, dried lipid extracts were reconstituted in 125 µL methanol and centrifuged at 12,000 rpm for 10 min to remove any particulates.
Preparation of packed capillary columns
Capillary LC columns were slurry-packed with 5 µm Jupiter C18-RP particles (Phenomenex, Torrance, CA) as previously described [25]. Briefly, the stationary phase was added to a stainless steel reservoir, to which an empty fused silica capillary (Polymicro Technologies, Phoenix, AZ) was connected. The opposite end of the capillary was connected to a stainless steel union (Valco, Houston, TX) containing a stainless steel screen (2 µm mesh, Valco) that served as a frit. Acetonitrile was used as the packing solvent and was delivered at constant pressure by syringe pump (ISCO, Lincoln, NE). Initially, a pressure of 100 psi was applied. The pressure was then increased stepwise to and held constant at 7,000 psi for 5–10 min under sonication.
RP-LC separation of lipids
An automated LC system with two 150 µm × 65 cm capillary columns was used. Detailed construction of similar LC systems has been previously described [26,27]. The mobile phases were (A) 10 mM ammonium acetate in 50:50 water/methanol and (B) 10 mM ammonium acetate in 50:50 methanol/acetonitrile. The LC system was equilibrated at 6,000 psi with mobile phase A prior to injecting 1 µL of sample. Exponential gradient elution was initiated 3 min after sample loading with an initial column flow of ~1 µL/min. After 90 min of gradient separation, the mobile phase mixer was purged with 3 mL of mobile phase B, followed by a 5 min column wash. Finally, the mobile phase mixer was purged with 10 mL of mobile phase A, which represented the end of one separation cycle. While gradient elution is performed on one column, the other column is equilibrated with mobile phase A.
Mass spectrometry
The capillary LC system was coupled to four different mass spectrometers during this study: a linear ion-trap MS (LTQ, ThermoFisher, San Jose, CA) a hybrid linear ion-trap-FTICR MS (LTQ-FT, ThermoFisher), a hybrid linear ion-trap-Orbitrap MS (LTQ-Orbitrap, ThermoFisher), and an orthogonal TOF MS (Agilent, Santa Clara, CA). The LTQ, LTQ-FT, and LTQ-Orbitrap instruments were utilized in lipid identification experiments, while the TOF was used in high-throughput analyses of individual samples. The capillary temperatures and electrospray (ESI) voltages for each instrument were 200°C and 2.2 kV (or −2.5 kV for negative ESI), respectively. High-mass measurement accuracy experiments were recorded over the m/z range 100–1,000 with a duty cycle of ~1.0 sec, 1.0 sec, and ~1.6 sec for the LTQ-FT, TOF, and LTQ-Orbitrap, respectively. Data-dependent MS/MS experiments on the LTQ-FT were performed for the top 5 ions when using both the FTICR and LTQ as the mass analyzers or for the top 10 ions when using the LTQ alone. In both cases, the FTICR was used as the mass analyzer during MS survey scans. In addition, neutral-loss-triggered data-dependent MS3 experiments were performed in order to identify certain classes of lipids (eg. glycerophosphoethanolamines and glycerophosphoserines). Dynamic exclusion in the LTQ during data-dependent MS/MS experiments was enabled as follows: repeat count of 2, repeat duration of 30 s, exclusion list size of 250, and exclusion duration of 60 s. For MS/MS scans, the normalized collision energy was set to 35% for positive ESI experiments and 45% for negative ESI experiments.
Isotopic distribution analysis
As part of our lipid identification strategy, we compared the theoretical and experimental isotopic distributions for tentatively identified species using the in-house developed software IsotopicDistributionModeler. This software first calculates the theoretical isotopic distribution for a molecule from its molecular formula using the Mercury algorithm [28]. Prior to isotopic distribution comparison, the theoretical profile is shifted in the m/z dimension to account for slight variances in the precision of the m/z values measured by the mass spectrometer. This is accomplished by initially matching the monoisotopic peak of the theoretical distribution to the peak in the experimental distribution with highest abundance within a mass tolerance of ±20 ppm. The theoretical isotopic distribution is then shifted in the m/z dimension by a value equal to the original difference between theoretical and experimental distributions. An initial scoring of the isotopic distribution fit is then made using a Euclidean scoring scheme in which all points in the distribution above a specified threshold are used. Then to achieve finer precision, small shifts in increments of ±0.1 to ±10 ppm are applied to the theoretical distribution and final scoring is performed to determine the best possible fit. It is important to note that consecutive points in the theoretical distribution are evenly spaced; however, consecutive points in the experimental distribution are not evenly spaced due to the fact that spectrum resolution varies as 1/(square root of m/z) and because spectral data from LTQ-FT and LTQ-Orbitrap instruments are subjected to an intensity threshold determined by the manufacturer. Therefore, interpolation is performed on the experimental isotopic distribution using a quadratic spline to produce evenly spaced points.
Population of the AMT tag database
Lipid species identified by manual interpretation of accurate mass measurements, isotopic distribution comparisons, and MS/MS data were stored as AMT tags in a Microsoft SQL Server database. The database contains the lipid name, empirical formula, monoisotopic mass (calculated based on the empirical formula), and observed NET. Lipid NETs ranging from 0 to 1 were determined by aligning several LC-MS datasets (here a dataset is equivalent to a single LC-MS analysis) to a common and arbitrarily chosen baseline dataset in the software MultiAlign, which is a stand-alone program that incorporates the LCMSWARP algorithm [29] for non-linear chromatographic alignment. Lipids reproducibly observed in multiple analyses were grouped by single linkage clustering in two dimensions, mass and NET, based on user-defined options. For LC-FTICR and LC-Orbitrap MS data, mass and NET tolerances of ±3 ppm and ±0.03 NET (representing ±3 min of a 100-min LC separation) were selected based on empirical observation of mass spectrometer and LC system performance.
Processing of quantitative LC-TOF MS datasets
LC-TOF MS datasets were processed using the PRISM Data Analysis system [30], a series of software tools (eg. Decon2LS, VIPER [31]; freely available at http://ncrr.pnl.gov/software/) developed in-house. The first step involved deisotoping of the raw MS data to give the monoisotopic mass, charge state, and intensity of the major peaks in each mass spectrum. The data were next examined in a 2-D fashion to identify groups of mass spectral peaks that were observed in sequential spectra using an algorithm that computes a Euclidean distance in n-dimensional space for combinations of peaks. Each group, generally ascribed to one detected species and referred to as a “feature”, has a median monoisotopic mass, central NET, and abundance estimate computed by summing the intensities of the MS peaks that comprise the entire LC-TOF MS feature.
The lipid identities of detected features in each LC-TOF MS dataset were determined by comparing their measured monoisotopic masses and NETs to the calculated monoisotopic masses and observed NETs of each of the lipids in the AMT tag database within search tolerances of ±20 ppm and ±0.03 NET for monoisotopic mass and elution time, respectively. The relatively high monoisotopic mass tolerance was used because the reference-spray mass-calibration option available on the TOF was not utilized in these experiments. Non-linear chromatographic alignment of LC-TOF MS datasets was also performed with the LCMSWARP algorithm during database matching by using the NETs of lipid AMT tags as retention time locks.
