Abstract
In this study, we report on the noninvasive identification of spectral markers of alveolar type II (ATII) cell differentiation in vitro using Raman microspectroscopy. ATII cells are progenitor cells for alveolar type I (ATI) cells in vivo, and spontaneously differentiate toward an ATI-like phenotype in culture. We analyzed undifferentiated and differentiated primary human ATII cells, and correlated Raman spectral changes to cellular changes in morphology and marker protein synthesis (surfactant protein C, alkaline phosphatase, caveolin-1). Undifferentiated ATII cells demonstrated spectra with strong phospholipid vibrations, arising from alveolar surfactant stored within cytoplasmic lamellar bodies (Lbs). Differentiated ATI-like cells yielded spectra with significantly less lipid content. Factor analysis revealed a phospholipid-dominated spectral component as the main discriminator between the ATII and ATI-like phenotypes. Spectral modeling of the data revealed a significant decrease in the spectral contribution of cellular lipids—specifically phosphatidyl choline, the main constituent of surfactant, as ATII cells differentiate. These observations were consistent with the clearance of surfactant from Lbs as ATII cells differentiate, and were further supported by cytochemical staining for Lbs. These results demonstrate the first spectral characterization of primary human ATII cells, and provide insight into the biochemical properties of alveolar surfactant in its unperturbed cellular environment.
INTRODUCTION
The unique environment of pulmonary alveoli is established and maintained by two highly specialized epithelial cell types, alveolar type I (ATI) and type II (ATII) cells. ATI cells dominate the alveolar epithelial lining, covering >90% of the alveolar surface area (1). ATII cells perform a multitude of indispensable functions, the most important being the production, storage, and secretion of surfactant, a phospholipid-rich, multifunctional lubricant that reduces alveolar surface tension. Surfactant prevents alveolar collapse during ventilation, aids in the maintenance of fluid homeostasis within the alveolus, and has also been linked to host defense through the binding of surfactant proteins to pathogens (2). Surfactant produced by ATII cells is stored in cytoplasmic organelles called lamellar bodies (Lbs), a distinguishing feature of type II cells.
ATII cells are also alveolar progenitor cells, and are believed to be the sole progenitor for ATI cells in vivo (3). In vitro, primary ATII cells lose their distinctive phenotype and express phenotypic features characteristic of ATI cells (4). The spontaneous differentiation is characterized by morphological changes, such as surfactant clearance, i.e., a decrease in the number of surfactant-containing Lbs, and changes in the expression of specific marker proteins, such as a decrease in surfactant protein C (Sp-C), which is unique to ATII cells (2). The parallels between mechanisms of ATII cell differentiation in vivo and in vitro have yet to be fully defined, and so the in vitro derived phenotype of differentiated alveolar epithelial cells is generally referred to as “ATI-like”. However, the differentiation process is believed to be accomplished by continuous transformation from ATII cells into ATI-like cells via an intermediate phenotype (3).
Electron microscopy techniques have previously been used to identify the main morphological and ultrastructural changes during ATII cell differentiation (5), whereas expression of important markers such as surfactant proteins (Sp-C, Sp-A, etc.), caveolin-1, and intracellular adhesion molecule-1 have been investigated with immunofluorescence confocal microscopy, reverse transcriptase polymerase chain reaction, flow cytometry, and immunoblotting (4,6). These morphological and immunocytochemical techniques provide insight into the differentiation process; however, they are invasive, requiring exogenous labeling and/or cell fixation or extraction, and therefore are unsuitable for studies on living cells.
Raman microspectroscopy is a laser-based analytical technique that enables chemical characterization of molecules within a sample. It is a nondestructive optical technique based on the inelastic scattering of photons by molecular bond vibrations (7). A small portion of incident photons are scattered by interaction with chemical bonds resulting in a shift toward lower frequencies (i.e., lower energies). Energy differences between incident and scattered photons correspond to specific vibrational energies of chemical bonds of the scattering molecules (8). The Raman spectrum of a cell represents an intrinsic biochemical fingerprint, containing molecular-level information about all cellular biopolymers, including DNA, RNA, proteins, lipids, and carbohydrates. The main advantages of Raman microspectroscopy over conventional cytochemical techniques include its capability for rapid, noninvasive sensing, and the weak Raman scattering of aqueous media enables the in vitro analysis of living cells in the absence of fixatives or labels (9,10). Furthermore, since Raman spectra are sensitive to changes in molecular composition, they can be used as cell-specific biochemical signatures to discriminate between different cellular phenotypes (11,12). Thus, Raman microspectroscopy offers the potential to noninvasively characterize biochemical changes in the phenotype of stem cells during differentiation. This technique has previously been used to monitor mRNA translation during embryonic stem cell differentiation (13), and to study enhanced differentiation of fetal osteoblastic progenitor cells with bioactive glass dissolution ions (14).
In a previous report, we described a novel procedure for isolation of human ATII cells that results in high ATII cell purity (∼95%) (15). Using this protocol, the undifferentiated type II phenotype can be maintained for ∼1 week in standard immersion cell culture (16), and the ATII to ATI-like cell transition occurs over a period of ∼8–14 days (S. J. Kemp and T. D. Tetley, unpublished observations). In this study, we report on the noninvasive biochemical characterization of in vitro differentiation of primary human ATII cells with Raman microspectroscopy. We compared the Raman spectra of live ATII cells measured at different points during a differentiation time course. Cellular changes in morphology and marker protein synthesis (Sp-C, alkaline phosphatase, caveolin-1) were monitored to verify the ATII to ATI-like transition of primary cells, and to correlate spectral changes with changes in cellular phenotype. We analyzed ATII cell spectra with several well-known chemometric techniques including principal component analysis (PCA), factor analysis, and spectral modeling to identify spectral markers describing important biochemical changes that occur during ATII cell differentiation.
MATERIALS AND METHODS
Isolation and culture of ATII cells
Primary human alveolar epithelial cells were isolated from a normal region of lung tissue after lobectomy for carcinoma. The ATII cells were isolated as described previously (16). Briefly, the tissue was perfused repeatedly with 0.15 M NaCl to remove leukocytes before digestion with trypsin (2.5 μl/ml Hanks' balanced salt solution; Sigma, Poole, UK) for 45 min at 37°C. The tissue was finely chopped in newborn calf serum (Invitrogen, Paisley, UK), filtered and the ATII cells isolated by differential adhesion of contaminating cells. For characterization via marker protein synthesis, the nonadherent ATII cells were cultured (0.75 × 106 cells/ml media) on collagen type-I coated plates in defined cell culture medium 1 medium (Biological Industries, Kibbutz, Israel) containing 10% newborn calf serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM glutamine. For characterization by Raman microspectroscopy, ATII cells were cultured (0.20 × 106 cells/ml media) on collagen type-I coated MgF2 coverslips (Global Optics, Bournemouth, UK) in defined cell culture medium 1, with supplements as above. ATII cells were plated at a lower seeding density on MgF2 to facilitate the collection of spectra of single, individual ATII cells.
