Abstract
Together with the NS5 polymerase, the NS3 helicase has a pivotal function in flavivirus RNA replication and constitutes an important drug target. We captured the dengue virus NS3 helicase at several stages along the catalytic pathway including bound to single-stranded (ss) RNA, to an ATP analogue, to a transition-state analogue and to ATP hydrolysis products. RNA recognition appears largely sequence independent in a way remarkably similar to eukaryotic DEAD box proteins Vasa and eIF4AIII. On ssRNA binding, the NS3 enzyme switches to a catalytic-competent state imparted by an inward movement of the P-loop, interdomain closure and a change in the divalent metal coordination shell, providing a structural basis for RNA-stimulated ATP hydrolysis. These structures demonstrate for the first time large quaternary changes in the flaviviridae helicase, identify the catalytic water molecule and point to a β-hairpin that protrudes from subdomain 2, as a critical element for dsRNA unwinding. They also suggest how NS3 could exert an effect as an RNA-anchoring device and thus participate both in flavivirus RNA replication and assembly.
Keywords: ATP analogues, dengue virus, flaviviruses, NS3 helicase structure, RNA complex
Introduction
Dengue, an endemic disease transmitted by Aedes aegypti mosquitoes, causes ∼25 000 deaths per year. No specific therapy exists to treat illnesses caused by flaviviruses, a family that also comprises yellow fever, Japanese encephalitis and West Nile virus, all significant human pathogens. Our understanding of the molecular basis for the various enzymatic events involved in viral RNA replication and packaging is scant for flaviviruses. Their genome—a single-stranded (ss) plus RNA segment of about 11 kb—has several functions during the virus life cycle: on capsid (C) disassembly, it is translated into a polypeptide precursor containing three structural and seven non-structural (NS) proteins. Following polyprotein cleavage by the viral protease activity present in the N-terminal domain of NS3 and also by host cell proteases, the NS5 RNA-dependent RNA polymerase (RdRp) is released, and, in tight association with NS3, synthesizes a genome-length minus strand RNA. In turn, this strand serves as a template for the synthesis of an excess of plus genomic RNA. The latter is specifically packaged in nascent viral particles in association with capsid proteins (Lindenbach et al, 2007). The C-terminal region of NS3 possesses RNA-stimulated nucleoside triphosphatase activity that provides chemical energy to unwind viral RNA replication intermediates into forms amenable to amplification by the RdRp (Warrener et al, 1993; Li et al, 1999). As a result of its RNA triphosphatase activity, the 5′ end of viral genomic RNA is dephosphorylated, before cap addition by the N-terminal methyl transferase region of NS5. Owing to their prominent roles in the virus life cycle, both NS3 and NS5 form antiviral targets of prime importance (Kuo et al, 1996; Wu et al, 2005; Xu et al, 2005; Yap et al, 2007). The presence of a set of conserved sequence motifs within its two N-terminal Rec-A-like subdomains qualifies the flavivirus NS3 as a superfamily 2 (SF2) helicase (Story and Steitz, 1992; Koonin, 1993). These motifs are important for ATP binding and hydrolysis (motifs I, II and VI), nucleic acid binding (motifs Ia, Ib, IV and V), or for coupling these two enzymatic activities (motif III) (Pyle, 2008). In addition, NS3 shares the DExD/H sequence (motif II) with a large family of proteins that participate in many aspects of cellular RNA metabolism, including ribosome biogenesis, mRNA splicing, RNA export or small RNA processing (Bleichert and Baserga, 2007). Several mechanistic aspects of the strand separation activity remain elusive for DExH viral helicases, including the molecular basis for RNA versus DNA recognition, for the 3′–5′ directionality of translocation along a tracking RNA strand and the structural basis for their RNA-stimulated ATPase activity. Moreover, a complete picture of the ATP hydrolytic cycle is still lacking for a flaviviridae helicase. Here, we report nine crystallographic structures that form a complete set of ligand-bound states for a RNA helicase and enable us to draw comparisons with the mechanism used by SF1 helicases to unwind double-stranded DNA (Lee and Yang, 2006).
Results
Structure determination
To decipher the molecular basis underlying the multiple functions of the DENV NS3 helicase domain (NS3h), we determined crystal structures for its apo form to a resolution of 1.7 Å, with a non-hydrolysable ATP analogue, with ADP–vanadate–Mn2+, with ADP–Pi–Mn2+ and ADP–Mn2+, providing snapshots along the complete hydrolytic cycle for a flavivirus ATPase (Tables I and II). Several structures of NS3h were resolved in the presence of a bound ssRNA: either with a 12-mer (RNA12) 5′-AGACUAACAACU-3′ or a 13-mer (RNA13) 5′-UAGACUAACAACU-3′, providing a structural basis for sequence-independent RNA recognition and giving insight at atomic resolution into mechanochemical events that couple ATP hydrolysis and RNA unwinding by the flavivirus NS3 helicase.