After database matching and chromatographic alignment, intensity normalization was applied using the expectation maximization algorithm. Briefly, this algorithm analyzes the histogram of log ratios of intensities of features common to two or more datasets and finds the peak apex of this distribution by assuming that the histogram is a mixture of a normal density corresponding to unchanged features and uniform density background corresponding to changed features. The expectation maximization algorithm calculates the normal and uniform parts of the histogram, and the shift in intensity is applied to all features in the aligned dataset. It is important to note that all lipid features (i.e. both identified and unidentified) were considered during intensity normalization. A matrix of normalized lipid feature intensities was then imported into Excel or MatLab for comparative data analysis using analysis of variance (ANOVA) or principal components analysis (PCA), respectively.
RESULTS AND DISCUSSION
Reversed-phase separation of lipids
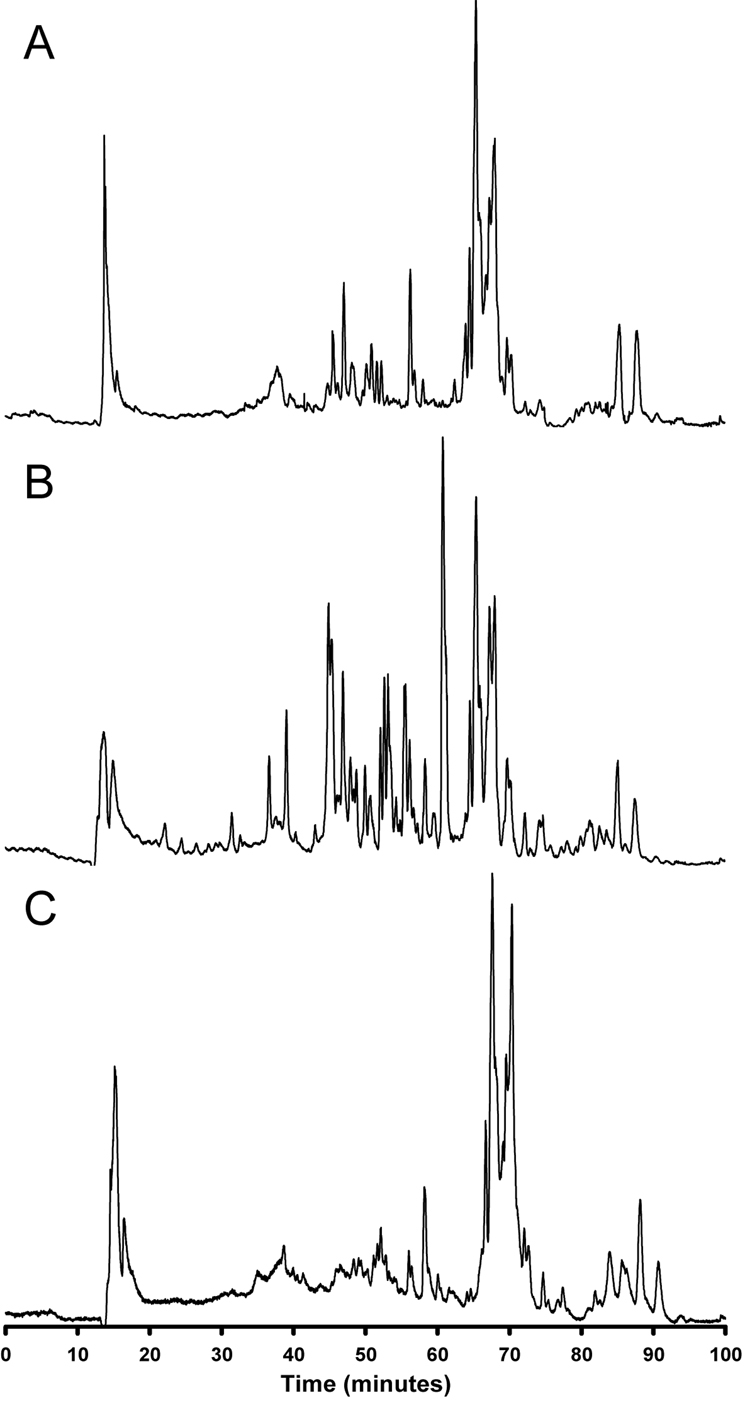
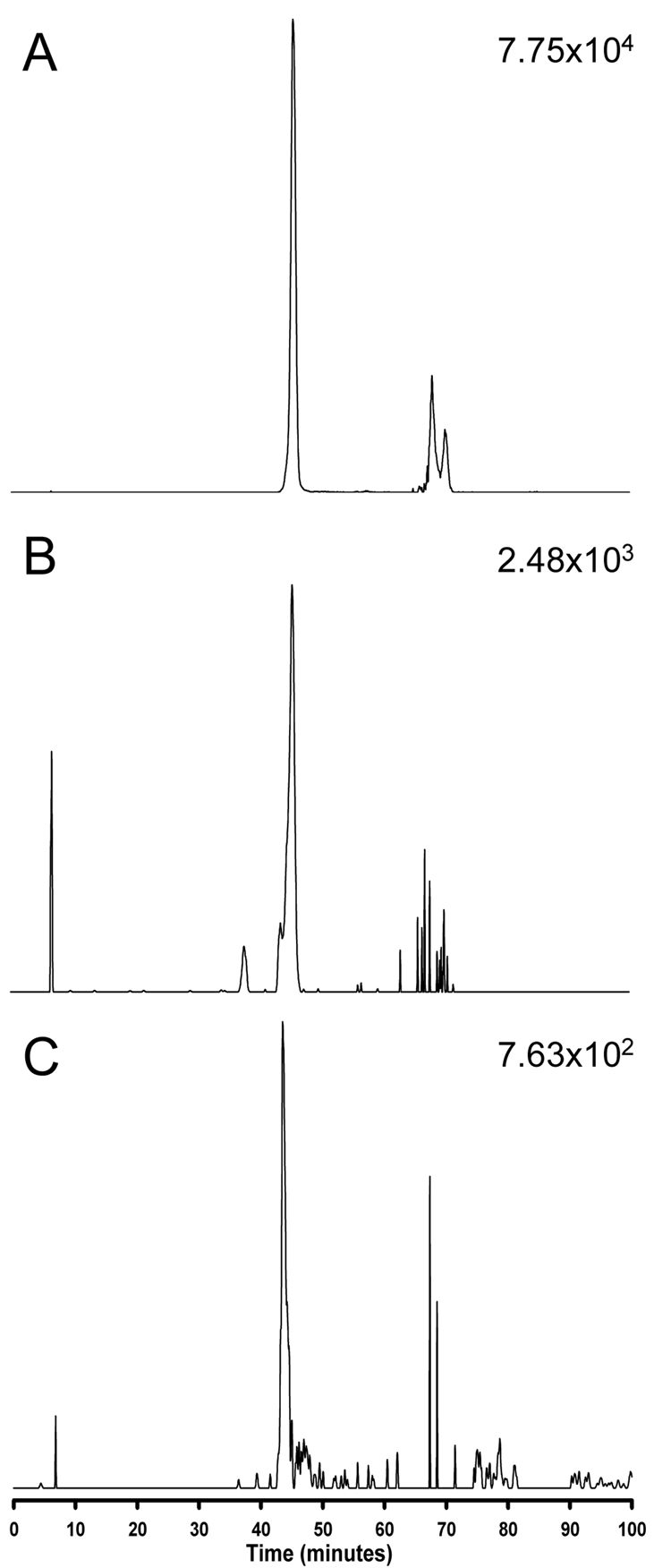
A mobile phase composition of (A) 10 mM ammonium acetate in 50:50 water/methanol (v/v) and (B) 10 mM ammonium acetate in 50:50 acetonitrile/methanol (v/v) was selected, with a LC system split flow of 35 µL/min. When methanol was added to the acetonitrile in mobile phase B, it was found that higher ratios of methanol resulted in faster elution of larger lipids, such as cholesterol esters and triacylglycerols. This is likely due to increased solubility of the lipids in methanol because of the greater hydrogen-bonding capacity of this solvent relative to acetonitrile, resulting in a faster mass transfer of analytes from the stationary phase to the mobile phase. Figure 1 shows the implementation of this mobile phase composition in the analysis of human plasma, erythrocyte, and lymphocyte lipid extracts. The hydrophobic interaction of lipid fatty acid chains and the C18 group of the reversed-phase stationary phase governs the separation of individual lipid species. Therefore, some glycerophospholipids containing identical acyl composition closely elute regardless of the difference in the head groups, particularly those containing phosphocholine and phosphoethanolamine. For example, three lysooctadecanoylglycerophospholipids all eluted at ~45 min. Their measured masses by FTICR MS are 524.3712 (18:0 LPC), 482.3261 (18:0 LPE), and 526.3158 (18:0 LPS). As illustrated in Figure 2, 18:0 LPC (A) and 18:0 LPE (B) coeluted at ~45.5 min, while 18:0 LPS eluted at ~43.5 min. The earlier elution of 18:0 LPS is expected because the phosphoserine head group is more polar than the phosphocholine and phosphoethanolamine head groups. Fortunately, the coeluting glycerophospholipids could still be identified by accurate mass measurements and tandem mass spectrometry because these species display characteristic MS/MS fragmentation patterns (Table 1) [21,22,32–38]. Thus, the combination of reversed-phase LC and MS can be effectively used to separate and identify individual lipid species.