Alkaline phosphatase staining
Staining for alkaline phosphatase (ALP) was performed as described previously (16). Washed cells were incubated with alkaline phosphatase stain (10 mg naphthol-AS-bisphosphate in 40 μl dimethylsuphoxide, made up to 10 ml with 0.625 M MgCl and 0.125 M amino methyl propanol in distilled water and filtered immediately before use) for 20 min at 37°C and washed with distilled water before viewing. Intense, pink staining identifies ATII cells.
Immunocytochemistry
Cell monolayers were washed, fixed in paraformaldehyde (40 μl/ml phosphate-buffered saline (PBS); Invitrogen), and pretreated by heat to facilitate antigen retrieval (15 min at 90°C in target retrieval solution; Dako, Ely, UK). Cells were probed with an antibody to prosurfactant protein-C (pro-Sp-C, 1 μg/ml rabbit polyclonal; Chemicon (Millipore), Chandlers Ford, UK) antibodies or an isotype specific control at the same concentration. Immunoreactivity of the antibody was detected using an indirect streptavidin-biotin method (LSAB kit; Dako) and horse radish peroxidase. Bound antibody was detected as a brown stain.
Caveolin-1 immunoblotting
Confluent cells were lysed in sample buffer (250 mM Tris, pH 6.8; 100 μl/ml glycerol; 40 μl/ml sodium dodecyl sulfate; 20 μl/ml 2-mercaptoethanol; 0.1 μl/ml bromophenol blue) and immediately boiled for 10 min. Samples were centrifuged (5 min, 7500 × g) before analysis of equal amounts of protein. The amount of protein was determined by lysis of additional equivalent wells of cells with PBS containing 20 μl/ml TX-100 and analysis by a bicinchoninic acid assay (Sigma). Proteins were resolved using a NuPAGE 10% Bis-Tris gel (Invitrogen) with 2-(N-morpholino)ethanesulfonic acid running buffer and transferred to a nitrocellulose membrane. The membrane was blocked (50 mg/ml skimmed milk powder in Tris NaCl; 1 μl/ml tween-20) for 1 h at room temperature before being probed with an antibody to caveolin-1 (1:2000, rabbit polyclonal; Transduction Laboratories, Lexington, KY) overnight at 4°C. Proteins were detected by incubation with horse radish peroxidase conjugated anti-rabbit IgG (Dako) and visualized using enhanced chemiluminescence (Visualizer; Upstate (Millipore), Chandlers Ford, UK). Human umbilical vein epithelial cells were used as a positive control.
Osmium tetroxide staining of lamellar bodies in ATII cells
Staining of Lbs within primary ATII cells was performed as described previously (17). Briefly, the cells were rinsed in PBS (pH 7.3) and fixed with glutaraldehyde (15 μl/ml PBS) for 15 min. The cells were then washed twice and treated with osmium tetroxide (10 mg/ml PBS; Sigma) for 90 min at room temperature. The cells were then washed twice and incubated in tannic acid (10 mg/ml PBS, pH 6.8; Sigma) overnight. After washing with PBS (pH 6.8), the cells were examined and photographed under light microscopy.
Raman microspectroscopy
Raman spectra were measured with a Renishaw InVia (Renishaw, Wotton-under-Edge, UK) spectrometer connected to a Leica (Wetzlar, Germany) microscope, as described previously (11). The spectrometer parameters were set as in Owen et al. (18), and a high power 785 nm diode line focus laser (∼120 mW before objective; Renishaw) was used for excitation. The laser was focused on individual cells by a 63× (NA = 0.9) long working distance (2 mm) water immersion objective (Leica). Under these conditions, the laser illuminates an elliptical region at the focal plane, with ∼10 μm × 20 μm lateral spatial resolution. Spectra of living ATII cells were measured in Dulbecco's modified Eagle's medium (Sigma) with 1% antibiotic/antimycotic (Invitrogen) maintained at 37°C with a heated stage. Cells were serum-starved overnight for synchronization in the G0/G1 phase of the cell cycle to minimize cell cycle related spectral variance (19). Spectra of the fingerprint region (550–1850 cm−1) were recorded at a resolution of ∼1–2 cm−1, with 40 s integration time per spectrum. Raman backscattered radiation was collected by the same objective, passed through a 785 nm holographic notch filter to block Rayleigh scattering and reflected laser light, before being directed through a 50 μm slit into the spectrometer equipped with a 1200 lines/mm grating, and finally detected by a deep-depletion charge-coupled device detector.
Raman spectral characterization of ATII cell phenotype was assessed at three time points postseeding: early (days 3–4), middle (days 10–11), and late (days 17–18). Our motivation for analyzing cells at these three particular time points was in hopes of observing the ATII and ATI-like cell phenotypes at the early and late time points, respectively, and an intermediate phenotype at the middle time point. Two-day time points were used to facilitate Raman analysis of sufficient numbers of cells.
Data analysis
All Raman data were processed and analyzed with software developed in-house for the MATLAB (The MathWorks, Natick, MA) environment, and with the multivariate statistical analysis toolbox PLS_Toolbox (Eigenvector Research, Manson, WA). Spectra were intensity-corrected for instrument response to minimize the effects of day-to-day spectral variation (laser intensity fluctuations, optical throughput, etc.) (11), and normalized with extended multiplicative signal correction (EMSC) (20). EMSC is an algorithm that corrects spectra for additive and multiplicative effects with respect to a common reference spectrum, usually the mean input spectrum. The open-source EMSC program was modified to incorporate fifth-order wavelength-dependent spectral variations to account for background effects. Spectra were smoothed using a Savitsky-Golay filter (five points, second order polynomial) (21), and the wavenumber axis of each spectrum was standardized and aligned to the sharp phenylalanine peak at 1003 cm−1, and truncated to the 600–1800 cm−1 spectral region. Where spectra have been baseline-corrected, the Modpoly algorithm described by Lieber and Mahadevan-Jansen (fifth order polynomial, 1000 iterations) (22) was used to eliminate fluorescence and other background effects. Fig. S1, Fig. S2, Fig. S3, Fig. S4, Fig. S5, Fig. S6, Fig. S7, and Fig. S8 in the Supplementary Material describe the effects of signal processing filters and multivariate analysis on Raman spectra of ATII cells.
Principal component analysis and factor analysis
More detailed descriptions of PCA and multivariate factor analysis have been published before (23,24). PCA is a well-established chemometric technique often used to compress high-dimensional spectra by expressing the data in terms of a small number of principal components (PCs) (12,25). PCA applied to Raman spectra generates PCs and scores. Formally, the PCs are orthogonal eigenvectors of the data covariance matrix, with the greatest variance captured by PC1, the second greatest variance by PC2, etc. The PCs contain spectral features corresponding to the main molecular species responsible for the statistical variation among spectra. The PCs can be considered as a reduced set of “basis” spectra, where a basis is a set of common spectra that describe the entire data set. The original spectra can be represented as a linear combination of the PC basis vectors, where score values are the weighting coefficients describing the contribution of the basis vectors to the original spectra. The preprocessed Raman spectra were analyzed using a singular value decomposition PCA algorithm.
