Abstract
Drug extrusion via efflux through a tripartite complex (an inner membrane pump, an outer membrane protein, and a periplasmic protein) is a widely used mechanism in Gram-negative bacteria. The outer membrane protein (TolC in Escherichia coli; OprM in Pseudomonas aeruginosa) forms a tunnel-like pore through the periplasmic space and the outer membrane. Molecular dynamics simulations of TolC have been performed, and are compared to simulations of Y362F/R367S mutant, and to simulations of its homolog OprM. The results reveal a complex pattern of conformation dynamics in the TolC protein. Two putative gate regions, located at either end of the protein, can be distinguished. These regions are the extracellular loops and the mouth of the periplasmic domain, respectively. The periplasmic gate has been implicated in the conformational changes leading from the closed x-ray structure to a proposed open state of TolC. Between the two gates, a peristaltic motion of the periplasmic domain is observed, which may facilitate transport of the solutes from one end of the tunnel to the other. The motions observed in the atomistic simulations are also seen in coarse-grained simulations in which the protein tertiary structure is represented by an elastic network model.
INTRODUCTION
The TolC family of proteins is central to type I secretion of toxins, small peptides, and drugs from Gram-negative bacteria. Type I secretion uses three proteins: AcrA, AcrB, and TolC. TolC is located in the outer membrane and extends into the periplasmic space (1–4). It interacts with a periplasmic protein (AcrA) and with an inner membrane efflux pump protein (AcrB). In this tripartite arrangement, the efflux complex enables the direct passage of the solute from the cytoplasm to the external medium (5,6). This observation is in agreement with the x-ray structures of the outer membrane protein TolC and of the inner membrane protein AcrB (7–10). TolC extends into the periplasmic region for ∼100 Å, whereas AcrB protrudes ∼70 Å. In combination with the third accessory protein AcrA (11), they form a long molecular tunnel.
The x-ray structure of TolC revealed the protein to be a trimer. The overall shape of TolC is that of a cylinder, which is 140 Å in length and contains a transmembrane (TM) domain and a periplasmic domain. The TM domain is a 12-stranded β-barrel domain formed by the three monomers. The periplasmic domain contains 12 α-helices, and is ∼100 Å in length and ∼30 Å in diameter. In the midsection of the periplasmic domain, there is a mixed α/β-domain, which is the equatorial domain. In the export of drugs and peptides, the main obstacle is at the periplasmic mouth. Indeed, even if the extracellular mouth of the TM pore domain is open (as is the case in the x-ray structure, where it forms a number of contacts with the periplasmic tip of an adjacent trimer in the crystal lattice), the overall pore is closed at its periplasmic mouth, with a pore radius of ∼2 Å. The mechanism of opening/closing i.e., gating) of TolC is a key aspect of the function of the efflux machinery. In vitro analyses of TolC in planar lipid bilayers have shown that channel opening is spontaneous (i.e., not induced by voltage); in addition, a closed state to drug translocation was reflected in the low conductance of TolC membrane pore in lipid bilayers (12). A mechanism of opening has been proposed that involves a twisting motion of the helices and the breaking of both intramonomer and intermonomer H-bond and electrostatic interactions (12). The residues Y326 and R367, located at the periplasmic mouth and involved in the latter interactions, are proposed to be involved in the gating (12). A double mutant of TolC (Y362F/R367S), in which some of these interactions were perturbed, resulted in an increase in conductance from 80 pS to 1000 pS (12). Eswaran and colleagues (13) introduced cysteine cross-links to validate the proposed mechanism in vivo; the cysteine cross-links served as constraining covalent links between the helices at the periplasmic mouth. Their study showed that locked TolC variants were still recruited by the other components of the exporting apparatus; however, the export function was abolished (13).
More recent studies using directed evolution of TolC suggest that the lower half (i.e., the part closer to the periplasmic mouth) of the α-helical domain of TolC interacts with AcrAB (14). A crystal structure of AcrB complexed with a single TM helix (YajC) shows a rotation of the periplasmic domain consistent with a twisting motion being conveyed to TolC upon activation of the latter (15); cross-linking studies also support a rotational mechanism of AcrB (16). It is, therefore, of some functional interest to further explore the conformational dynamics of TolC, particularly because two recent x-ray structures of a leaky mutant of TolC indicate a twisting displacement of the periplasmic α-helices as a possible gating mechanism (17).
An understanding of the intrinsic flexibility of TolC when in a lipid bilayer environment may provide further insights into its gating mechanism. Such flexibility may be explored on a nanosecond timescale via molecular dynamics (MD) simulations (18,19). MD simulations have been used as a computational tool to study the conformational dynamics of a wide range of membrane proteins (20–22). For example, they have been applied with some success to analysis of possible transport mechanisms of, e.g., ABC transporters (23) of the SecY translocon (24) and of lactose permease (25,26), and have also been used to explore possible channel gating mechanisms, e.g., of MscL (27–29)
In this study, we use multiple MD simulations to explore the dynamic behavior of TolC (and of its homolog OprM (30)) embedded in a phospholipid bilayer. Analysis of the simulation data suggests that the dynamic behavior of this protein is perhaps more complex than previously expected. In particular, our simulations suggest that the iris-like mechanism is insufficient to describe the overall motion of the protein. Instead, we suggest that gating transitions may occur at both the extracellular and periplasmic mouths of TolC and that a peristaltic motion may play a role in enabling passage of the solute through the channel.