Table 1.
Crystallographic data collection statistics
| Parameters | Native | AMPPNP | ADP | RNA12 | RNA12-AMPPNPa | RNA12-ADP-VO4 | RNA12-ADP-PO4 | RNA12-ADP | RNA13 |
|---|---|---|---|---|---|---|---|---|---|
| Space group | P212121 | P21 | P43 | C2 | C2 | C2 | C2 | C2 | C2 |
| Cell parameters | |||||||||
| a, b, c (Å) | a=52.8, b=92.6, c=103.0 | a=52.6, b=88.7, c=54.7 | a=b=52.3, c=192.9 | a=132.0, b=105.5, c=72.5 | a=132.8, b=105.2, c=72.6 | a=132.5, b=105.2, c=72.4 | a=131.7, b=105.6, c=72.7 | a=133.0, b=104.8, c=73.0 | a=132.4, b=104.6, c=72.3 |
| α, β, γ (deg) | α=β=γ=90 | α=γ=90, β=105.9 | α=β=γ=90 | α=γ=90, β=117.5 | α=γ=90, β=117.9 | α=γ=90, β=117.6 | α=γ=90, β=117.7 | α=γ=90, β=115.9 | α=γ=90, β=117.7 |
| Resolution range (Å) | 21.2–1.7 (1.8–1.7) | 26.5–1.9 (2.0–1.9) | 35.4–2.2 (2.3–2.2) | 29.8–2.0 (2.1–2.0) | 64.3–1.9 (2.0–1.9) | 78.3–2.2 (2.3–2.2) | 40.8–2.4 (2.5–2.4) | 31.9–2.2 (2.3–2.2) | 43.4–2.6 (2.7–2.6) |
| Observed reflectionsb | 445 134 (59 415) | 138 360 (16 030) | 176 639 (24 346) | 352 858 (44 388) | 294 791(15 191) | 127 207 (18 691) | 188 211 (24 876) | 193 997 (27 739) | 119 823 (16 942) |
| Unique reflections | 59 272 (8383) | 34 139 (4241) | 24 904 (3512) | 54 798 (7126) | 60 795 (7262) | 44 015 (6458) | 34 344 (4870) | 45 701 (6666) | 26 932 (3907) |
| Completeness (%) | 99.5 (97.8) | 93.6 (79.9) | 98.3 (95.0) | 96.5 (86.4) | 87.7 (72.6) | 98.8 (99.7) | 99.6 (97.5) | 100 (100) | 100 (100) |
| Multiplicity | 7.5 (7.1) | 4.1 (3.8) | 7.1 (6.9) | 6.4 (6.2) | 4.8 (2.1) | 2.9 (2.9) | 5.5 (5.1) | 4.2 (4.2) | 4.4 (4.3) |
| Rmerge c (%) | 4.5 (75.8) | 11.5 (69.9) | 6.6 (37.2) | 8.7 (37.7) | 14.4 (41.4) | 10.3 (49.7) | 14.0 (72.3) | 8.3 (58.1) | 15.2 (57.9) |
| Mean I/σ(I) | 22.4 (2.7) | 11.1 (1.3) | 21.3 (5.3) | 17.4 (4.2) | 11.5 (1.9) | 11.5 (3.3) | 13.8 (1.7) | 13.3 (2.3) | 11.0 (1.9) |
| Monomers (a.u.) | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 |
| a.u., asymmetric unit. | |||||||||
| aFinal dataset from two crystals. | |||||||||
| bThe numbers in parentheses refers to the last (highest) resolution shell. | |||||||||
| cRmerge=∑h∑i∣Ihi−〈Ih〉∣/∑h,iIhi, where Ihi is the ith observation of the reflection h, whereas 〈Ih〉 is its mean intensity. | |||||||||
Table 2.