Figure 1.
Human plasma, erythrocyte, and lymphocyte lipid extracts separated by reversed-phase capillary LC and analyzed by TOF MS.
Figure 2.
LC-FTICR MS extracted ion chromatograms of lymphocyte lipids. (A) 18:0 lyso-glycerophosphocholine, (B) 18:0 lyso-glycerophosphoethanolamine, and (C) 18:0 lyso-glycerophosphoserine.
Table 1.
MS and MS/MS information for individual classes of lipids under both positive and negative ion modes.
| Positive mode | Negative mode | |||||
|---|---|---|---|---|---|---|
| Class | Head group | observed m/z in MS | observed m/z in MS/MS | observed m/z in MS | observed m/z in MS/MS | |
| monoacylphosphocholine (LPC), diacylphosphocholine (PC), alkenyl-acyl phosphocholine (p-PC), alkyl-acyl phosphocholine (o-PC), sphingomyelin (SM) | Phosphocholine | [M+H]+ | 184 (C5H15NO4P), [M+H-59]+, [M+H-183]+, [M+H-R2’CH=C=O]+, [M+H-R1’CH=C=O]+, [M+H-R2COOH]+, [M+H-R1COOH]+ | [M+CH3COO]− | [M-CH3]−, [M-H]−, carboxylate anion | |
| [M+Na]+ | 184, [M+Na-N(CH3)3]+ | N/D | ||||
| monoacylphosphoethanolamine (LPE), diacylphosphoethanolamine (PE), alkenyl-acyl phosphoethanolamine (p-PE), alkyl-acyl phosphoethanolamine (o-PE) | phosphoethanolamine | [M+H]+, [M+Na]+ | [M+H-141]+, Neutral loss of 141 (C2H8NO4P) | [M-H]− | carboxylate anion | |
| Oxidized phosphatidylcholine | phosphocholine | [M+H]+ | loss of water, oxygen atom, methanol | [M+CH3COO]− | [M-CH3]−, [M-H]−, [M-CH3OH]−, carboxylate anion, etc | |
| Oxidized phosphatidylethanolamine | phosphoethanolamine | [M-H]− | [M-H-H2O]−, [M-HCH3OH]−, carboxylate anion, etc. | |||
| Phosphatidylserine (PS) | phosphoserine | [M+H]+ | Neutral loss of 185 (C3H8NO6P) | [M-H]− | Neutral loss of 87 (C3H5NO2) | |
| Phosphatidylinositol (PI) | phosphoinositol | [M+NH4]+ | Neutral loss of 277 (C6H16NO9P) | [M-H]− | 241 (C6H10O8P−) | |
| Triacylglycerol (TAG) | N/A | [M+NH4]+ | Netural loss of fatty acid | N/D | ||
| Cholesterol, Cholestrol esters | N/A | [M+NH4]+ | 369 (C27H45+) | N/D | ||
Identification of lipid species
Our lipid identification strategy was to obtain data-dependent MS/MS information of human plasma, erythrocyte, and lymphocyte lipids in both positive and negative ESI modes via LTQ, LTQ-FTICR, or LTQ-Orbitrap MS in order to assign lipid class and fatty acid composition; it is important to note that high-mass accuracy and isotopic distribution information is also obtained when using LTQ-FTICR or LTQ-Orbitrap MS. Identification of lipid molecular species was then based on manual interpretation of this data, and, in some cases, comparisons of this data to that archived and freely available as part of the Lipid Maps Initiative in Lipidomics (www.lipidmaps.org) [39].
The first step in our lipid identification approach was to collect MS/MS data. In our experiments, molecular ion adducts [M+H]+, [M+Na]+, and [M+NH3]+ or [M−H]−, [M−CH3]−, and [M+CH3COO]− were the primary species formed during positive and negative ESI, respectively [21,22,32–38]. Data dependent MS/MS experiments in positive ESI provided information on the nature of the head-group for glycerophospholipids, with formation of an ion corresponding to the head group itself (e.g., phosphocholine head-group at m/z 184) or a neutral loss of the head group from the molecular ion adduct (e.g., loss of 141 Da corresponding to phosphoethanolamine head-group). Alternatively, negative ESI experiments provided information on the fatty acid composition of the lipids through the formation of RCOO− ions. Table 1 lists the common MS/MS fragment ions for individual classes of lipids in both positive and negative ESI. For some species, the sn-1 and sn-2 position of the fatty acid substituents were tentatively assigned based on the intensities of fragment ions in the MS/MS scan. For example, the sn-2 carboxylate anion is reported to be more abundant than the sn-1 carboxylate anion in negative ESI [40]. Alternatively, the [M+H-R2 ’CH=C=O]+ ion, arising from elimination of the sn-2 fatty acid moiety as a ketene, is described as more intense than the [M+H-R1 ’CH=C=O]+ ion in positive ESI [35]. Unfortunately, the positions of fatty acid unsaturation could not be determined in these studies. Representative MS/MS spectra for the lipid classes and molecular species identified in this study are available as Supplemental Information.