The PCs are abstract mathematical vectors, and in general do not represent pure biochemical components, but rather combinations of both positive and negative spectral peaks from multiple components. Factor analysis can be used as an extension of PCA to obtain a more interpretable form of the PC vectors. In this approach, the eigenvectors generated in the covariance matrix diagonalization are “‘rotated” (linearly combined by weighted addition or subtraction) to give “factors” that describe the spectral signatures of biochemical species that vary within the data set. Score values are also rotated, so that factor scores indicate the relative amount of each factor present in the original spectra. The multivariate curve resolution program in the PLS_Toolbox was used to rotate the significant PCs using a constrained iterative least-squares solver subject to nonnegativity constraints.
Spectral modeling
A nonnegative least-squares fitting procedure was used to estimate the signal contribution of biochemical components to the Raman spectral signatures of ATII and ATI-like cells. The basis spectra generated in PCA and factor analysis are mathematically derived spectral vectors based on the variance-covariance structure of the data (12). Spectral modeling, in contrast, imposes a preselected set of basis spectra on the data. Each mean ATII cell spectrum was modeled by a linear combination of basis spectra representative of cellular biochemical components (26). Details of the reference biochemicals used in spectral modeling can be found elsewhere (18). Briefly, the basis components included spectra of nucleic acids (DNA and RNA), proteins (actin, human serum albumin, chymotrypsin, and collagen), carbohydrates (glycogen), lipids (phosphatidyl choline and cholesterol), and a fifth-order polynomial (to model the background signal). The quality of the fitting was assessed by comparing model spectra to the empirical data by examining residuals, and by computing the correlation coefficient (R) between each original spectrum and its model spectrum. The correlation coefficient provides a statistical measure of the similarity among spectra (27) and was used here as a goodness-of-fit parameter (R = 1 for identical spectral profiles).
RESULTS
Characterization of ATII cell phenotype
Early time-point ATII cells (days 3–4) were small and cuboidal in shape, grouped together to form small islands of cells when cultured on MgF2 coverslips, and demonstrated positive staining for numerous Lbs (Fig. 1 A)—the main morphological feature of ATII cells. These cells formed monolayers when cultured on tissue culture plates, and demonstrated intense positive staining for ALP (Fig. 2 A) and pro-Sp-C (Fig. 2 B), both markers for the ATII phenotype. Cells cultured on MgF2 coverslips were slightly larger than those on tissue culture plates, most likely because they were plated at a lower seeding density and thus had more room to spread out. ATII cells cultured on MgF2 also stained positive for ALP and pro-Sp-C markers (Fig. S9).
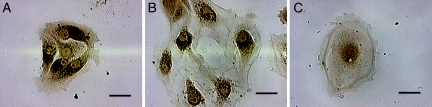
FIGURE 1.
Morphology of primary ATII cells cultured on MgF2 coverslips. Light microscope images of ATII cells stained with osmium tetroxide and tannic acid for the presence of Lbs. Shown are typical staining pictures of cells collected at the (A) early time point (days 3–4), (B) middle time point (days 10–11), and (C) late time point (days 17–18). The dark osmiophilic granules indicate cytoplasmic Lbs. Scale bar, 20 μm.
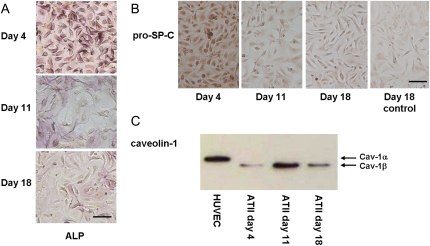
FIGURE 2.
Characterization of primary ATII cells. (A) Positive ALP and (B) pro-Sp-C staining of human ATII cell monolayers. (C) Western blot of caveolin-1 (α- and β-isoforms) expression from ATII cells over 18 days. HUVEC (human umbilical vein epithelial cells) represents the positive control for caveolin-1. Scale bar, 50 μm.
Over the 18-day culture period, the ATII cells flattened and spread out, demonstrating a roughly circular, elevated, central perinuclear region, surrounded by large, thin, and featureless cytoplasmic attenuations (5). In some regions, the squamous cells formed a continuous epithelial monolayer, but several individual, isolated cells were also present. The clearance of cellular surfactant during differentiation was evident by the reduction in the number of cells containing Lbs as well as the number of Lbs per cell (Fig. 1). This is consistent with the secretion of surfactant from ATII cells into the cell culture media, as demonstrated in previous in vitro investigations of ATII cell differentiation (28). Late-culture ATII cells (days 17–18) contained few Lbs, and when present, were observed throughout the perinuclear region but were scarcely observed in the flattened, peripheral attenuations. The decrease in staining intensity for both ALP and pro-Sp-C (Fig. 2, A and B) over time indicates an increase in the number of differentiated cells in culture. Positive expression of the ATI-like phenotype was confirmed by examining caveolin-1 protein expression by immunoblotting (Fig. 2 C). Caveolin-1 is a major structural protein of caveolae, plasma membrane invaginations involved in membrane transport, which are features of ATI cells, but are absent or only weakly expressed in ATII cells (6). The western blot indicates low caveolin-1 levels for early culture cells (day 4), and higher expression for middle and late culture cells (days 11 and 18). The decrease in caveolin-1 expression from day 11 to day 18 may be due to fewer numbers of cells at the late time point, possibly reflecting loss of cells through cell death or loss of phenotype over time in vitro.
Spectral variability within single living ATII cells
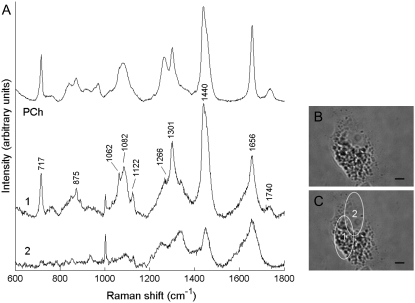
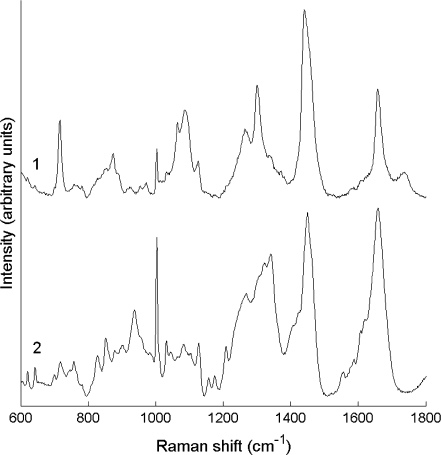
For each cell, Raman spectra were measured at different positions to study spectral variation with respect to spatial location. Significant spectral heterogeneity was observed between spectra collected from the nucleus and cytoplasm of early-culture ATII cells (Fig. 3). Cytoplasm spectra (e.g., spectrum 1, Fig. 3) were very intense and dominated by molecular vibrations from phospholipids (Table 1) (29). The high signal intensity and prominent phospholipid features are due to the presence of Lbs, cytoplasmic organelles densely packed with surfactant, which is rich in phosphatidyl choline. The strong similarity between this cytoplasm spectrum and the Raman spectrum of phosphatidyl choline is also highlighted in Fig. 3.
FIGURE 3.