Refinement statistics
| Parameters | Native | AMPPNP | ADP | RNA12 | RNA12- AMPPNP | RNA12- ADP-VO4 | RNA12- ADP-PO4 | RNA12-ADP | RNA13 |
|---|---|---|---|---|---|---|---|---|---|
| Resolution range (Å) | 20.1–1.7 | 19.7–2.0 | 19.8–2.2 | 20.0–2.0 | 20.0–1.9 | 20.0–2.2 | 20.0–2.4 | 20.0–2.2 | 20.0–2.6 |
| No. of reflections (working/test) | 56 198/2989 | 29 652/1555 | 23 586/1275 | 51 971/2779 | 57 632/3105 | 41 743/2213 | 32 553/1726 | 43 344/2304 | 25 521/1348 |
| Rfactor a,b (%) | 21.1 (44.1) | 20.1 (29.6) | 18.6 (21.6) | 19.7 (22.1) | 19.0 (25.0) | 19.6 (30.7) | 19.9 (27.8) | 19.2 (26.3) | 19.9 (28.3) |
| Rfree c (%) | 23.4 (46.6) | 25.1 (38.3) | 24.0 (27.1) | 24.2 (29.1) | 24.8 (33.0) | 24.5 (38.3) | 25.5 (33.3) | 23.8 (35.6) | 27.0 (36.1) |
| No. of non-H atoms | 4030 | 3918 | 3854 | 7898 | 8296 | 7977 | 7942 | 7914 | 7564 |
| Protein | 3675 | 3575 | 3569 | 3605/3603 | 3622/3622 | 3609/3611 | 3604/3584 | 3609/3602 | 3591/3572 |
| Ligands | — | 32 | 28 | 128/128 | 160/160 | 185/185 | 181/181 | 166/166 | 128/128 |
| Water | 348 | 311 | 242 | 410 | 718 | 386 | 383 | 357 | 129 |
| Mean B-factor (Å2) | 28.5 | 33.2 | 32.3 | 24.2 | 25.2 | 21.6 | 35.6 | 35.3 | 33.3 |
| Main chain | 27.0 | 32.1 | 31.7 | 23.2/23.2 | 23.6/23.5 | 20.4/20.4 | 35.1/35.2 | 34.8/34.9 | 32.8/32.7 |
| Side chain | 28.4 | 33.1 | 32.2 | 24.4/24.4 | 25.0/24.6 | 21.4/21.3 | 35.5/35.4 | 35.1/35.1 | 33.2/33.1 |
| Ligands | |||||||||
| ssRNA | 28.3/28.8 | 26.1/24.3 | 29.8/29.4 | 45.0/46.7 | 36.2/38.9 | 45.3/45.3 | |||
| AMPPNP | 33.5 | 23.9/25.1 | |||||||
| ADP | 36.2 | 36.0/38.4 | 36.9/33.5 | 34.4/35.9 | |||||
| VO4 | 27.0/26.8 | ||||||||
| PO4 | 47.6/47.9 | 60.7/60.5 | |||||||
| Mn2+ | 27.6 | 36.8 | 14.5/18.1 | 17.4/17.7 | 24.2/28.1 | 27.5/26.0 | |||
| Water | 37.7 | 40.6 | 37.6 | 30.6 | 35.4 | 27.1 | 33.7 | 36.9 | 32.5 |
| RMSDs | |||||||||
| Bond lengths (Å) | 0.002 | 0.003 | 0.007 | 0.011 | 0.010 | 0.009 | 0.007 | 0.009 | 0.008 |
| Bond angles (deg) | 0.410 | 0.506 | 0.981 | 1.329 | 1.338 | 1.223 | 1.134 | 1.218 | 1.212 |
| Ramachandran plot | |||||||||
| Most favoured (%) | 91.3 | 90.0 | 91.9 | 92.6 | 91.4 | 92.1 | 91.9 | 90.7 | 90.3 |
| Allowed regions (%) | 8.7 | 10.0 | 8.1 | 7.3 | 8.4 | 7.9 | 8.1 | 9.2 | 9.7 |
| Overall G factord | 0.13 | 0.10 | 0.16 | 0.07 | 0.07 | 0.09 | 0.12 | 0.11 | 0.10 |
| PDB code | 2JLQ | 2JLR | 2JLS | 2JLU | 2JLV | 2JLX | 2JLY | 2JLZ | 2JLW |
| aRfactor=∑∥Fobs∣−∣Fcalc∥/∑∣Fobs∣. | |||||||||
| bThe numbers in parentheses refers to the last (highest) resolution shell. | |||||||||
| cRfree was calculated with 5% of reflections excluded from the whole refinement procedure. | |||||||||
| dG factor is the overall measure of structure quality from PROCHECK (Laskowski et al, 1993). | |||||||||
RNA recognition
We obtained five isomorphous crystal forms with a 12-mer (RNA12) and one with a 13-mer (RNA13) at a resolution between 1.9 and 2.4 Å (Table I). A comparison of the contacts established by the ssRNA with NS3h in its different nucleotide-bound states does not reveal any major difference, and RNA recognition appears similar regardless of the ATP-bound state, given the resolution limit of this study.