METHODS
Simulation systems
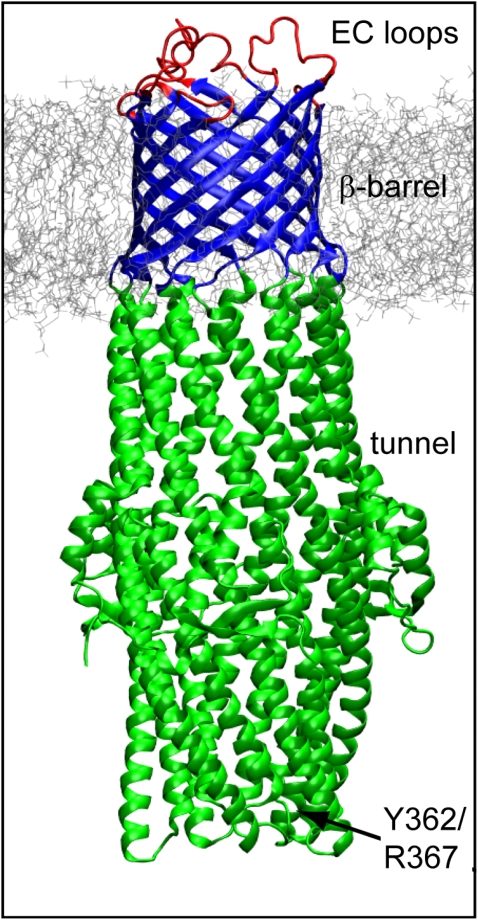
Four MD simulations were performed: MD1, MD2, MD3, and MD4 (Table 1). MD1 was of just the TM β-barrel domain (residues 40–77 and 246–296) of TolC (Protein Data Bank (PDB) code 1EK9). MD2 was of the complete TolC molecule (Fig. 1), and MD3 was of a TolC mutant (Y362F/R367S). To generate the starting structure of TolC Y362F/R367S, the two residues were mutated using the computer software Insight II (Accelyrs, San Diego, CA). MD4 was a simulation of OprM, a homolog of TolC from Pseudomonas aeruginosa (PDB code 1WP1).
TABLE 1.
Summary of simulations
| Simulation | System | No. of atoms | Final Cα RMSD (Å)* |
|---|---|---|---|
| MD1 | TolC, TM domain only | ∼55,000 | 3.3 ± 0.1 |
| MD2 | TolC, intact | ∼120,000 | 3.8 ± 0.1 |
| MD3 | TolC Y362F/R367S, intact | ∼120,000 | 4.0 ± 0.1 |
| MD4 | OprM, intact | ∼120,000 | 4.7 ± 0.1 |
Each simulation was of 20 ns duration.
Final Cα RMSD is calculated for all residues of the protein, relative to the starting coordinates and averaged over the last 5 ns of the simulation.
FIGURE 1.
TolC in a DMPC bilayer. In simulation MD1, only the TM domain (blue) was used; all other simulations (MD2–MD4) used the intact protein.
Each protein was embedded in a preequilibrated DMPC buffer (1,2-dimyristoyl-3-sn-glycero-phosphatidylcholine) bilayer containing 288 lipid molecules (177 after insertion of the protein), and solvated with SPC (31) water. Na+ and Cl− ions were added in numbers equivalent to a ∼0.1 M solution.
Simulation methods
The simulations were performed using GROMACS, version 3 (32,33), and a modified GROMOS96 force field (34). The LINCS (35) algorithm was used to constrain all bond lengths. A cutoff of 10 Å for Lennard-Jones interactions was used, and the particle mesh Ewald method was used to calculate longer range electrostatic contributions on a grid with a 1.2 Å spacing and a cutoff of 10 Å (36,37). The simulation was conducted at constant temperature (310 K), coupling each component separately to a temperature bath, with a coupling constant τT of 0.1 ps, using the Berendsen coupling method. A constant pressure of 1 bar was applied in all three directions, with a coupling constant of 1.0 ps. A time step of 2 fs was used, with coordinates stored every 2 ps. Before each simulation was run, an energy minimization was performed. The first 1 ns of simulation was performed imposing positional restraints on the non-H atoms of the protein, applying a force constant of 10 kJ mol−1 Å−2. The restraints were then released, and 20 ns production runs were obtained and analyzed for each system. Pore radius profiles were calculated using HOLE (39). Molecular graphics images were generated using visual molecular dynamics (40). Coarse-grained (CG) simulations were performed as described previously (41), using a modified version (42,43) of the force field developed by Marrink and colleagues (44); the results were deposited in the CG Database, located at http://sbcb.bioch.ox.ac.uk/cgdb.
RESULTS
Conformational stability and fluctuations
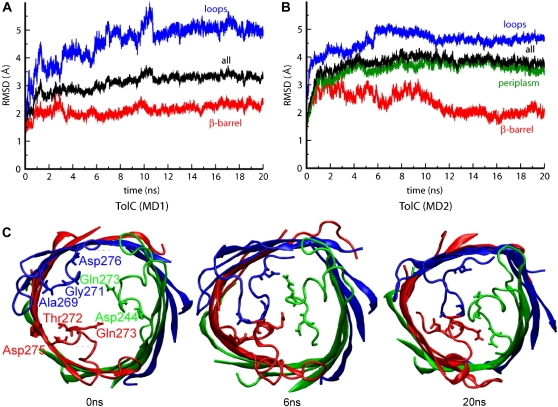
Analysis of the root mean-square deviation (RMSD) of the Cα atoms with respect to their starting coordinates provides a measure of overall conformational drift from the initial (that is, x-ray) structures. This analysis revealed that simulation MD1 was overall somewhat more stable than MD2 (Cα RMSDs for all residues of ∼3.3 and ∼3.8 Å, respectively; Table 1). For the TM domains, however, the two simulations were of comparable stability, with final Cα RMSDs of ∼2 Å for their β-barrels. In both simulations, the extracellular loops exhibited substantial conformational drift (Fig. 2, A and B).
FIGURE 2.
(A and B) The RMSD of the Cα atoms from their initial coordinates as function of time, for MD1 (a) and MD2 (b). The black line shows the RMSD for all the Cα atoms; the other lines show the RMSD of the extracellular loops (blue), periplasmic region (green), and β-barrel domain (red). (C) Collapse of extracellular loops in TolC. Top view of snapshots of MD1 at t = 0 ns, t = 6 ns, and t = 20 ns, showing the closure of the pore at the extracellular region.