(A) Raman spectra collected from the cytoplasm (spectrum 1) and nucleus (spectrum 2) of an early-culture ATII cell. The spectrum of phosphatidyl choline (PCh) is included for comparison with spectrum 1. The spectra have been corrected for instrument response, baseline-corrected, the wavenumber axis standardized and aligned, and the PCh spectrum scaled to represent equal maximum intensity to spectrum 1. (B) Light microscope picture of an early-culture ATII cell. (C) Same cell as in B, with sites of spectral acquisition labeled 1 and 2. Ellipses indicate the region probed by each scan. The spectra are offset for clarity. The correlation coefficient between spectrum 1 and the PCh spectrum is R = 0.97. Scale bar, 5 μm.
TABLE 1.
Assignments for prominent Raman shifts arising from molecular vibrations in cellular lipids
| Raman shift (cm−1) | Lipid assignment |
|---|---|
| 717 | Choline headgroup: CN+(CH3)3 stretch |
| 875 | C-C-N+ symmetric stretch (phosphatidyl ethanolamine) |
| 1062, 1122 | Out-of-phase and in-phase modes of all-trans chain |
| 1082 | Gauche conformation |
| 1266 | C=C-H in-plane bend of the cis -CH=CH- linkage |
| 1301 | In-phase CH2 twisting mode |
| 1449 | CH bending modes, including CH2 scissors and CH3 degenerate deformation of hydrocarbon chains |
| 1656 | C=C stretch of the cis -CH=CH- linkage of unsaturated lipid chains |
| 1740 | Carbonyl C=O ester stretch |
In contrast, spectra collected from cellular regions including the nucleus (e.g., spectrum 2, Fig. 3), had much lower intensity and resembled typical, protein-dominated cell spectra with minor contributions from nucleic acids and lipids (30,31). Spectra 1 and 2 exhibit clear differences in several bands specific to lipids, amino acids, and nucleotides, as well as bands that contain information from some or all of these biopolymers, including symmetric  stretch (1095 cm−1), C-H deformation (1449 cm−1), amide I (1656 cm−1), and amide III (1250–1350 cm−1) envelopes (26). For example, the broadening of the 1656 cm−1 C=C stretching band from unsaturated lipid chains compared to the pure phosphatidyl choline (PCh) spectrum is due to amide I modes of proteins (C=O stretching mode, peptide linkage), which feature much more prominently in spectrum 2, where the 1656 cm−1 band is broader and weaker than in spectrum 1.
stretch (1095 cm−1), C-H deformation (1449 cm−1), amide I (1656 cm−1), and amide III (1250–1350 cm−1) envelopes (26). For example, the broadening of the 1656 cm−1 C=C stretching band from unsaturated lipid chains compared to the pure phosphatidyl choline (PCh) spectrum is due to amide I modes of proteins (C=O stretching mode, peptide linkage), which feature much more prominently in spectrum 2, where the 1656 cm−1 band is broader and weaker than in spectrum 1.
Biomolecular characterization of single living ATII cells
Early-culture ATII cells had typical dimensions of ∼20 μm × 30 μm, and we found that three spectra were adequate to probe a single cell. Middle and late-culture ATII cells were much larger, with the main cellular features confined to a central perinuclear region. Perinuclear spectra exhibited features of proteins, lipids, and nucleic acids, but lacked the intense lipid peaks that dominated the spectra of early-culture ATII cells. Spectra collected from thin cytoplasmic attenuations were extremely weak and noisy, and did not display significant features above the signal of the culture medium (Fig. S10). Therefore, we focused our attention on the perinuclear region of these larger, flattened cells, which helped to expedite live cell analysis and preserve signal quality. Furthermore, we observed that the perinuclear region of such cells was similar in size to early-culture ATII cells, and could be scanned with three spectra.
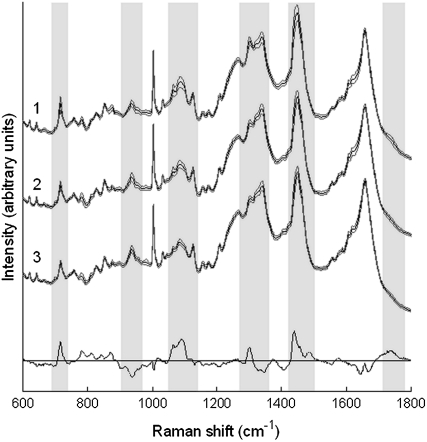
For each cell, the spectral measurements were averaged, with the mean Raman spectrum representing the cell's biomolecular fingerprint. The average processed Raman spectra of ATII cells from each of the three time points are shown in Fig. 4. Solid lines denote average spectra, which represent the mean of 183, 204, and 171 spectra collected from 61 early culture ATII cells (spectrum 1), 68 middle time point cells (spectrum 2), and 57 late time point cells (spectrum 3), respectively. The shaded lines represent ± 1 SD from the mean. All of the spectra exhibit similar general features, and consist of peaks corresponding to molecular vibrations of all cellular biopolymers. Detailed tables of peak assignments are available in the literature (11). The small standard deviations within each group indicate that Raman spectral characterization of ATII cell phenotypes is highly reproducible. The difference spectrum between the mean spectra of early and late time point ATII cells is also included to indicate the main differences between the cells at these time points (magnified by a factor of 2.5).
FIGURE 4.
Average Raman spectra of individual ATII cells measured at early (days 3–4, spectrum 1), middle (days 10–11, spectrum 2), and late (days 17–18, spectrum 3) time points. The spectra represent the mean of 183, 204, and 171 spectra collected from 61, 68, and 57 cells, respectively. The solid lines indicate the average spectra and the shaded lines delineate one standard deviation. Spectra have been offset for clarity. The difference spectrum (bottom spectrum) between the mean of early time point and late time point cells is also included, and has been magnified by a factor of 2.5 to highlight the main differences among cells at these time points.
PCA-based factor analysis
PCA was performed on the preprocessed, average Raman spectra of ATII cells. The first six PCs were identified as the main descriptors of spectral variance (accounting for 83%), whereas the remaining PCs were noisy and each accounted for <1% of the total variance. The PCs are abstract vectors, containing both positive and negative peaks from multiple biochemical components. PC1, for example, displayed prominent lipid features, and was very similar to the difference spectrum between the mean spectra of early and late time-point ATII cells presented in Fig. 4 (R = 0.84, Fig. S11).
We rotated the significant PCs in a constrained factor analysis to obtain physically realistic factors representing biochemical species that differ among the spectral profiles of ATII and ATI-like cells. Two main factors, describing phospholipid and protein spectral components, are shown in Fig. 5. The phospholipid factor (spectrum 1, Fig. 5) strongly resembles spectrum 1 of Fig. 3, collected from the cytoplasm of early-culture (days 3–4) ATII cells (R = 0.98). The protein factor (spectrum 2, Fig. 5) mostly describes cellular proteins, confirmed by accurate spectral modeling of the protein factor with the protein basis spectra (R = 0.98, Fig. S12). The four additional factors did not describe pure-component spectra, but rather minor peak shifts, correlated spectral features from multiple components (e.g., protein and lipid, protein and nucleic acids), minor peak shifts, and residual noise.
FIGURE 5.