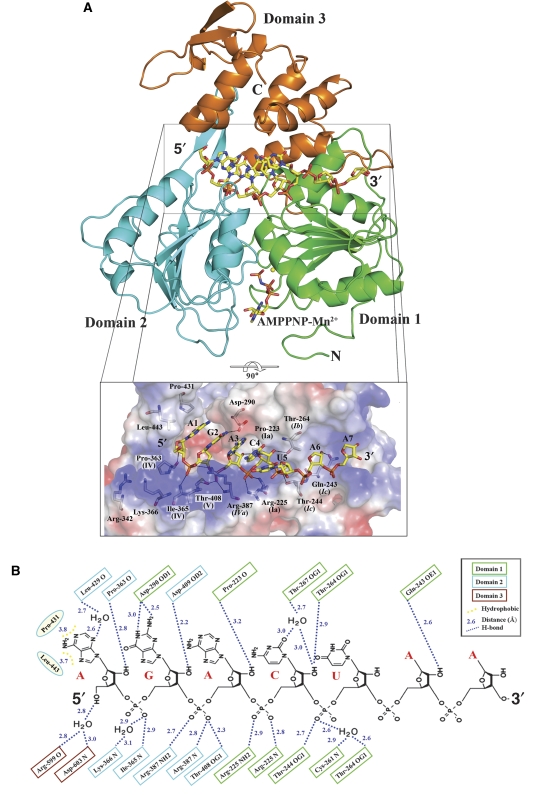
As surmised earlier (Wu et al, 2005; Xu et al, 2005), the ssRNA is accommodated in an extended conformation in the tunnel that separates the ATP-binding subdomains 1 and 2 of NS3h from its subdomain 3 (Figure 1A). Subdomain 1 binds to the 3′ end of ssRNA, whereas the 5′ end mainly interacts with subdomain 2 (Figure 1B). Both the ssRNA location and polarity follow that of a deoxyuridine octamer bound to the HCV helicase (Kim et al, 1998). The exact path of the sugar–phosphate backbone, however, differs markedly between these two structures, with the ssRNA conforming to one strand of A-form dsRNA in the flavivirus helicase, whereas it is more extended in the HCV helicase. In this respect, ssRNA recognition by DENV NS3h is strikingly similar to that found in eukaryotic DEAD box proteins eIF4AIII or Vasa in complex with polyU, with the RNA moieties closely superimposable among the three structures (see below) (Andersen et al, 2006; Bono et al, 2006; Sengoku et al, 2006). It is noted that the HCV helicase is more active on DNA templates (Pang et al, 2002), whereas the DENV helicase exhibits higher activity on RNA duplexes (Xu et al, 2005). Five bases of the RNA located at the 5′ end are stacked to each other (Figure 1). Only the sugar–phosphate backbone is well ordered for nucleotides 6 and 7, and only weak electron density is visible for the corresponding bases. As in the Vasa–RNA complex (Sengoku et al, 2006), the RNA structure makes a sharp bend between bases 5 and 6. At the 3′ end, the remaining five nucleotides are disordered. No aromatic side chain seems properly positioned for base stacking as was proposed for HCV or for PcrA (Kim et al, 1998; Velankar et al, 1999). Residues from the three domains of the protein (but in majority from subdomains 1 and 2) contact the sugar–phosphate backbone either directly or through water molecules (Figure 1), explaining how NS3h can accommodate diverse viral genomic sequences as it translocates towards the 5′ end. As seen in the HCV helicase complexed with a dU8 oligonucleotide (Kim et al, 1998), several evolutionarily conserved threonine residues make direct (Thr-244 and Thr-408) or water-mediated (Thr-267) contacts to the phosphoryl oxygens of ssRNA (Figure 1B). The specificity of DENV NS3h for RNA should derive, in part, from several hydrogen bonds that are established with the 2′-OH moieties by carbonyl oxygens of Pro-363 and Pro-233 and by side chain atoms from Asp-409 and Thr-264 (Figure 1B). As expected, RNA recognition appears to largely occur in a sequence-independent manner as a 13-mer oligoribonucleotide (RNA13) having a different sequence binds to NS3h in essentially the same way as the 12-mer oligoribonucleotide (Supplementary Figure 1). Only the two bases at the 5′ end of the 12 mer make hydrophobic and polar interactions with NS3h. The N2 atom of the guanine at position 2 establishes the sole direct hydrogen bond with the protein through the carboxylic group of Asp-290 (Figure 1B). A basic patch (Figure 1A, inset) shaped by side chains protruding from Arg-342 and Lys-366 leaves ample space to accommodate the triphosphate moiety of flaviviral genomic-capped RNA, an intriguing possibility that might relate to the presence of a strictly conserved AG dinucleotide sequence that is conserved at the 5′ end of all flaviviruses genomes.