The elevated RMSDs of the loops are mainly due to their inward collapse with consequent closure of the pore at its extracellular mouth. This may reflect relaxation from crystal lattice contacts, and can be seen, e.g., in snapshots of the extracellular mouth from MD1 at t = 0 ns, t = 6 ns, and t = 20 ns (Fig. 2 C). A network of H-bonds between the side chains and the backbones of residues located on the loops generates a constriction of the pore, reducing the pore radius to < 2 Å. If one measures the distances between representative residues involved in these H-bonds, a mixture of both stable interactions (Fig. 3, A–C, upper plots), reached after 2 ns, and of elevated flexibility of some side chains (Fig. 3, D–F, lower plots) is observed. The flexibility of the extracellular loops in TolC is to be expected (45), yet it remains of interest in comparison with similar behavior in a number of outer membrane proteins. For example, a possible role of loop regions in opening/closing a putative pore has also been seen in simulation studies of OpcA (46). Furthermore, two recent x-ray structures of OmpG (at different pH values) also indicate a role of an extracellular loop in pore gating (47), paralleling loop mobility seen in recent simulation studies (48).
FIGURE 3.
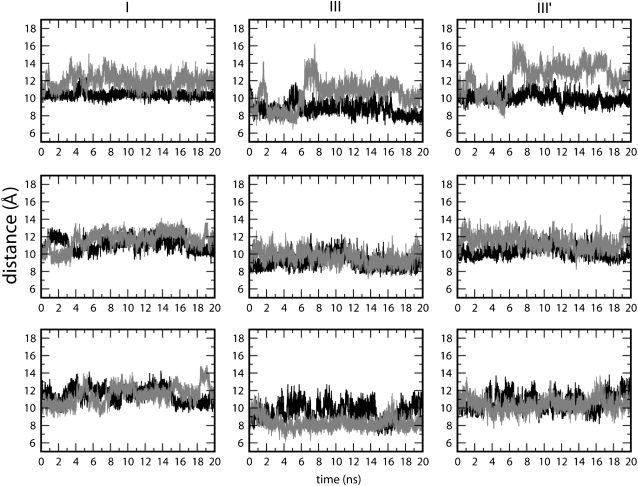
Intermonomeric distances between residues involved in the formation of H-bonds among the loops.
An overall higher RMSD is observed in the simulation of OprM (Table 1) due to a distortion of the loops connecting the β-barrel and the α-helical periplasmic domain. Collapse of the extracellular loops is observed, similar to that seen for TolC, strengthening the suggestion of a possible role in a gating process.
β-Barrel (TM domain) distortions
The TM domain of TolC differs from the domains of many other outer membrane proteins in that it consists of a single β-barrel formed by three monomers. Comparison of the TM domain of TolC with that of its homolog OprM suggests that there may be two alternative conformations for such a β-barrel. In TolC, the barrel is approximately cylindrical; in OprM, however, it is more like a triangular prism (Fig. 4). Interestingly, during the simulation of TolC, the shape of the β-barrel changes, switching from a cylindrical to a prism conformation while retaining the overall transbilayer pore. Thus, the intact TolC molecule switches to a conformation very similar to the x-ray structure of OprM within the first nanosecond of the simulation. It seems that the more stable conformation of TolC, once released from its crystallographic environment, may be the triangular prism, because this state is accessed by the protein during all the simulations. By way of comparison, the triangular prism conformation is maintained throughout the OprM simulation (MD4). The triangular prismatic shape is also seen in the x-ray structure of VceC (not simulated in this study). It is possible that spin-label studies (49,50), for example, could be used to probe for the cylindrical to triangular prismatic conformational transition in vitro.
FIGURE 4.
X-ray versus MD: conformational changes in the TM β-barrel viewed from the extracellular face. The cylindrical conformation is seen in the x-ray structure of TolC, whereas the triangular prism conformation is seen in the final structures from simulations MD1 and MD2 and in the x-ray structure of OprM.
Intriguingly, the change in β-barrel conformation for TolC appears to be more marked in the simulation of the intact protein (MD2) than in the simulation of the isolated TM domain (MD1). Thus, the transition from the cylindrical to triangular prism seems to be facilitated by the presence of the periplasmic domain, that is, of the α-helical tunnel domain.
The periplasmic tunnel
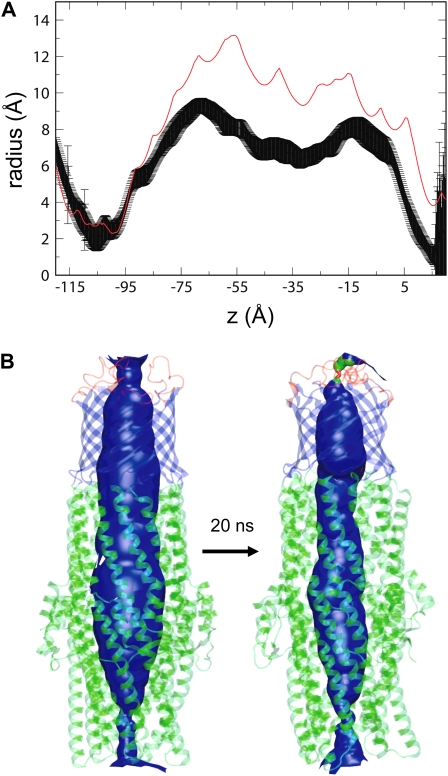
A major characteristic of both TolC and OprM is the presence of the α-helical domain, which protrudes for ∼100 Å into the periplasmic space. This domain interacts directly with the inner membrane transport protein (for example, AcrB), thus forming a continuous exit pathway for drugs. In the x-ray structure of TolC, the tunnel forms an internal pore that is wide enough to allow a solute to pass through unimpeded. However, the results of our simulations suggest that the conformational dynamics of this region may be rather complex. In particular, a constriction is observed, involving the whole pore. This starts at the link region between the β-barrel and the α-helical domain, and by way of a breathing-like motion it progresses along the length of the tunnel during the simulation. This can be seen in the pore radius profile of the protein (Fig. 5), where the average pore radius profile is compared with that at the start of the simulation. In addition to this global contraction of the pore, an iris-like (i.e., twisting) motion is also observed. This involves not only the periplasmic mouth of the α-helical domain, but the whole protein (see Movie S1 in the Supplementary Material), with the equatorial domain possibly functioning as a hinge-like region.
FIGURE 5.
Possible functional consequences of TolC motions. (A) Pore radius profile of TolC based on the x-ray structure (red line) compared with the average pore radius profile (± SD) derived from simulations MD2. (B) Pore lining surface for TolC simulation MD2 at the start and end of the simulation.