Factors generated from eigenvector rotation. Factor 1 describes changes in cellular phospholipids during ATII cell differentiation, and is extremely similar to the cytoplasm spectrum displayed in Fig. 1 (spectrum 1, R1 = 0.98). Factor 2 describes protein-related differentiation changes, and demonstrated significant correlation with a model protein spectrum generated by least-squares fitting Factor 2 with a basis set of protein spectra (R2 = 0.98). Factors have been baseline-corrected and normalized to maximum intensity values, and are offset for clarity.
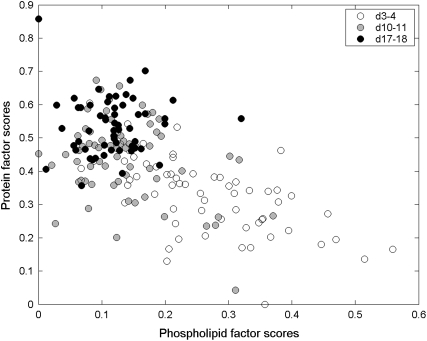
The score values generated in the factor analysis were plotted to investigate clustering of the data. PC and factor score plots can be used to determine whether or not spectra are related: clustering or grouping of spectra within these plots indicates systematic differences among the spectra (32). Overlap of time point clusters (Fig. 6) indicates that each group represents a mixed population of ATII and ATI-like cells. The small degree of overlap between clusters of early and late time point cells (<10%) indicates that most early culture cells express an ATII phenotype, and most late time point cells express a differentiated ATI-like phenotype. The close proximity of the middle time point cluster to the late time point cluster suggests that most of these cells have already differentiated and express an ATI-like phenotype. However, the scattering of ∼¼ of the middle time point cells throughout the early time point cluster indicates that the differentiation process is not homogeneous, and that some cells differentiate faster than others.
FIGURE 6.
Factor scores plot. Phospholipid factor scores versus protein factor scores, generated by constrained eigenvector rotation. The time point clusters indicate a decrease in lipid and an increase in protein contribution to the Raman signal of ATII cells during differentiation in vitro.
Spectral modeling
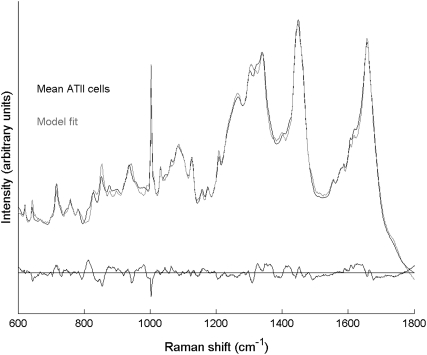
The mean spectra of ATII cells were individually fit with a set of basis spectra to investigate changes in spectral contributions of cellular biochemical components during ATII cell differentiation. An example of the fitting results is displayed in Fig. 7, where the mean spectrum of early time point cells is plotted along with the model-predicted spectrum and residual. The good quality of the fit is indicated by the small residual spectrum and high correlation coefficient R = 0.996. The small number and magnitude of negative peaks in the residual spectrum indicates minimal overestimation in the contribution of reference spectra. Positive residual peaks point to some unmodeled signal, likely due to lipids and proteins (small positive peaks at 717 cm−1 and 815, 1325, 1607 cm−1, respectively). High quality modeling results were obtained for all 61 early time point cells (Rmean = 0.995 ± 0.002), 68 middle time point cells (Rmean = 0.994 ± 0.003), and 57 late time point cells (Rmean = 0.995 ± 0.002). All correlation coefficients were statistically significant. Least-squares model fitting of the mean spectra of cells collected at the middle and late time points is included in Fig. S13.
FIGURE 7.
Spectral modeling of ATII cells with Raman microspectroscopy: The mean spectrum of the 61 early time point cells (solid), the model fit (shaded), and the residual spectrum (R = 0.996).
The model fit coefficients are weighting/scaling factors that, when normalized to unity sum, indicate the relative percent signal contribution of each component to the original Raman spectrum (Table 2). The results are grouped together in terms of component classification, i.e., nucleic acids, proteins, carbohydrates, and lipids, rather than individual components. Thus the coefficients for DNA and RNA were added to indicate the total nucleic acid composition, the separate protein fit coefficients added together to give the overall protein composition, etc. This grouping is necessary because the basis components contain overlapping spectral features (i.e., are not orthogonal), and hence similar components such as DNA and RNA are not completely spectrally resolved (18,33). Differences in normalized fit coefficients of components between time points were tested for statistical significance using the Kruskal-Wallis test.
TABLE 2.
Relative percent signal contribution of model basis components to the Raman spectra of early, middle, and late time point ATII cells
| Time point | Nucleic acid (%) | Protein (%) | CHO* (%) | Lipid (%) |
|---|---|---|---|---|
| Early (days 3–4) | 11.8 ± 2.0 | 51.8 ± 7.7 | 8.5 ± 1.5 | 27.9 ± 8.8 |
| Middle (days 10–11) | 10.8 ± 2.8 | 61.2 ± 5.6† | 9.2 ± 1.3 | 18.8 ± 6.9† |
| Late (days 17–18) | 8.4 ± 2.4 | 63.0 ± 4.6‡ | 9.5 ± 1.0 | 19.1 ± 5.9‡ |
CHO, carbohydrate.
and ‡ denote statistically significant differences between early/middle and early/late time points, respectively, computed using Kruskal-Wallis test (p < 0.05).
DISCUSSION
The application of Raman microspectroscopy to distinguish among different cell types and phenotypes has been reported by several research groups (11,12,25,34). Multivariate analysis techniques have made possible systematic (and often unsupervised) analysis of large, high-dimensional spectral data sets, and can identify important sources of spectral variation across a broad frequency range. Few studies have focused on the analysis of live cells, or on phenotypic discrimination of stem/progenitor cells and their differentiated progeny. A previous study by Notingher et al. (13) demonstrated the potential of Raman microspectroscopy for biomolecular characterization of live embryonic stem cells, and identified spectral markers describing mRNA translation as indicators of stem cell differentiation. In this study, we have taken full advantage of the noninvasive sensing capabilities of Raman microspectroscopy to characterize the differentiation of live, primary human ATII cells in vitro.
Cellular changes in morphology (Fig. 1) and marker protein synthesis (Fig. 2) were consistent with the known differentiation pattern of ATII cells (4,16), confirming that an ATII phenotype was dominant among early culture cells (d3-4). We did not detect a separate, intermediate phenotype at the middle time point (days 10–11), and instead found that most of the cells expressed an ATI-like phenotype by the middle time point. Thus, spectrum 1 in Fig. 4 represents the spectral profile corresponding to an ATII phenotype, whereas an ATI-like phenotype is associated with spectral profiles 2 and 3. It is possible that the time resolution of our experiment was too long to detect an intermediate phenotype, which has previously been reported to appear transiently during the differentiation process (2).
For the Raman spectra of ATII cells (Fig. 4), the differences at the various time points in the differentiation process manifest themselves mostly as changes in phospholipid vibrations (717, 1062, 1082, 1301, 1740 cm−1). Investigation of spectral heterogeneity revealed that a phospholipid-dominated spectral component (spectrum 1, Fig. 3), commonly measured from the cytoplasm of early-culture ATII cells but absent in late-culture cells, was primarily responsible for these differences. Changes in several other bands ascribed to proteins or multiple components were also detected, most notably the medium band at 937 cm−1 with contributions from C-C α-helix protein stretch, C-C-N+ symmetric stretch in lipids, and C-O-C glycosidic linkage of carbohydrates; the CH2 deformation in proteins at 1340 cm−1; the 1449 cm−1 band encompassing C-H bending modes in all cellular biopolymers; and the 1665-1680 cm−1 band that includes the protein amide I mode as well as the 1656 cm−1 C=C stretch of the cis-CH=CH linkage of unsaturated lipid chains (11,35).