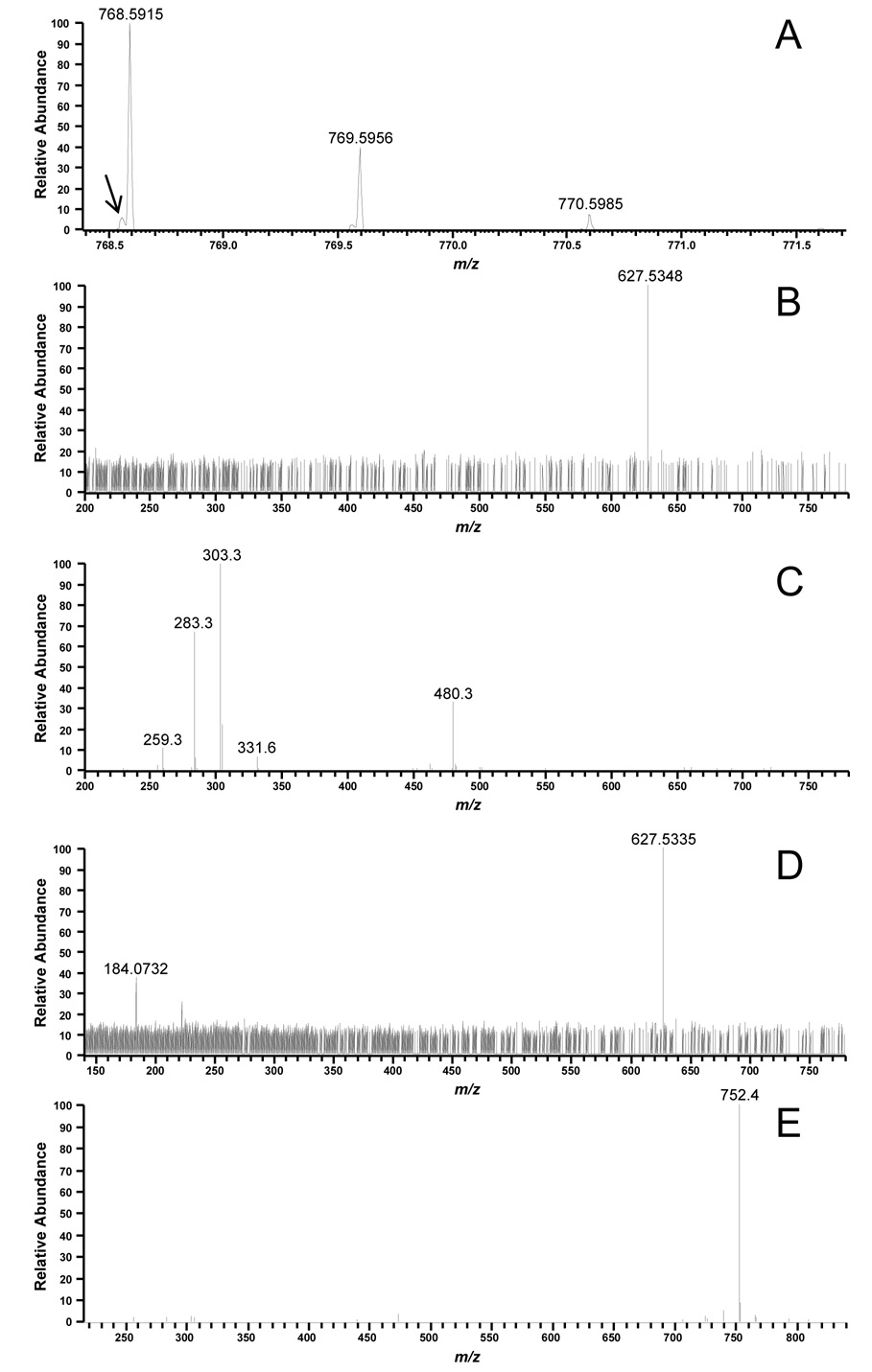
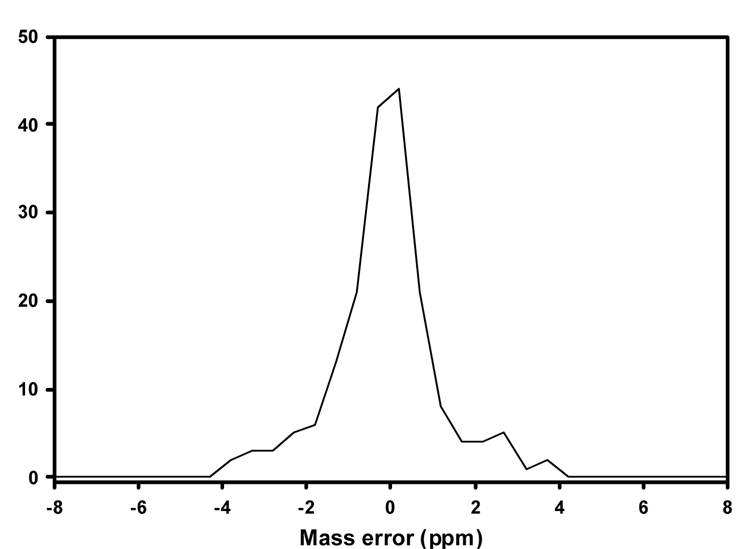
The second step in our lipid identification approach was to examine accurate mass data. High MMA is an invaluable tool when an unambiguous identification cannot be made due to coelution of isobaric species in low mass accuracy MS/MS data. Figure 3 illustrates an example of two coeluting lipid species each with a nominal mass of 768 that were detected in a human erythrocyte lipid extract. The FTICR MS spectrum in Figure 3A shows a major species with m/z 768.5915 and a minor species (arrow) with m/z 768.5558 that differ by a Δm of 46.4 ppm. High mass measurement accuracy MS/MS analysis of m/z 768 (Figure 3B) produced a product ion at 627.5348, corresponding to a neutral loss of 141 and characteristic of the PE lipid class. The calculated mass of the phosphoethanolamine head-group (C2H8NO4P) is 141.0191. Adding the mass of the phosphoethanolamine head-group to the mass of the fragment ion indicates a parent mass of 768.5534, which is 3 ppm from the mass of the minor species detected in the FTICR MS. Thus, the 768.5558 ion is the most likely candidate for having produced the 627.5343 fragment ion. The 768.5558 ion can then be identified as 38:4 PE (C43H78NO8P) having a theoretical m/z of 768.5543 ([M+H]+), which is 1.9 ppm from the observed m/z of the minor species in Figure 3A. Subsequent low mass measurement accuracy MS/MS experiments in negative ESI (Figure 3C) resulted in the detection of the carboxylate anions for 20:4 (m/z 303) and 18:0 (m/z 283). Because the 303 ion was observed in higher abundance than the 283 ion, it was assumed that the 20:4 acyl group occupied the sn-2 position and the 18:0 occupied the sn-1 position [40]. The 38:4 PE species was thus further identified as 18:0/20:4 PE. To identify the major species in Figure 3A with m/z 768.5915, we lowered the ion-trap activation Q value from 0.25 (default value) to 0.18 for positive ESI MS/MS experiments. The activation Q value is the RF frequency used to fragment ions, and a lower activation Q results in lower energy deposition and allows one to observe lower m/z fragment ions. By lowering this value, we shift the mass range of detected fragment ions to lower m/z during MS/MS experiments. In addition to the 627.5335 fragment ion, an ion with m/z 184.0732 was detected, corresponding to the phosphocholine head-group (Figure 3D). Based on this observation, it was assumed that the species with m/z 768.5916 was a glycerophosphocholine. We then examined the corresponding negative ESI MS/MS spectrum (Figure 3E), which showed a neutral loss of 74 (methyl acetate). This confirmed that the major species was indeed a glycerophosphocholine. This species was then tentatively identified as an alkenyl-acyl PC (e.g.16:0p/20:3) or an alkyl-acyl PC (16:0e/20:4). In general, excellent MMA was obtained for the 250 lipid species identified in this study when the FT or Orbitrap MS was used as the mass detector, as illustrated by the mass error histogram in Figure 4.