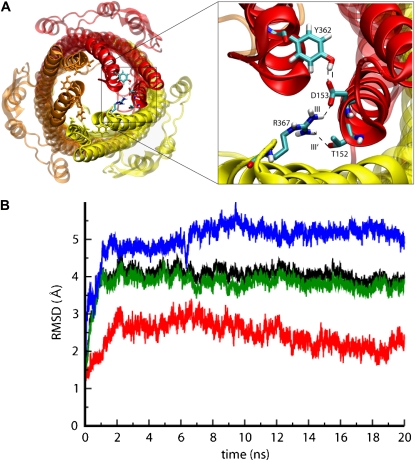
Simulations of the Y362F/R367S mutant
In the TolC opening mechanism proposed on the basis of the initial x-ray structure (7), a key role is played by the periplasmic mouth of the tunnel at the tip of the α-helical domain (Fig. 6 A). Residues including Y362, D153, and R367 form an H-bond network that stabilizes the closure of the pore and probably regulates the opening of the periplasmic mouth. Significantly, mutants of, for example, R367 seem to result in a constitutively open TolC pore, thus allowing antibiotic influx and resulting in increased antibiotic sensitivity (51). The H-bonds involving Y362, D153, and R367 are both intramonomeric (link I) and intermonomeric (link III and III′; Fig. 6 A). All of these links are missing in the Y362F/R367S mutant. The Cα RMSD values of the mutant simulation (MD3; Fig. 6 B) appear to be very similar to those of the wild-type, showing a plateau for the RMSD calculated for the whole protein. Breaking down the RMSDs into those of the constituent domains confirms the large mobility of the loops in the TM region and of the periplasmic helices. Some fluctuations are observed for the β-barrel and, again, a triangular cross-sectional β-barrel is formed (data not shown). A further comparison between the dynamics of TolC wild-type (MD2) and Y362F/R367S (MD3) can be obtained from the average atomic displacement of the Cα atoms with respect to the starting structure (Fig. 7). Comparing the results of such an analysis for TolC and for Y362F/R367S reveals similarities and differences. In both the wild-type and mutant simulations, a closure of the pore by the collapse of the extracellular loops is observed, as well as the mobility of the equatorial domain. Interestingly, the Y362F/R367S system reveals a significantly higher mobility at the tips of the periplasmic helices, i.e., in the region of the periplasmic mouth, where the mutations destabilizing the closed state are located.
FIGURE 6.
(A) Snapshot of TolC showing the position of residues Tyr-362 and Arg-367 at the tip of the periplasmic region of TolC. (B) These residues form intra- and intermonomeric interactions responsible for the stability of the closed state. (C) The RMSD of the Cα atoms from their initial coordinates as a function of time for Y362F/R367S. The black line shows the RMSD for all the Cα atoms; the other lines show the RMSD of the extracellular loops (blue), periplasmic region (green), and β-barrel domain (red).
FIGURE 7.
Average atomic displacement of the Cα atoms of (A) TolC and (B) Y362F/R367S with respect to the x-ray structure. A gradient of color from blue (0.08 nm) to red (1.29 nm) is used to show a greater displacement.
From the plots of the distances between the Cα atoms of the residues involved in the mutations (Fig. 8) and from comparison between the wild-type simulation and Y362F/R367S, the largest fluctuations are observed in the latter, particularly in link III and link III'. Therefore, the mutation of residues involved in these interactions leads to an unstable network of side-chain H-bonds. Simulations of the mutant revealed an enhanced mobility at the periplasmic mouth compared to simulations of the wild-type TolC. This finding is what could be expected, given the loss of H-bonding; it is also in agreement with the experimental data that propose these residues to be involved in the control of the outer membrane tunnel opening.
FIGURE 8.
Distance between the Cα atoms of residues involved in the proposed opening mechanism. The black line shows the analysis of MD2, and the red line shows the results for Y362F/R367S. (I) Intramonomeric distance between Tyr/Phe-362 and Asp-153 for each monomer. (III) Intermonomeric distance between Arg/Thr-367 and Asp-153 for each pair of adjacent monomers. (III′) Intermonomeric distance between Arg/Thr-367 and Thr-152 for each pair of adjacent monomers.
PRINCIPAL COMPONENTS AND ESSENTIAL DYNAMICS (ED)
The MD simulations of wild-type TolC and of the Y362F/R367S mutant indicate that flexibility of the protein involves the tips of the helices and the whole protein. In particular, the iris-like motion appears to be combined with a twisting motion of the upper half of the periplasmic domain (Fig. 9). The equatorial domain may therefore play the role of a hinge point in the overall motion of the protein. It is important to note that the observed simulated motions did not yield an open state. This finding is not unexpected, given the requirement of the interaction with the other components of the complex to open the channel and the relatively short timescale (20 ns) of the simulations.
FIGURE 9.
Superimposed snapshots from the TolC (MD2) simulation. (A) Superimposition of the x-ray structure (cyan) and a 3 ns snapshot (red). (B–D) Superimpositions of simulation snapshots at 3 ns (red), 9 ns (green), 15 ns (orange), and 20 ns (blue). The arrows follow the same color scheme and indicate the motion observed in the corresponding time range.
In Fig. 9, sequential snapshots of the TolC (MD2) simulation are presented to enable a better visualization of the motions of the protein (Movie S1). The first rearrangements of the structure involve a constriction at the top part of the helices. Following the movements of TolC by examining the sequential superimpositions of snapshots, the twisting (iris-like) motion can be seen. An iris-like motion can be observed from 3 to 9 ns; this motion involves the whole periplasmic domain rather than just the inner helices at the tip of the domain as previously proposed (7). The equatorial domain seems therefore to act as a hinge point for the change in direction of the swivel motion of the helices that are 100 Å in length. Nevertheless, it is also possible that the two halves of the helices move independently, as indicated by a superimposition of the structures at 9 and 15 ns, in which case only the lower regions of the helices move in an iris-like fashion. In the simulation, such movement can be clockwise (Fig. 9 C) or anticlockwise (Fig. 9 D).