The observed changes in ATII cell spectra were consistent with the spectral factors generated by unsupervised PCA and subsequent constrained eigenvector rotation. We identified two independent factors, one describing cellular phospholipids and the other cellular proteins (Fig. 5) as the main discriminating factors between ATII and ATI-like cells. The phospholipid factor (spectrum 1, Fig. 5) was remarkably similar to the cytoplasm spectra collected from days 3–4 ATII cells (e.g., spectrum 1, Fig. 3), and the intense vibrations at 1266 cm−1 and 1656 cm−1 indicate the presence of unsaturated phospholipids (e.g., phosphatidyl glycerol) within cytoplasmic Lbs of ATII cells, which are known to be rich in saturated phospholipids (e.g., dipalmitoyl phosphatidyl choline) (2). This observation is supported by the ratio of the trans-to-gauche intensity of the skeletal C-C stretch modes in the 1000-1150 cm−1 region, which are sensitive to the conformation of hydrocarbon chains. Specifically, the bands at 1062 cm−1 and 1122 cm−1 are assigned to the all-trans chain configuration, whereas the 1082 cm−1 band is attributed to the gauche conformation (36). The trans-to-gauche ratio for Factor 1 is intermediate of that previously reported for fully saturated dipalmitoyl phosphatidyl choline and for unsaturated dioleoyl phosphatidyl choline (37), indicating that the conformational order of the hydrocarbon chains within cytoplasmic Lbs of ATII cells is intermediate of that for pure saturated and unsaturated phospholipids, which is consistent for a vesicle containing both saturated and unsaturated phospholipids.
The phospholipid factor also contained some minor protein features (1003, 1336 cm−1, amide I broadening of the 1656 cm−1 band), which likely arise from both surfactant proteins (Sp-A, B, C, and D) and nonsurfactant cellular proteins. Apart from the much higher intensity of the choline peak at 717 cm−1 observed here, the phospholipid factor is very similar to the cluster-averaged spectrum of cellular vesicles reported by Krafft et al. (26), collected from a high spatial resolution Raman map of a fixed human lung fibroblast.
The protein factor (spectrum 2, Fig. 5) described mainly α-helix rather than β-sheet proteins, inferred from the presence of peaks characteristic of the α-helix protein structure (937, 952 cm−1), the absence of key β-sheet protein vibrations (760, 975 cm−1), and an amide I center frequency of ∼1658 cm−1 (11,35,38). The protein factor also contained minor signal contributions from lipids (717, 1082, 1122 cm−1) and carbohydrates (855, 937, 1045 cm−1), indicating that these minor spectral features are correlated to the main protein features.
The factor scores plot (Fig. 6) illustrates how the spectral contribution of the phospholipid and protein factors change as ATII cells differentiate. Spectra of early time point (days 3–4) cells were characterized by high phospholipid content relative to protein, whereas late time point cells (days 17–18) yield spectra with much higher protein/lipid ratios. The decrease in phospholipid signal contribution to ATII cell spectra correlates well with the clearance of phospholipid-rich surfactant from Lbs as ATII cells differentiate (Fig. 1). This observation is further supported by the spectral modeling results (Table 2), which indicate a significant 9% relative decrease in the Raman signal contribution from cellular lipids from ∼28% to ∼19% as ATII cells differentiate (p < 0.05 for both early/middle and early/late time point comparisons). Of this decrease, 8% is attributed to a decrease in phosphatidyl choline, and only 1% to cholesterol. The reduction of spectral signal from lipids is offset by a significant 10% relative increase in the signal contribution from cellular proteins, from ∼52% in early culture cells to ∼62% in middle/late culture cells (p < 0.05 for both early/middle and early/late time point comparisons). The negative correlation between relative signal contribution of lipids and proteins to the spectra of ATII cells was further confirmed by plotting the protein model fit coefficients against those for lipids for all primary ATII cells analyzed (Fig. S14). Thus, the model-predicted spectral contribution from phospholipids can be used as a spectroscopic marker for the identification of cellular phenotype (i.e., ATII versus ATI-like), and hence the differentiation state of primary ATII cells in vitro.
This experiment complements previous ATII cell differentiation studies by offering a noninvasive, spectroscopic perspective of the biochemical changes that live ATII cells undergo as they differentiate toward an ATI-like phenotype. It also complements a coherent anti-Stokes Raman scattering (CARS) vibrational imaging study by Nan et al., in which they demonstrated selective, high-speed imaging of lipid droplets in unstained, live cells (39). CARS is a nonlinear optical process that provides resonant enhancement of the weak spontaneous Raman scattering signal, typically by a factor of ∼105. The chemical selectivity, speed, and real-time imaging features of CARS microscopy make it an attractive alternative to fluorescence microscopy for cellular imaging. Using CARS, Nan et al. were able to generate chemical images describing the cellular distribution of lipids by tuning the excitation laser to interrogate a lipid vibration at 2845 cm−1. By collecting chemical images at different time points, the authors investigated the dynamics of lipid droplets during the differentiation of 3T3-L1 fibroblast cells to adipocytes. Their experiments revealed an initial clearance of lipid droplets in the early stages of differentiation, followed by an unsynchronized reaccumulation of lipid droplets at later stages. This is in contrast to our findings for ATII cells, where no reaccumulation of Lbs was observed at later stages of differentiation.
The focus of this study was to noninvasively characterize the in vitro differentiation of single, living ATII cells based on the collection of Raman scattering from the entire cellular volume. This was facilitated by using a high-power near infrared line-focus laser offering nondiffraction limited spatial resolution to excite Raman scattering from large subcellular regions. Raman microspectroscopy can also be used to study specific cellular organelles, e.g., mitochondria (40), nucleus, and endoplasmic reticulum (26), as well as the composition and physical properties of phospholipids within model protein-surfactant systems, including the conformational order of hydrocarbon chains, interchain interactions, intrachain isomerizations, and phospholipid headgroup packing order (41). Takai et al. have previously demonstrated the application of confocal Raman microspectroscopy to characterize the lipid structure of triacylglycerol-based cytotoxic granules within single living human lymphocytes (42). We hope to use this technology to noninvasively analyze the composition and physical properties of surfactant phospholipids stored in Lbs within the cytoplasm of live ATII cells at different stages of differentiation. In a comprehensive lipidomic analysis of surfactant produced by primary human ATII cells, Postle et al. demonstrated that ATII cell differentiation has a profound effect on composition of surfactant phospholipids, especially on acyl chain length (43). However, the significance of these changes is not yet understood. Our aim is to apply confocal Raman microspectroscopy with diffraction-limited spatial resolution to probe individual Lbs (typically 1–2 μm in diameter), which would complement existing ultrastructural information on Lbs within fixed ATII cells (44). This line of future research could also be applied to the development of synthetic surfactant systems, whereby Raman microspectroscopic data on the unperturbed cellular system could aid in the design of functional, biologically-relevant, and therapeutic surfactants (45).