Figure 3.
Utilization of accurate mass information in the identification of coeluting species with identical nominal mass (A) FTICR MS survey scan showing a major species at m/z 768.5915 and a minor (arrow) species with m/z 768.5558; (B) positive ESI high mass accuracy MS/MS spectrum for the 768 parent ion obtained with an ion-trap Q value of 0.20; (C) negative ESI low mass accuracy MS/MS spectrum for the 768 parent ion showing detection of fatty acid anions at m/z 283.3 and 303.3 (D) positive ESI high mass accuracy MS/MS spectrum for the 768 parent ion obtained with an ion-trap Q value of 0.18 showing detection of the phosphocholine head-group.
Figure 4.
Mass error histogram for the 250 identified lipid species. The mass error was calculated as (theoretical mass-experimental mass)/theoretical mass and expressed in ppm.
The final step in our lipid identification approach was to compare theoretical and experimental isotopic distributions for all lipid species identified by MS/MS and accurate mass information. The mass spectral peaks for each lipid were exported from the MS spectra and saved in tab-delimited format. Comparison of theoretical and experimental isotopic distributions was then performed in an automated manner using IsotopicDistributionModeler, which generated a summary file containing the empirical formula, isotopic fit score, theoretical monoisotopic mass, mass delta (ppm), and peak intensity for all distributions modeled. Based on our experience comparing theoretical and experimental isotopic distributions of peptides, an isotopic fit score ≤ 0.05 is considered highly confident (unpublished observations). The isotopic fit scores for the lipid species identified in this study were all ≤ 0.05 with a median fit score of 0.008. Graphs of the isotopic distribution comparison for each identified lipid are available as Supplemental Information. In general, lipid species with high MS intensity or well-resolved spectra displayed correspondingly low isotopic fit scores. In contrast, species with low MS intensity or poorly-resolved spectra displayed correspondingly high isotopic fit scores.
Lipid AMT tag database
Empirical formulae, calculated monoisotopic masses, and observed NETs for lipid species identified based on MS/MS, accurate mass, and isotopic distribution information were placed in two separate AMT tag databases (available at http://ncrr.pnl.gov/data/) depending on whether they were observed during positive or negative ESI. Twenty-three species observed in negative ESI that could be expected to be ionized under positive ESI conditions (e.g. some glycerophosphoethanolamines and oxidized glycerophosphocholines) were also added to the positive ESI AMT tag database. In addition, 12 saturated lysoglycerophosphocholines and 10 saturated fatty acids were added to the positive and negative ESI AMT tag databases, respectively, using their predicted NETs after plotting observed NETs of species in the same series (i.e. 14:0, 15:0, 16:0, etc.) versus the corresponding m/z. The positive ESI AMT tag database contains 250 entries (the negative ESI database contains 45 entries), including glycerophosphoethanolamines, glycerophosphocholines, lyso-glycerophosphoethanolamines, lyso-glycerophosphocholines, triacylglycerols, sphingomyelins, cholesterol and its esters, and fatty acids. While this is a relatively modest number of identifications, significantly higher coverage of the lipidome may be obtained in future studies through fractionation of lipids during sample processing or utilization of 2-D LC (eg. offline normal-phase fractionation of lipids by class followed by online reversed-phase separation and analysis of each fraction). Similarly, development and implementation of software capable of identifying detected lipids in an automated fashion from tandem mass spectra, accurate mass, and isotopic distribution information will increase the throughput, number, and accuracy of lipid identifications. Progress towards the latter goal is already being reported [41,42].
Quantitative profiling of human plasma, erythrocyte, and lymphocyte lipid extracts using the AMT tag approach
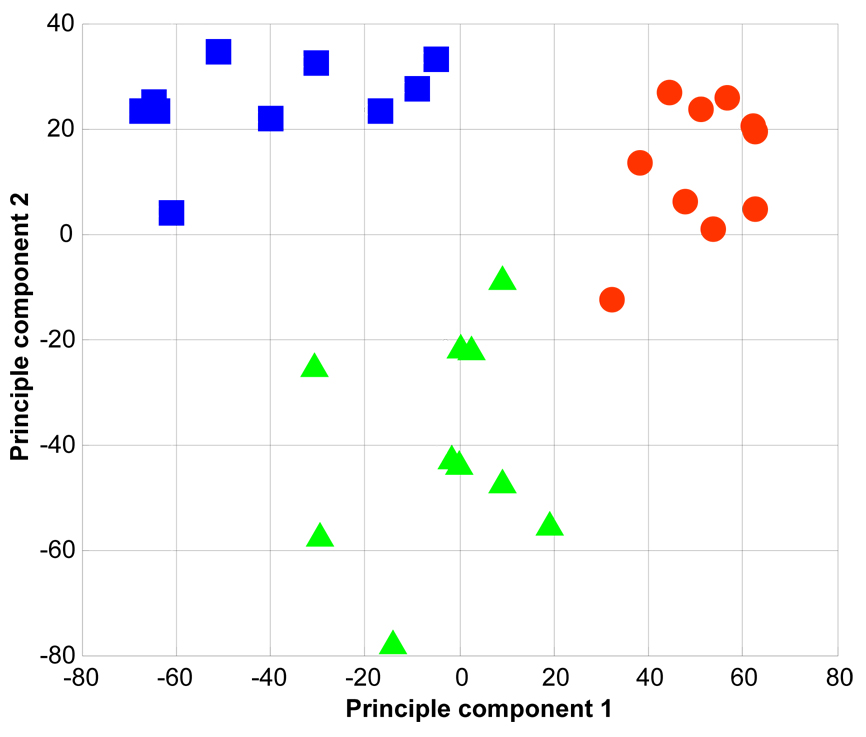
After establishing lipid AMT tag databases based on analyses of pooled samples, we performed quantitative (based on the absolute abundance of detected species) LC-TOF MS analyses of individual plasma, erythrocyte, and lymphocyte lipid extracts using positive ESI. LC-TOF MS datasets were then processed as described under the Experimental section in order to identify detected lipid species and to normalize MS intensities for downstream comparative data analysis. Averages of 8,005 ± 853, 9,221 ± 932, and 7,622 ± 384 features (mean ± standard deviation; n = 30 each) were detected during LC-TOF MS analyses of individual lipid extracts of human plasma, erythrocytes, and lymphocytes, respectively. It is important to note that these feature numbers include contributions from chemical noise that are not accounted for by the peak-picking algorithm currently used in our downstream data processing approach. Therefore, these numbers are an overestimate of the true number of lipid features present in each dataset. Of the features reported above, 158 ± 24, 168 ± 13, and 162 ± 16 (mean ± standard deviation) from plasma, erythrocyte, and lymphocyte datasets, respectively, matched entries in the positive ESI AMT tag database within ±20 ppm and ±0.03 NET. A total of 244 AMT tags were matched to lipid features detected in LC-TOF MS datasets, and, to place emphasis on those species frequently detected, we required that each AMT tag be identified in at least 11 of 30 datasets per sample type. This resulted in each species being observed in ~75 of 90 total datasets. Normalized abundances for identified lipids were subsequently averaged within their respective replicates, followed by utilization of principal components analysis (PCA) to identify lipid profiles representative of human plasma, erythrocytes, and lymphocytes. Figure 5 is a PCA scores plot showing segregation of the three sample types based on both qualitative and quantitative differences in those lipid features matching the AMT tag database. We next performed a statistical analysis to identify significant differences in lipid abundances, which resulted in 66 differentially expressed (ANOVA, p < 0.01) lipids among the three sample types (Table 2 and Table 3). Plasma samples showed a higher relative abundance of all triacylglycerols (p<0.01), consistent with previous reports describing erythrocytes and lymphocytes as containing negligible amounts of this class of lipid [43]. In addition, erythrocytes showed relatively higher concentrations of sphingomyelins and some lyso-glycerophosphoethanolamines than either plasma or lymphocytes (p<0.01), which is again consistent with data reported by Schwarz et al. [44] and Diagne et al. [45].