We also used principal components analysis (PCA) (52,53) of the MD2 simulation to explore these motions more fully. PCA showed an asymmetric motion of the monomers, indicating a breakdown of the symmetry of TolC between the first two eigenvectors (Fig. 10, A and B). Thus, PCA confirmed the iris-like (that is, twisting) motion observed by visual examination of the simulations (described above). This is important not only in terms of the possible opening mechanism of TolC but also because it is in broad agreement with the rotational motion proposed for the periplasmic region of AcrB (10,15,16,54), and suggests a possible link between the interaction of the two proteins and opening export “tunnel”.
FIGURE 10.
ED on TolC. (A and B) Three-dimensional projections of the MD simulation of TolC on the first and the second eigenvector, respectively. (C) Projection of the trajectory (black dots) of the x-ray structure (green circle) and of the open model (black square) on the first essential plane. In red and blue, respectively, are the projections of preliminary ED sampling in target mode from the x-ray structure and from a 2 ns snapshot from the simulation.
The passage from the closed state to the open state of TolC is equivalent to the passage from one energetically stable state (the closed one) to another stable state (the open one). Interchange between such states is likely to have a large energy barrier unless facilitated by, for example, interactions with another protein. A possible solution to the problem of exploring this conformational transition in simulations may be the use of essential dynamics (ED) (52). This technique, previously used to investigate the dynamics of protein, e.g., the mechanism of domain closure in citrate synthase (55), may allow the exploration of the energy surface of a protein, crossing energy barriers that cannot be crossed by classical MD. Using the first principal motions of the protein, obtained by MD, it is, in principle, possible to direct the protein conformation toward a particular target e.g., the open state). In Fig. 10 C, preliminary results of ED applied to TolC are shown. These calculations were conducted in a target mode, where the target was the open model (as proposed by Koronakis and colleagues (7)). The conformational dynamics were driven along the first two eigenvectors obtained from the covariance matrix of the tips of the helices. These eigenvectors confirmed the asymmetry of motion of the three monomers that has already been noted. Preliminary attempts at opening the periplasmic mouth of TolC by ED did not reach the target (Fig. 10 C). This may be due to the choice of the target, an open state model (7) and not an experimentally obtained structure. It would be necessary to identify the domains involved in the opening and to remove any internal motions within the domain to obtain a rigid-body trajectory (56). This may be facilitated in future studies by the recent x-ray structure of a partly open mutant of TolC (17).
CG simulations
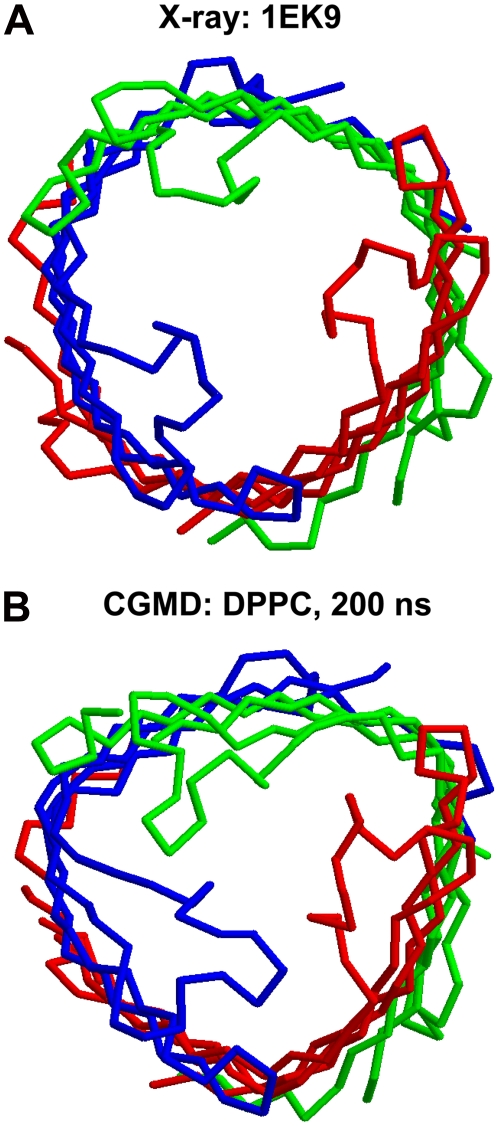
The atomistic simulations of wild-type TolC and of the Y362F/R367S mutant provide information on the pattern of flexibility within the protein. However, as with most atomistic simulations of membrane proteins, the simulations present issues of incomplete convergence (57,58). Thus, we also explored TolC via CG-MD simulations (41–43) in which the internal dynamics of the protein were modeled via an elastic network (59). These simulations, which extended to 200 ns, provided a more approximate, but better, sampled picture of the protein dynamics. In total, eight simulations of TolC (using different lipid bilayer components) and simulations of OprM and VceC were performed (see the CG Database at http://sbcb.bioch.ox.ac.uk/cgdb for a detailed summary). In terms of the TM domain, the CG-MD simulations revealed both the collapse of the extracellular loops and the transition to triangular prismatic cross-section β-barrel seen in the atomistic simulations (Fig. 11). In the periplasmic domain, there was a degree of loss of exact 3× symmetry, with the periplasmic α-helices of one subunit moving to partially open the periplasmic mouth (not shown). Thus, although this is only a preliminary comparison, we can be reasonably confident that the motions of TolC captured in the relatively short atomistic MD simulations and also seen in the longer (but more approximate) CG-MD simulations are representative of the protein when in a lipid bilayer.
FIGURE 11.
CG simulations of TolC. The Cα trace of the β-barrel domain of TolC at (A) the start and (B) the end of a 200 ns simulation of the intact TolC protein in a DPPC bilayer.
DISCUSSION
The outer membrane protein TolC plays an essential role in the drug resistance exhibited by Gram-negative bacteria. It forms a stable tripartite complex with an inner membrane protein, the efflux pump, and an accessory periplasmic protein, allowing the drugs to be expelled directly from the inside to the outside of the bacteria. Although the mechanism of recruitment of TolC by the other two components of the machinery and of the drug extrusion are not yet known, a model for the open state of the former has been proposed (7). In this model, the opening involves an iris-like motion of the tip of the periplasmic helices where a network of inter- and intramonomeric H-bonds can provide a structural gating. In this work, we analyzed the results of simulations of TolC in a lipid bilayer environment, and we compared them with the dynamics of the homolog OprM and of a mutant Y362F/R367S, where the structural links at the periplasmic mouth of the protein had been removed.