CONCLUSIONS
We have demonstrated that Raman microspectroscopy can be used to characterize and identify phenotypic differences between alveolar epithelial cells at different stages of differentiation in vitro. The spectral profile of undifferentiated, ATII cells exhibited intense phospholipid peaks, corresponding to the presence of surfactant Lbs in the cytoplasm. In contrast, the spectra of differentiated, ATI-like cells demonstrated significantly less lipid character. By correlating cytochemical staining results with multivariate spectral analysis, we were able to systematically detect spectroscopic markers indicating a reduction in cellular surfactant during differentiation. PCA-based factor analysis identified a phospholipid spectral component as the main discriminator between the spectra of ATII and ATI-like cells, and a decrease in the contribution of this factor to ATII cell spectra during differentiation was observed in the factor scores. Spectral modeling revealed a significant 9% decrease in relative phospholipid Raman signal contribution as ATII cells differentiate. These observations are consistent with the clearance of surfactant from cytoplasmic Lbs in the transition from an ATII to an ATI-like phenotype. This study demonstrates the advantages of Raman microspectroscopy over invasive cytochemical techniques, and the potential of this technology to further enhance our understanding of surfactant phospholipid modifications during ATII cell differentiation.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
We thank Dr. Kees Maquelin for assistance with modifying the EMSC code for signal processing, and Dr. Gavin Jell for helpful comments and proof-reading.
We thank the Leverhulme Trust for financial support, and acknowledge the Rothermere Trust Foundation, the National Science and Engineering Research Council (Canada), and the Canadian Centennial Scholarship Fund for fellowship support to R.J.S. S.J.K. acknowledges funding from the Department of Health, UK.
Editor: David W. Piston.
References
- 1.Crapo, J. D., B. E. Barry, P. Gehr, M. Bachhofen, and E. R. Weibel. 1982. Cell number and cell characteristics of normal human lung. Am. Rev. Respir. Dis. 125:332–337. [DOI] [PubMed] [Google Scholar]
- 2.Fehrenbach, H. 2001. Alveolar epithelial type II cell: defender of the alveolus revisited. Respir. Res. 2:33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uhal, B. D. 1997. Cell cycle kinetics in the alveolar epithelium. Am. J. Physiol. 272:L1031–L1045. [DOI] [PubMed] [Google Scholar]
- 4.Fuchs, S., A. J. Hollins, M. Laue, U. F. Schaefer, K. Roemer, M. Gumbleton, and C.-M. Lehr. 2003. Differentiation of human alveolar epithelial cells in primary culture: morphological characterization and synthesis of caveolin-1 and surfactant protein-C. Cell Tissue Res. 311:31–45. [DOI] [PubMed] [Google Scholar]
- 5.Cheek, J. M., M. J. Evans, and E. D. Crandall. 1989. Type I cell-like morphology in tight alveolar epithelial monolayers. Exp. Cell Res. 184:375–387. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, L., A. J. Hollins, A. Al Eid, G. R. Newman, C. von Ruhland, and M. Gumbleton. 1999. Caveolin expression and caveolae biogenesis during transdifferentiation in lung alveolar epithelial primary cultures. Biochem. Biophys. Res. Commun. 262:744–751. [DOI] [PubMed] [Google Scholar]
- 7.Raman, C. V., and K. S. Krishnan. 1928. A new type of secondary radiation. Nature. 121:501–502. [Google Scholar]
- 8.Ellis, D. I., and R. Goodacre. 2006. Metabolic fingerprinting in disease diagnosis: biomedical applications of infrared and Raman spectroscopy. Analyst. 131:875–885. [DOI] [PubMed] [Google Scholar]
- 9.Puppels, G. J., F. F. de Mul, C. Otto, J. Greve, M. Robert-Nicoud, D. J. Arndt-Jovin, and T. M. Jovin. 1990. Studying single living cells and chromosomes by confocal Raman microspectroscopy. Nature. 347:301–303. [DOI] [PubMed] [Google Scholar]
- 10.Swain, R. J., and M. M. Stevens. 2007. Raman microspectroscopy for non-invasive biochemical analysis of single cells. Biochem. Soc. Trans. 35:544–549. [DOI] [PubMed] [Google Scholar]
- 11.Notingher, I., G. Jell, U. Lohbauer, V. Salih, and L. L. Hench. 2004. In situ non-invasive spectral discrimination between bone cell phenotypes used in tissue engineering. J. Cell. Biochem. 92:1180–1192. [DOI] [PubMed] [Google Scholar]
- 12.Chan, J. W., D. S. Taylor, T. Zwerdling, S. M. Lane, K. Ihara, and T. Huser. 2006. Micro-Raman spectroscopy detects individual neoplastic and normal hematopoietic cells. Biophys. J. 90:648–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Notingher, I., I. Bisson, A. E. Bishop, W. L. Randle, J. M. Polak, and L. L. Hench. 2004. In situ spectral monitoring of mRNA translation in embryonic stem cells during differentiation in vitro. Anal. Chem. 76:3185–3193. [DOI] [PubMed] [Google Scholar]
- 14.Jell, G., I. Notingher, O. Tsigkou, P. Notingher, J. M. Polak, L. L. Hench, and M. M. Stevens. 2008. Bioactive glass-induced osteoblastic differentiation: A noninvasive spectroscopic study. J. Biomed. Mater. Res. A. 86A:31–40. [DOI] [PubMed] [Google Scholar]
- 15.Witherden, I. R., and T. D. Tetley. 2001. Isolation and culture of human type II pneumocytes. In Human Airway Inflammation: Sampling Techniques and Analytical Protocols. D. F. Rogers and L. E. Donnelly, editors. Humana Press, Totawa, NJ. 127–136.
- 16.Witherden, I. R., E. J. Vanden Bon, P. Goldstraw, C. Ratcliffe, U. Pastorino, and T. D. Tetley. 2004. Primary human alveloar type II epithelial cell chemokine release: effects of cigarette smoke and neutrophil elastase. Am. J. Respir. Cell Mol. Biol. 30:500–509. [DOI] [PubMed] [Google Scholar]
- 17.Alcorn, J. L., M. E. Smith, J. F. Smith, L. R. Margraf, and C. R. Mendelson. 1997. Primary cell culture of human Type II pneumocytes: maintenance of a differentiated phenotype and transfection with recombinant adenoviruses. Am. J. Respir. Cell Mol. Biol. 17:672–682. [DOI] [PubMed] [Google Scholar]
- 18.Owen, C. A., J. Selvakumaran, I. Notingher, G. Jell, L. L. Hench, and M. M. Stevens. 2006. In vitro toxicology evaluation of pharmaceuticals using Raman micro-spectroscopy. J. Cell. Biochem. 99:178–186. [DOI] [PubMed] [Google Scholar]
- 19.Swain, R. J., G. Jell, and M. M. Stevens. 2008. Non-invasive analysis of cell cycle dynamics in single living cells with Raman micro-spectroscopy. J. Cell. Biochem. 104:1427–1438. [DOI] [PubMed] [Google Scholar]
- 20.Martens, H., J. P. Nielsen, and S. B. Engelsen. 2003. Light scattering and light absorbance separated by extended multiplicative signal correction: application to near-infrared transmission analysis of powder mixtures. Anal. Chem. 75:394–404. [DOI] [PubMed] [Google Scholar]
- 21.Savitsky, A., and M. J. E. Golay. 1964. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 36:1627–1639. [DOI] [PubMed] [Google Scholar]
- 22.Lieber, C. A., and A. Mahadevan-Jansen. 2003. Automated method for subtraction of fluorescence from biological Raman spectra. Appl. Spectrosc. 57:1363–1367. [DOI] [PubMed] [Google Scholar]
- 23.Wold, S., K. Esbensen, and P. Geladi. 1987. Principal components analysis. Chemom. Intell. Lab. Syst. 2:37–52. [Google Scholar]
- 24.Shaver, J. M. 2001. Chemometrics for Raman spectroscopy. In Handbook of Raman Spectroscopy: From the Research Laboratory to the Process Line. I. R. Lewis and H. G. M. Edwards, editors. Marcel Dekker, New York. 275–306.