Figure 5.
Principal components analysis scores plot showing segregation of human plasma (blue squares), erythrocyte (red circles), and lymphocyte (green triangles) samples.
Table 2.
Lipids identified as having significantly higher abundance (ANOVA, p < 0.01) in plasma versus erythrocytes and lymphocytes. A total of 36 molecular species were retained after ANOVA and subsequent filtering. Shown are the molecular species, class, empirical formula (including charge carrier), m/z (M+H for glycerophospholipids and M+NH4 for triacylglycerols), and normalized elution time (NET; retention time can be calculated by multiplying NETs by total time of separation) information for identified lipids.
| Molecular Species | Lipid class | Formula | m/z | NET |
|---|---|---|---|---|
| LPC 16:0 [48,50,52] | Lyso-glycerophosphocholine | C24H51NO7P | 496.3403 | 0.264 |
| LPC 16:1 [48,50] | Lyso-glycerophosphocholine | C24H49NO7P | 494.3246 | 0.225 |
| PC 16:0p/20:4 or 16:0e/20:5 [47] | Choline plasmalogen or ether | C44H81NO7P | 766.5750 | 0.545 |
| PC 18:0p/22:5 or 18:0e/22:6 [47] | Choline plasmalogen or ether | C48H87NO7P | 820.6220 | 0.585 |
| PC 35:2 [47] | Glycerophosphocholine | C43H83NO8P | 772.5856 | 0.549 |
| PC 36:3 [47–50,53] | Glycerophosphocholine | C44H83NO8P | 784.5856 | 0.530 |
| PC 37:3 | Glycerophosphocholine | C45H85NO8P | 798.6012 | 0.559 |
| PC 37:4 [47] | Glycerophosphocholine | C45H83NO8P | 796.5856 | 0.545 |
| PC 38:4 [47–50] | Glycerophosphocholine | C46H85NO8P | 810.6012 | 0.572 |
| PC 40:5 [47,49] | Glycerophosphocholine | C48H87NO8P | 836.6169 | 0.572 |
| *PE 18:1p/20:4 [47,49] | Ethanolamine plasmalogen | C43H77NO7P | 750.5437 | 0.542 |
| TAG 48:0 [51] | Triacylglycerol | C51H102NO6 | 824.7707 | 0.855 |
| TAG 48:2 [51] | Triacylglycerol | C51H98NO6 | 820.7394 | 0.773 |
| TAG 48:3 [51,52] | Triacylglycerol | C51H96NO6 | 818.7237 | 0.737 |
| TAG 49:1 [51] | Triacylglycerol | C52H102NO6 | 836.7707 | 0.836 |
| TAG 49:2 [51] | Triacylglycerol | C52H100NO6 | 834.7550 | 0.796 |
| TAG 50:2 [51,52] | Triacylglycerol | C53H102NO6 | 848.7707 | 0.819 |
| TAG 50:3 [51] | Triacylglycerol | C53H100NO6 | 846.7550 | 0.776 |
| TAG 50:4 [51] | Triacylglycerol | C53H98NO6 | 844.7394 | 0.742 |
| TAG 50:5 | Triacylglycerol | C53H96NO6 | 842.7237 | 0.718 |
| TAG 51:2 [51] | Triacylglycerol | C54H104NO6 | 862.7863 | 0.842 |
| TAG 51:3 [51] | Triacylglycerol | C54H102NO6 | 860.7707 | 0.803 |
| TAG 51:4 | Triacylglycerol | C54H100NO6 | 858.7550 | 0.765 |
| TAG 52:2 [51] | Triacylglycerol | C55H106NO6 | 876.8020 | 0.854 |
| TAG 52:3 [51,52] | Triacylglycerol | C55H104NO6 | 874.7863 | 0.823 |
| TAG 52:4 [51] | Triacylglycerol | C55H102NO6 | 872.7707 | 0.785 |
| TAG 52:5 [51,52] | Triacylglycerol | C55H100NO6 | 870.7550 | 0.750 |
| TAG 52:6 | Triacylglycerol | C55H98NO6 | 868.7394 | 0.724 |
| TAG 54:2 [51,52] | Triacylglycerol | C57H110NO6 | 904.8333 | 0.870 |
| TAG 54:4 [51] | Triacylglycerol | C57H106NO6 | 900.8020 | 0.828 |
| TAG 54:6 [52] | Triacylglycerol | C57H102NO6 | 896.7707 | 0.752 |
| TAG 54:7 | Triacylglycerol | C57H100NO6 | 894.7550 | 0.731 |
| TAG 56:5 [51] | Triacylglycerol | C59H108NO6 | 926.8176 | 0.836 |
| TAG 56:6 [51,52] | Triacylglycerol | C59H106NO6 | 924.8020 | 0.810 |
| TAG 56:7 [51,52] | Triacylglycerol | C59H104NO6 | 922.7863 | 0.779 |
| TAG 56:8 | Triacylglycerol | C59H102NO6 | 920.7707 | 0.746 |
Note: this species was detected in higher abundance in plasma versus erythrocytes but in lower abundance versus lymphocytes. Those species that have been previously identified in blood or blood constituents contain the corresponding references after the molecular species.
Table 3.
Lipids identified as having significantly higher abundance (ANOVA, p < 0.01) in erythrocytes and lymphocytes versus plasma. A total of 30 molecular species were retained after ANOVA and subsequent filtering. Shown are the molecular species, class, empirical formula (including charge carrier), m/z (M+H), and normalized elution time (NET; retention times can be calculated by multiplying NETs by total time of separation) information for identified lipids.