The overall dynamics of these proteins suggest that four different regions can be distinguished, each of which has a role in the global activity of the outer membrane protein. The most flexible region appears to be the one containing the extracellular loops. A flexible collapsing of the latter is shown by the dynamic interactions between residues located on the loops. These may therefore act as a “gating” region able to restrict access to external agents and/or to open the pore for solutes being expelled from the cytoplasm by the IM protein. The importance of the loop region in the activity of OM proteins has also been previously pointed out by computational studies (46,48,60,61) and by, e.g., a recent x-ray structure of the OM porin OmpG obtained at different pH, suggesting the role of one extracellular loop in the pH-dependent pore gating (47).
A second dynamic region can be identified in the β-barrel, as reflected in a difference between the structures of TolC and of its homolog OprM in this region. Starting from a cylindrical shape, the β-barrel of TolC undergoes an initial distortion that leads to a stable triangular shape, which is more similar to the OprM β-barrel. It seems, therefore, that the most stable conformation of the TM region of these outer membrane proteins is the triangular prism. Interestingly, the magnitude of the distortion from the cylindrical cross section appears to depend on the presence of the perisplamic helices, being less evident in the simulation of the TM region only. This finding suggests that the change in the overall shape of the β-barrel is due to more than only a relaxation from the crystallographic environment. The change in shape also reveals an intrinsic dynamic property of the whole protein that is transmitted from the periplasmic mouth; at this location, the protein would interact with the inner membrane protein and the accessory protein through the periplasmic helices toward the TM region. This dynamic motion of the whole protein, especially of the α-helices, observed in our simulation studies can help in swallowing the substrate toward the external medium; thus, the final main obstacle to the process is the opening of the periplasmic mouth. This opening may occur after an interaction with the efflux pump and the accessory protein; the former would provide the energy for the process, and the latter would function both by clamping the inner membrane protein together with the outer membrane protein and by mediating the gating of the outer membrane protein (62).
Therefore, the conformational dynamics of the TolC appear to be more complex than may have been expected. In particular, a model of TolC in which its dynamics are restricted to an iris-like motion of the periplasmic mouth appears to be a little too simplistic. It is undeniable that the pore must be open at this region for transport to occur, and it is likely that this opening occurs alongside an interaction with the periplasmic protein. However, the intrinsic flexibility of TolC during simulations suggests that the iris-like motion is combined with a twisting motion of the upper half of the periplasmic tunnel. The latter undergoes a constriction that may be involved in a breathing-like (or peristaltic) motion that may catalyze transport of the solute along the tunnel. Interestingly, the equatorial region (surrounding the middle of the periplasmic tunnel) has been suggested to play a key role in the transport activity of TolC on the basis of functional measurements on chimeric TolC proteins (63).
At the opposite end of the protein, the extracellular mouth seems to act as a gating region, where the mobility of the loops may further regulate the passage of the substrate. Fluctuations in the conformation of the transmembrane β-barrel may serve to couple the extracellular region to the periplasmic tunnel. Thus, far from being a passive tube, TolC seems to be gated at either end and to also undergo complex breathing motions in the domains in between. In particular, the β-barrel switches dynamically between two conformations, that with a circular cross section (seen in the x-ray structures of TolC (5) and of VceC, its homolog from Vibrio cholerae (64)) and that with a triangular cross section (seen in the x-ray structure of OprM).
It is useful to reflect critically on the limitations of this study. As noted above, we do not expect 20 ns simulations to converge in terms of the motions of the protein (57) and, indeed, sampling is still likely to be incomplete for substantially longer atomistic simulations (58). However, both comparison of multiple atomistic simulations (TolC, TolC mutant, and OprM) and analysis of CG simulations indicate the generality of the motions analyzed. Of course, in the bacterial membrane, it is likely that the intrinsic motions of TolC will be modulated via formation of a complex with AcrA and AcrB. We might anticipate that AcrA is more likely to modulate the twisting, rather than the breathing, motion as the latter occurs mostly in the upper region of the helical domain, mostly at the interface between helices and β-barrel, whereas AcrA seems to interact with residues close to the periplasmic mouth (65). However, this remains speculative in the absence of a structure for and simulations of the TolC/AcrA/AcrB complex.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
The authors thank all our colleagues and especially Vassilis Koronakis for his interest in this work.
This work was supported by grants from the Wellcome Trust and the Biotechnology and Biological Sciences Research Council.
Loredana Vaccaro's present address is the Institute de Biologie Structurale, 41, rue Jules Horowitz, 38027 Grenoble, cedex 1, France.
Editor: Peter C. Jordan.