- 25.Crow, P., B. Barrass, C. Kendall, M. Hart-Prieto, M. Wright, R. Persad, and N. Stone. 2005. The use of Raman spectroscopy to differentiate between different prostatic adenocarcinoma cell lines. Br. J. Cancer. 92:2166–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krafft, C., T. Knetschke, R. H. W. Funk, and R. Salzer. 2005. Identification of organelles and vesicles in single cells by Raman microspectroscopic mapping. Vib. Spectrosc. 38:85–93. [Google Scholar]
- 27.Diem, M., M. Romeo, S. Boydston-White, M. Miljkovic, and C. Matthaus. 2004. A decade of vibrational micro-spectroscopy of human cells and tissue (1994–2004). Analyst. 129:880–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rooney, S. A., S. L. Young, and C. R. Mendelson. 1994. Molecular and cellular processing of lung surfactant. FASEB J. 8:957–967. [DOI] [PubMed] [Google Scholar]
- 29.Huang, Y.-S., T. Karashima, M. Yamamoto, and H. Hamaguchi. 2005. Molecular-level investigation of the structure, transformation, and bioactivity of single living fission yeast cells by time- and space-resolved Raman spectroscopy. Biochemistry. 44:10009–10019. [DOI] [PubMed] [Google Scholar]
- 30.Matthäus, C., S. Boydston-White, M. Miljkovic, M. Romeo, and M. Diem. 2006. Raman and infrared microspectral imaging of mitotic cells. Appl. Spectrosc. 60:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krafft, C., T. Knetschke, R. H. Funk, and R. Salzer. 2006. Studies on stress-induced changes at the subcellular level by Raman microspectroscopic mapping. Anal. Chem. 78:4424–4429. [DOI] [PubMed] [Google Scholar]
- 32.Boydston-White, S., M. Romeo, T. Chernenko, A. Regina, M. Miljkovic, and M. Diem. 2006. Cell-cycle-dependent variations in FTIR micro-spectra of single proliferating HeLa cells: principal component and artificial neural network analysis. Biochim. Biophys. Acta. 1758:908–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mourant, J. R., K. W. Short, S. Carpenter, N. Kunapareddy, L. Coburn, T. M. Powers, and J. P. Freyer. 2005. Biochemical differences in tumorigenic and nontumorigenic cells measured by Raman and infrared spectroscopy. J. Biomed. Opt. 10:031106–1-031106–15. [DOI] [PubMed] [Google Scholar]
- 34.Krishna, C. M., G. Kegelaer, I. Adt, S. Rubin, V. B. Kartha, M. Manfait, and G. D. Sockalingum. 2006. Combined Fourier transform infrared and Raman spectroscopic approach for identification of multidrug resistance phenotype in cancer cell lines. Biopolymers. 82:462–470. [DOI] [PubMed] [Google Scholar]
- 35.Krishna, C. M., G. D. Sockalingum, J. Kurien, L. Rao, L. Venteo, M. Pluot, M. Manfait, and V. B. Kartha. 2004. Micro-Raman spectroscopy for optical pathology of oral squamous cell carcinoma. Appl. Spectrosc. 58:1128–1135. [DOI] [PubMed] [Google Scholar]
- 36.Lippert, J. L., and W. L. Peticolas. 1971. Laser Raman investigation of the effect of cholesterol on conformational changes in dipalmitoyl lecithin multilayers. Proc. Natl. Acad. Sci. USA. 68:1572–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cherney, D. P., J. C. Conboy, and J. M. Harris. 2003. Optical-trapping Raman microscopy detection of single unilamellar lipid vesicles. Anal. Chem. 75:6621–6628. [DOI] [PubMed] [Google Scholar]
- 38.Guan, Y., E. N. Lewis, and I. W. Levin. 1999. Biomedical applications of Raman spectroscopy: tissue differentiation and potential clinical usage. In Analytical Applications of Raman Spectroscopy. M. J. Pelletier, editor. Blackwell Science, Oxford. 276–321.
- 39.Nan, X., J.-X. Cheng, and X. S. Xie. 2003. Vibrational imaging of lipid droplets in live fibroblast cells with coherent anti-Stokes Raman scattering microscopy. J. Lipid Res. 44:2202–2208. [DOI] [PubMed] [Google Scholar]
- 40.Matthäus, C., T. Chernenko, J. A. Newmark, C. M. Warner, and M. Diem. 2007. Label-free detection of mitochondrial distribution in cells by nonresonant Raman microspectroscopy. Biophys. J. 93:668–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vincent, J. S., S. D. Revak, C. G. Cochrane, and I. W. Levin. 1991. Raman spectroscopic studies of model human pulmonary surfactant systems: phospholipid interactions with peptide paradigms for the surfactant protein SP-B. Biochemistry. 30:8395–8401. [DOI] [PubMed] [Google Scholar]
- 42.Takai, Y., T. Masuko, and H. Takeuchi. 1997. Lipid structure of cytotoxic granules in living human killer T lymphocytes studied by Raman microspectrocopy. Biochim. Biophys. Acta. 1335:199–208. [DOI] [PubMed] [Google Scholar]
- 43.Postle, A. D., L. W. Gonzales, W. Bernhard, G. T. Clark, M. H. Godinez, R. I. Godinez, and P. L. Ballard. 2006. Lipidomics of cellular and secreted phospholipids from differentiated human fetal type II alveolar epithelial cells. J. Lipid Res. 47:1322–1331. [DOI] [PubMed] [Google Scholar]
- 44.Weaver, T. E., C.-L. Na, and M. Stahlman. 2002. Biogenesis of lamellar bodies, lysosome-related organelles involved in storage and secretion of pulmonary surfactant. Semin. Cell Dev. Biol. 13:263–270. [DOI] [PubMed] [Google Scholar]
- 45.Bernhard, W., J. Mottaghian, A. Gebert, G. A. Rau, H. von der Hardt, and C. F. Poets. 2000. Commercial versus native surfactants: surface activity, molecular components, and the effect of calcium. Am. J. Respir. Crit. Care Med. 162:1524–1533. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.