| Molecular Species | Lipid class | Formula | m/z | NET |
|---|---|---|---|---|
| *LPC 18:2 [48,50,52] | Lyso-glycerophosphocholine | C26H51NO7P | 520.3403 | 0.238 |
| *LPC 19:0 [50] | Lyso-glycerophosphocholine | C27H57NO7P | 538.3872 | 0.345 |
| LPE 16:0 | Lyso-glycerophosphoethanolamine | C21H45NO7P | 454.2933 | 0.262 |
| *LPE 18:2 | Lyso-glycerophosphoethanolamine | C23H45NO7P | 478.2933 | 0.237 |
| *LPE 20:3 | Lyso-glycerophosphoethanolamine | C25H47NO7P | 504.3090 | 0.254 |
| *LPE 22:6 | Lyso-glycerophosphoethanolamine | C27H45NO7P | 526.2933 | 0.254 |
| *Oxidized PC (16:0/Hydroxy-18:1) | Oxidized glycerophosphocholine | C42H83NO9P | 776.5805 | 0.484 |
| Oxidized PC (18:0/Hydroxy-16:3) | Oxidized glycerophosphocholine | C42H79NO9P | 772.5492 | 0.428 |
| Oxidized PC (18:0/Hydroxy-18:3) | Oxidized glycerophosphocholine | C44H83NO9P | 800.5805 | 0.460 |
| PC (16:0e/18:0) [47] | Choline ether | C42H87NO7P | 748.6220 | 0.634 |
| PC (16:0p/20:0;16:0e/20:0) [47] | Choline plasmalogen or ether | C44H89NO7P | 774.6376 | 0.638 |
| *PC (18:0p/22:2;18:0e/22:3) | Choline plasmalogen or ether | C48H93NO7P | 826.6689 | 0.649 |
| PC 32:0 [47–50] | Glycerophosphocholine | C40H81NO8P | 734.5699 | 0.554 |
| PC 33:2 | Glycerophosphocholine | C41H79NO8P | 744.5543 | 0.562 |
| PC 34:0 [47–50,53] | Glycerophosphocholine | C42H85NO8P | 762.6012 | 0.609 |
| *PC 35:1 [47] | Glycerophosphocholine | C43H85NO8P | 774.6012 | 0.587 |
| PC 36:0 [47,49] | Glycerophosphocholine | C44H89NO8P | 790.6325 | 0.646 |
| PE 34:3 | Glycerophosphoethanolamine | C39H73NO8P | 714.5073 | 0.478 |
| PE 36:4 [47–50,53] | Glycerophosphoethanolamine | C41H75NO8P | 740.5230 | 0.512 |
| PE 36:5 [47,48] | Glycerophosphoethanolamine | C41H73NO8P | 738.5073 | 0.478 |
| PE 38:5 [47–50,53] | Glycerophosphoethanolamine | C43H77NO8P | 766.5386 | 0.518 |
| *SM (d16:1/24:1) [47,53] | Sphingomyelin | C45H90N2O6P | 785.6536 | 0.622 |
| *SM (d17:1/24:0) [47] | Sphingomyelin | C46H94N2O6P | 801.6849 | 0.668 |
| *SM (d17:1/24:1) [47] | Sphingomyelin | C46H92N2O6P | 799.6693 | 0.634 |
| SM (d18:1/24:0) [47,53] | Sphingomyelin | C47H96N2O6P | 815.7006 | 0.679 |
| SM (d18:1/26:0) | Sphingomyelin | C49H100N2O6P | 843.7319 | 0.708 |
| SM (d18:1/26:1) | Sphingomyelin | C49H98N2O6P | 841.7162 | 0.680 |
| SM (d18:1/26:2) | Sphingomyelin | C49H96N2O6P | 839.7006 | 0.659 |
| *SM (d19:1/24:1) | Sphingomyelin | C48H96N2O6P | 827.7006 | 0.664 |
| *SM (d20:1/22:3) | Sphingomyelin | C47H90N2O6P | 809.6536 | 0.584 |
Note: these species were detected in higher abundance in erythrocytes versus plasma but in lower abundance in lymphocytes versus plasma. Those species that have been previously identified in blood or blood constituents contain the corresponding references after the molecular species.
These experiments were based on LC-TOF MS analyses only without tandem MS, allowing the instrument duty cycle to be exploited entirely for quantitation. And while the comparison of lipid profiles from plasma, erythrocytes, and lymphocytes is unfair, the application of the AMT tag approach in lipidomic studies of these tissues illustrates the increased sample analysis throughput (i.e. simultaneous identification and quantitation) obtained in a proof-of-principle study.
CONCLUSIONS
In a preliminary demonstration of the AMT tag approach in lipid profiling studies, we have used capillary LC in conjunction with high-mass measurement accuracy MS and MS/MS in order to structurally identify 272 unique lipid species in human plasma, erythrocytes, and lymphocytes, which were then used to populate AMT tag databases. The majority of these species have been previously reported in studies of these tissue types [46–51]. After identification of lipid species were made, further MS/MS analyses were unnecessary, and high-throughput LC-TOF MS analyses were utilized to maximize analysis throughput and quantitative sensitivity. The future development of a comprehensive lipid AMT tag database should result in highly confident lipid identifications with corresponding quantitative information from high-throughput LC-TOF MS or LC-FTICR MS experiments.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Lawrence S. Phillips of Emory University for providing the plasma, erythrocyte, and lymphocyte samples via the NIDDK Central Repository. This research was supported by NIH grants DK070146 and DK071283. Work was performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility located at Pacific Northwest National Laboratory (PNNL) and sponsored by the U.S. Department of Energy Office (DOE) of Biological and Environmental Research. PNNL is operated by Battelle for the DOE under Contract No. DE-AC06-76RLO-1830.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ostrowski SG, Van Bell CT, Winograd N, Ewing AG. Science. 2004;305:71. doi: 10.1126/science.1099791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim D. Trends Pharmacol Sci. 2003;24:648. doi: 10.1016/j.tips.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Movahed P, Jonsson BA, Birnir B, Wingstrand JA, Jorgensen TD, Ermund A, Sterner O, Zygmunt PM, Hogestatt ED. J Biol Chem. 2005;280:38496. doi: 10.1074/jbc.M507429200. [DOI] [PubMed] [Google Scholar]
- 4.Honore E. Nat Rev Neurosci. 2007;8:251. doi: 10.1038/nrn2117. [DOI] [PubMed] [Google Scholar]
- 5.Miller B, Sarantis M, Traynelis SF, Attwell D. Nature. 1992;355:722. doi: 10.1038/355722a0. [DOI] [PubMed] [Google Scholar]
- 6.Kloda A, Lua L, Hall R, Adams DJ, Martinac B. Proc Natl Acad Sci U S A. 2007;104:1540. doi: 10.1073/pnas.0609649104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaus K, Gratton E, Kable EP, Jones AS, Gelissen I, Kritharides L, Jessup W. Proc Natl Acad Sci U S A. 2003;100:15554. doi: 10.1073/pnas.2534386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobson K, Mouritsen OG, Anderson RG. Nat Cell Biol. 2007;9:7. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
- 9.Piomelli D, Volterra A, Dale N, Siegelbaum SA, Kandel ER, Schwartz JH, Belardetti F. Nature. 1987;328:38. doi: 10.1038/328038a0. [DOI] [PubMed] [Google Scholar]