References
- 1.Paulsen I. T., J. H. Park, P. S. Choi, and M. H. Saier Jr. 1997. A family of gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from gram-negative bacteria. FEMS Microbiol. Lett. 156:1–8. [DOI] [PubMed] [Google Scholar]
- 2.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803–5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchanan, S. K. 2001. Type I secretion and multidrug efflux: transport through the TolC channel-tunnel. Trends Biochem. Sci. 26:3–6. [DOI] [PubMed] [Google Scholar]
- 4.Poole, K., K. Krebes, C. McNally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koronakis, V., J. Eswaran, and C. Hughes. 2004. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 73:467–489. [DOI] [PubMed] [Google Scholar]
- 6.Eswaran, J., E. Koronakis, M. K. Higgins, C. Hughes, and V. Koronakis. 2004. Three's company: component structures bring a closer view of tripartite drug efflux pumps. Curr. Opin. Struct. Biol. 14:717–747. [DOI] [PubMed] [Google Scholar]
- 7.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 405:914–919. [DOI] [PubMed] [Google Scholar]
- 8.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 419:587–593. [DOI] [PubMed] [Google Scholar]
- 9.Murakami, S., R. Nakashima, E. Yamashita, T. Matsumoto, and A. Yamaguchi. 2006. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 443:173–179. [DOI] [PubMed] [Google Scholar]
- 10.Seeger, M. A., A. Schiefer, T. Eicher, F. Verrey, K. Diederichs, and K. M. Pos. 2006. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 313:1295–1298. [DOI] [PubMed] [Google Scholar]
- 11.Mikolosko, J., K. Bobyk, H. I. Zgurskaya, and P. Ghosh. 2006. Conformational flexibility in the multidrug efflux system protein AcrA. Structure. 14:577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersen, C., E. Koronakis, E. Bokma, J. Eswaran, D. Humphreys, C. Hughes, and V. Koronakis. 2002. Transition to the open state of the TolC periplasmic tunnel entrance. Proc. Natl. Acad. Sci. USA. 99:11103–11108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eswaran, J., C. Hughes, and V. Koronakis. 2003. Locking TolC entrance helices to prevent protein translocation by the bacterial type I export apparatus. J. Mol. Biol. 327:309–315. [DOI] [PubMed] [Google Scholar]
- 14.Bokma, E., E. Koronakis, S. Lobedanz, C. Hughes, and V. Koronakis. 2006. Directed evolution of a bacterial efflux pump: adaptation of the E. coli TolC exit duct to the Pseudomonas MexAB translocase. FEBS Lett. 580:5339–5343. [DOI] [PubMed] [Google Scholar]
- 15.Tornroth-Horsefield, S., P. Gourdon, R. Horsefield, L. Brive, N. Yamamoto, H. Mori, A. Snijder, and R. Neutze. 2007. Crystal structure of AcrB in complex with a single transmembrane subunit reveals another twist. Structure. 15:1663–1673. [DOI] [PubMed] [Google Scholar]
- 16.Seeger, M. A., C. von Ballmoos, T. Eicher, L. Brandstatter, F. Verrey, K. Diederichs, and K. M. Pos. 2008. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat. Struct. Mol. Biol. 15:199–205. [DOI] [PubMed] [Google Scholar]
- 17.Bavro, V. N., Z. Pietras, N. Furnham, L. Pérez-Cano, J. Fernández-Recio, X. Y. Pei, R. Misra, and B. Luisi. 2008. Assembly and channel opening in a bacterial drug efflux machine. Mol. Cell. 30:114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karplus, M. J., and J. A. McCammon. 2002. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 9:646–652. [DOI] [PubMed] [Google Scholar]
- 19.Adcock, S. A., and J. A. McCammon. 2006. Molecular dynamics: survey of methods for simulating the activity of proteins. Chem. Rev. 106:1589–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ash, W. L., M. R. Zlomislic, E. O. Oloo, and D. P. Tieleman. 2004. Computer simulations of membrane proteins. Biochim. Biophys. Acta. 1666:158–189. [DOI] [PubMed] [Google Scholar]
- 21.Roux, B., and K. Schulten. 2004. Computational studies of membrane channels. Structure. 12:1343–1351. [DOI] [PubMed] [Google Scholar]
- 22.Lindahl, E., and M. S. P. Sansom. 2008. Membrane proteins: molecular dynamics simulations. Curr. Opin. Struct. Biol. 18:425–431. [DOI] [PubMed] [Google Scholar]
- 23.Oloo, E. O., and D. P. Tieleman. 2004. Conformational transitions induced by the binding of MgATP to the vitamin B12 ATP-binding cassette (ABC) transporter BtuCD. J. Biol. Chem. 279:45013–45019. [DOI] [PubMed] [Google Scholar]
- 24.Gumbart, J., and K. Schulten. 2006. Molecular dynamics studies of the archael translocon. Biophys. J. 90:2356–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin, Y., M. Jensen, E. Tajkhorshid, and K. Schulten. 2006. Sugar binding and protein conformational changes in lactose permease. Biophys. J. 91:3972–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holyoake, J., and M. S. P. Sansom. 2007. Conformational change in an MFS Protein: MD simulations of LacY. Structure. 15:873–884. [DOI] [PubMed] [Google Scholar]
- 27.Gullingsrud, J., and K. Schulten. 2003. Gating of MscL studied by steered molecular dynamics. Biophys. J. 85:2087–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gullingsrud, J., and K. Schulten. 2004. Lipid bilayer pressure profiles and mechanosensitive channel gating. Biophys. J. 86:3496–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeon, J., and G. A. Voth. 2008. Gating of the mechanosensitive channel protein MscL: the interplay of membrane and protein. Biophys. J. In press. [DOI] [PMC free article] [PubMed]
- 30.Akama, H., M. Kanemaki, T. Tsukihara, A. Nakagawa, and T. Nakae. 2004. Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end. J. Biol. Chem. 279:52816–52819. [DOI] [PubMed] [Google Scholar]
- 31.Hermans, J., H. J. C. Berendsen, W. F. van Gunsteren, and J. P. M. Postma. 1984. A consistent empirical potential for water-protein interactions. Biopolymers. 23:1513–1518. [Google Scholar]
- 32.Lindahl, E., B. Hess, and D. van der Spoel. 2001. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 7:306–317. [Google Scholar]
- 33.van der Spoel, D., E. Lindahl, B. Hess, G. Groenhof, A. E. Mark, and H. J. Berendsen. 2005. GROMACS: fast, flexible, and free. J. Comput. Chem. 26:1701–1718. [DOI] [PubMed] [Google Scholar]
- 34.Scott, W. R. P., P. H. Hunenberger, I. G. Tironi, A. E. Mark, S. R. Billeter, J. Fennen, A. E. Torda, T. Huber, P. Kruger, and W. F. van Gunsteren. 1999. The GROMOS biomolecular simulation program package. J. Phys. Chem. A. 103:3596–3607. [Google Scholar]
- 35.Hess, B., H. Bekker, H. J. C. Berendsen, and J. G. E. M. Fraaije. 1997. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463–1472. [Google Scholar]
- 36.Darden, T., D. York, and L. Pedersen. 1993. Particle mesh Ewald: an N · log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. [Google Scholar]
- 37.Essmann, U., L. Perera, M. L. Berkowitz, T. Darden, H. Lee, and L. G. Pedersen. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 103:8577–8593. [Google Scholar]
- 38.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, A. DiNola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- 39.Smart, O. S., J. G. Neduvelil, X. Wang, B. A. Wallace, and M. S. P. Sansom. 1996. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14:354–360. [DOI] [PubMed] [Google Scholar]
- 40.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38. [DOI] [PubMed] [Google Scholar]
- 41.Scott, K. A., P. J. Bond, A. Ivetac, A. P. Chetwynd, S. Khalid, and M. S. P. Sansom. 2008. Coarse-grained MD simulations of membrane protein-bilayer self-assembly. Structure. 16:621–630. [DOI] [PubMed] [Google Scholar]
- 42.Bond, P. J., and M. S. P. Sansom. 2006. Insertion and assembly of membrane proteins via simulation. J. Am. Chem. Soc. 128:2697–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bond, P. J., J. Holyoake, A. Ivetac, S. Khalid, and M. S. P. Sansom. 2007. Coarse-grained molecular dynamics simulations of membrane proteins and peptides. J. Struct. Biol. 157:593–605. [DOI] [PubMed] [Google Scholar]
- 44.Marrink, S. J., A. H. de Vries, and A. E. Mark. 2004. Coarse grained model for semiquantitative lipid simulations. J. Phys. Chem. B. 108:750–760. [Google Scholar]
- 45.Cox, K., P. J. Bond, A. Grottesi, M. Baaden, and M. S. P. Sansom. 2008. Outer membrane proteins: comparing X-ray and NMR structures by MD simulations in lipid bilayers. Eur. Biophys. J. 37:131–141. [DOI] [PubMed] [Google Scholar]
- 46.Bond, P. J., J. P. Derrick, and M. S. P. Sansom. 2007. Membrane simulations of OpcA: gating in the loops? Biophys. J. 92:L23–L25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yildiz, O., K. R. Vinothkumar, P. Goswami, and W. Kühlbrandt. 2006. Structure of the monomeric outer-membrane porin OmpG in the open and closed conformation. EMBO J. 25:3702–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen, M., S. Khalid, M. S. P. Sansom, and H. Bayley. 2008. Outer membrane protein G: engineering a quiet pore for biosensing. Proc. Natl. Acad. Sci. USA. 105:6272–6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perozo, E., D. M. Cortes, and L. G. Cuello. 1999. Structural rearrangements underlying K+-channel activation gating. Science. 285:73–78. [DOI] [PubMed] [Google Scholar]
- 50.Kim, M., Q. Xu, G. E. Fanucci, and D. S. Cafiso. 2006. Solutes modify a conformational transition in a membrane transport protein. Biophys. J. 90:2922–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Augustus, A. M., T. Celaya, F. Husain, M. Humbard, and R. Misra. 2004. Antibiotic-sensitive TolC mutants and their suppressors. J. Bacteriol. 186:1851–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amadei, A., A. B. M. Linssen, and H. J. C. Berendsen. 1993. Essential dynamics of proteins. Proteins Struct. Funct. Genet. 17:412–425. [DOI] [PubMed] [Google Scholar]
- 53.Koshi, J. M., and W. J. Bruno. 1999. Major structural determinants of transmembrane proteins identified by principal components analysis. Proteins Struct. Funct. Genet. 34:333–340. [PubMed] [Google Scholar]
- 54.Lu, W. C., C. Z. Wang, E. W. Yu, and K. M. Ho. 2006. Dynamics of the trimeric AcrB transporter protein inferred from a B-factor analysis of the crystal structure. Proteins. 62:152–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daidone, I., D. Roccatano, and S. Hayward. 2004. Investigating the accessibility of the closed domain conformation of citrate synthase using essential dynamics sampling. J. Mol. Biol. 339:515–525. [DOI] [PubMed] [Google Scholar]
- 56.Roccatano, D., A. E. Mark, and S. Hayward. 2001. Investigation of the mechanism of domain closure in citrate synthase by molecular dynamics simulation. J. Mol. Biol. 310:1039–1053. [DOI] [PubMed] [Google Scholar]
- 57.Faraldo-Gómez, J. D., L. R. Forrest, M. Baaden, P. J. Bond, C. Domene, G. Patargias, J. Cuthbertson, and M. S. P. Sansom. 2004. Conformational sampling and dynamics of membrane proteins from 10-nanosecond computer simulations. Proteins. 57:783–791. [DOI] [PubMed] [Google Scholar]
- 58.Grossfield, A., S. E. Feller, and M. C. Pitman. 2007. Convergence of molecular dynamics simulations of membrane proteins. Proteins. 67:31–40. [DOI] [PubMed] [Google Scholar]
- 59.Atilgan, A. R., S. R. Durell, R. L. Jernigan, M. C. Demirel, O. Keskin, and I. Bahar. 2001. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 80:505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tieleman, D. P., and H. J. C. Berendsen. 1998. A molecular dynamics study of the pores formed by Escherichia coli OmpF porin in a fully hydrated palmitoyloleoylphosphatidylcholine bilayer. Biophys. J. 74:2786–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Varma, S., S. W. Chiu, and E. Jakobsson. 2006. The influence of amino acid protonation states on molecular dynamics simulations of the bacterial porin OmpF. Biophys. J. 90:112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vaccaro, L., V. Koronakis, and M. S. P. Sansom. 2006. Flexibility in a drug transport accessory protein: molecular dynamics simulations of MexA. Biophys. J. 91:558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamanaka, H., S. Tadokoro, M. Miyano, E. Takahashi, H. Kobayashi, and K. Okamoto. 2007. Studies on the region involved in the transport activity of Escherichia coli TolC by chimeric protein analysis. Microb. Pathog. 42:184–192. [DOI] [PubMed] [Google Scholar]
- 64.Federici, L., D. Du, F. Walas, H. Matsumura, J. Fernandez-Recio, K. S. McKeegan, M. I. Borges-Walmsley, B. F. Luisi, and A. R. Walmsley. 2005. The crystal structure of the outer membrane protein VceC from the bacterial pathogen Vibrio cholerae at 1.8 Å resolution. J. Biol. Chem. 280:15307–15314. [DOI] [PubMed] [Google Scholar]
- 65.Lobedanz, S., E. Bokma, M. F. Symmons, E. Koronakis, C. Hughes, and V. Koronakis. 2007. A periplasmic coiled-coil interface underlying TolC recruitment and the assembly of bacterial drug efflux pumps. Proc. Natl. Acad. Sci. USA. 104:4612–4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.