- 10.Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD. Annu Rev Biochem. 2001;70:535. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 11.Fitzgerald KA, Chen ZJJ. Cell. 2006;125:834. doi: 10.1016/j.cell.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Shemarova IV. Crit Rev Microbiol. 2007;33:141. doi: 10.1080/10408410701415927. [DOI] [PubMed] [Google Scholar]
- 13.Vane JR, Bakhle YS, Botting RM. Annu Rev Pharmacol Toxicol. 1998;38:97. doi: 10.1146/annurev.pharmtox.38.1.97. [DOI] [PubMed] [Google Scholar]
- 14.Milne GL, Morrow JD. Antioxid Redox Signal. 2006;8:1379. doi: 10.1089/ars.2006.8.1379. [DOI] [PubMed] [Google Scholar]
- 15.Morrow JD. Curr Pharm Des. 2006;12:895. doi: 10.2174/138161206776055985. [DOI] [PubMed] [Google Scholar]
- 16.Kopitz J, Holz FG, Kaemmerer E, Schutt F. Biochimie. 2004;86:825. doi: 10.1016/j.biochi.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 17.Requena JR, Fu MX, Ahmed MU, Jenkins AJ, Lyons TJ, Thorpe SR. Nephrol Dial Transplant. 1996;11 Suppl 5:48. doi: 10.1093/ndt/11.supp5.48. [DOI] [PubMed] [Google Scholar]
- 18.Wolf C, Quinn PJ. Prog Lipid Res. 2008;47:15. doi: 10.1016/j.plipres.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Schwudke D, Liebisch G, Herzog R, Schmitz G, Shevchenko A. Methods Enzymol. 2007;433:175. doi: 10.1016/S0076-6879(07)33010-3. [DOI] [PubMed] [Google Scholar]
- 20.Han XL, Gross RW. J Lipid Res. 2003;44:1071. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- 21.Murphy RC, Fiedler J, Hevko J. Chem Rev. 2001;101:479. doi: 10.1021/cr9900883. [DOI] [PubMed] [Google Scholar]
- 22.Pulfer M, Murphy RC. Mass Spectrom Rev. 2003;22:332. doi: 10.1002/mas.10061. [DOI] [PubMed] [Google Scholar]
- 23.Zimmer JS, Monroe ME, Qian WJ, Smith RD. Mass Spectrom Rev. 2006;25:450. doi: 10.1002/mas.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Folch J, Lees M, Stanley GHS. J Biol Chem. 1957;226:497. [PubMed] [Google Scholar]
- 25.Shen YF, Zhao R, Belov ME, Conrads TP, Anderson GA, Tang KQ, Pasa-Tolic L, Veenstra TD, Lipton MS, Udseth HR, Smith RD. Anal Chem. 2001;73:1766. doi: 10.1021/ac0011336. [DOI] [PubMed] [Google Scholar]
- 26.Shen YF, Tolic N, Zhao R, Pasa-Tolic L, Li LJ, Berger SJ, Harkewicz R, Anderson GA, Belov ME, Smith RD. Anal Chem. 2001;73:3011. doi: 10.1021/ac001393n. [DOI] [PubMed] [Google Scholar]
- 27.Belov ME, Anderson GA, Wingerd MA, Udseth HR, Tang K, Prior DC, Swanson KR, Buschbach MA, Strittmatter EF, Moore RJ, Smith RD. J Am Soc Mass Spectrom. 2004;15:212. doi: 10.1016/j.jasms.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Rockwood AL, Vanorden SL, Smith RD. Anal Chem. 1995;67:2699. [Google Scholar]
- 29.Jaitly N, Monroe ME, Petyuk VA, Clauss TR, Adkins JN, Smith RD. Anal Chem. 2006;78:7397. doi: 10.1021/ac052197p. [DOI] [PubMed] [Google Scholar]
- 30.Kiebel GR, Auberry KJ, Jaitly N, Clark DA, Monroe ME, Peterson ES, Tolic N, Anderson GA, Smith RD. Proteomics. 2006;6:1783. doi: 10.1002/pmic.200500500. [DOI] [PubMed] [Google Scholar]
- 31.Monroe ME, Tolic N, Jaitly N, Shaw JL, Adkins JN, Smith RD. Bioinformatics. 2007;23:2021. doi: 10.1093/bioinformatics/btm281. [DOI] [PubMed] [Google Scholar]
- 32.Kerwin JL, Tuininga AR, Ericsson LH. J Lipid Res. 1994;35:1102. [PubMed] [Google Scholar]
- 33.Hsu FF, Turk J. J Mass Spectrom. 2000;35:596. doi: 10.1002/(SICI)1096-9888(200005)35:5<595::AID-JMS965>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 34.Hsu FF, Turk J. J Am Soc Mass Spectrom. 2000;11:437. doi: 10.1016/S1044-0305(99)00150-6. [DOI] [PubMed] [Google Scholar]
- 35.Hsu FF, Turk J. J Am Soc Mass Spectrom. 2003;14:352. doi: 10.1016/S1044-0305(03)00064-3. [DOI] [PubMed] [Google Scholar]
- 36.Hsu FF, Turk J, Thukkani AK, Messner MC, Wildsmith KR, Ford DA. J Mass Spectrom. 2003;38:752. doi: 10.1002/jms.491. [DOI] [PubMed] [Google Scholar]
- 37.Hsu FF, Turk J. J Am Soc Mass Spectrom. 2005;16:1510. doi: 10.1016/j.jasms.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 38.Taguchi R, Houjou T, Nakanishi H, Yamazaki T, Ishida M, Imagawa M, Shimizu T. J Chromatogr B Analy Technol Biomed Life Sci. 2005;823:26. doi: 10.1016/j.jchromb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 39.Schmelzer K, Fahy E, Subramaniam S, Dennis EA. Methods Enzymol. 2007;432:171. doi: 10.1016/S0076-6879(07)32007-7. [DOI] [PubMed] [Google Scholar]
- 40.Smith PBW, Snyder AP, Harden CS. Anal Chem. 1995;67:1824. doi: 10.1021/ac00107a011. [DOI] [PubMed] [Google Scholar]
- 41.Schwudke D, Oegema J, Burton L, Entchev E, Hannich JT, Ejsing CS, Kurzchalia T, Shevchenko A. Anal Chem. 2006;78:585. doi: 10.1021/ac051605m. [DOI] [PubMed] [Google Scholar]
- 42.Song H, Hsu FF, Ladenson J, Turk J. J Am Soc Mass Spectrom. 2007;18:1848. doi: 10.1016/j.jasms.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson GJ. Blood lipids and lipoproteins: quantitation, composition, and metabolism. New York: Wiley-Interscience; 1972. [Google Scholar]
- 44.Schwarz HP, Dahlke MB, Dreisbach L. Clinical Chemistry. 1977;23:1548. [PubMed] [Google Scholar]
- 45.Diagne A, Fauvel J, Record M, Chap H, Douste-Blazy L. Biochim Biophys Acta. 1984;793:221. doi: 10.1016/0005-2760(84)90324-2. [DOI] [PubMed] [Google Scholar]
- 46.Riley C, Nunn RF. Biochem J. 1960;74:56. doi: 10.1042/bj0740056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Myher JJ, Kuksis A, Pind S. Lipids. 1989;24:408. doi: 10.1007/BF02535148. [DOI] [PubMed] [Google Scholar]
- 48.Wang C, Xie S, Yang J, Yang Q, Xu G. Anal Chim Acta. 2004;525:1. [Google Scholar]
- 49.Malavolta M, Bocci F, Boselli E, Frega NG. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;810:173. doi: 10.1016/j.jchromb.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 50.Uran S, Larsen A, Jacobsen PB, Skotland T. J Chromatogr B. 2001;758:265. doi: 10.1016/s0378-4347(01)00188-8. [DOI] [PubMed] [Google Scholar]
- 51.Mares P, Rezanka T, Novak M. J Chromatogr. 1991;568:1. doi: 10.1016/0378-4347(91)80335-a. [DOI] [PubMed] [Google Scholar]
- 52.Bijlsma S, Bobeldijk I, Verheij ER, Ramaker R, Kochhar S, Macdonald IA, van Ommen B, Smilde AK. Anal Chem. 2006;78:567. doi: 10.1021/ac051495j. [DOI] [PubMed] [Google Scholar]
- 53.Wang C, Yang J, Gao P, Lu X, Xu G. Rapid Commun Mass Spectrom. 2005;19:2443. doi: 10.1002/rcm.2080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.