Figure 1.
The flavivirus NS3h–RNA–AMPPNP ternary complex. (A) Cartoon representation of NS3h bound to ssRNA and AMPPNP, a non-hydrolysable ATP analogue shown as sticks. The Rec-A-like subdomains 1 and 2 are coloured in green and cyan, respectively. The manganese ion is represented as a yellow sphere. Inset: magnified view of the interactions between the protein and its RNA ligand, using a colour-coded representation of the NS3h electrostatic surface potential. The colour scale is −10 kT (red) to +10 kT (blue). Residues that contact RNA are displayed as sticks and labelled. Also indicated their motif (see also Figure 5). Domain 3 was omitted for clarity. (B) Schematic representation of the interactions observed between NS3h and ssRNA (RNA12). Notice several contacts established with the 2′-hydroxyl groups of the ribose moieties.
Conformational changes on ssRNA binding
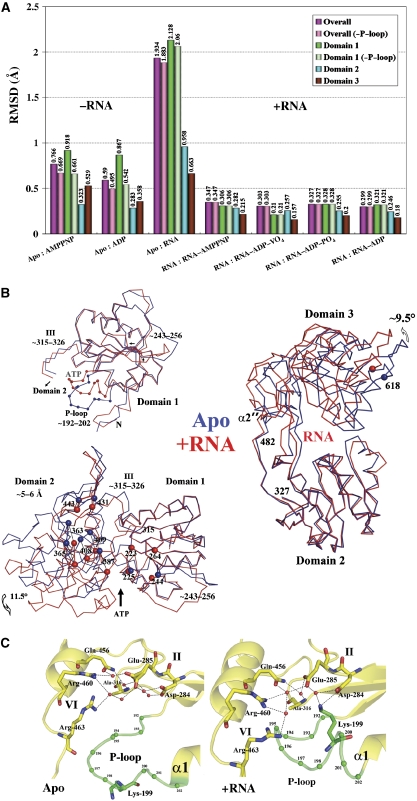
NS3h undergoes several major conformational changes on RNA binding (Figure 2A). We provide the first description of quaternary changes in the flaviviridae RNA helicases that are likely to accompany RNA translocation: subdomain 3 rotates approximately 11° away from the ATP-binding domains leading to a widening of the RNA-binding tunnel and allowing the insertion of several water molecules (Figure 2B). Helix α2′ that functions as the hinge for this movement is deformed into a coil. On ssRNA binding, we also observe closure between subdomains 1 and 2 by ca 12°, leading to a narrower ATP-binding cleft (Figure 2B). This closure leads to a translation of about 5 Å of the RNA-binding motifs within subdomain 2 relative to subdomain 1. This distance, that approximately spans two adjacent bases, might correspond to one translocation step as NS3h tracks along ssRNA in the tunnel in the 3′–5′ direction (Figure 2B). It is noted that such pronounced domain rearrangements have not been observed in the HCV NS3 helicase on oligonucleotide binding (Kim et al, 1998). For DENV4 NS3h, these movements in the quaternary structure are accompanied by an inward movement of the P-loop leading to a reduction of the volume of the ATP-binding cleft and the formation of a salt bridge between Lys-199 and Asp-284 (Figure 2C). Thus, in the absence of ATP, allosteric conformational changes triggered by ssRNA binding convert the P-loop into its substrate-bound conformation, which is poised to react with ATP.
Figure 2.
Structural rearrangements on RNA binding. (A) RMSDs between the various NS3h structures (listed in Table I) or their individual domains after superposition. (−P-loop) indicates values with residues 195–202 omitted from the calculation. A clear partition can be seen between the RNA-bound (‘+RNA') and the RNA-free NS3h structures (‘−RNA'). (B) Comparison between the apo-NS3h structure (blue) and NS3h bound to RNA (red). Top left: superposition of subdomain 1 highlighting P-loop movements. Left lower panel: closure of the ATP-binding cleft on RNA binding (both subdomains 1 were superposed to generate this figure). The position of the α-carbon atoms of residues that come in contact with RNA is shown as blue (apo-NS3h) or red (+RNA) spheres and labelled. Right panel: reorientation of subdomain 3 on RNA binding (subdomain 2 was superposed). (C) Close-up view of the ATP-binding site highlighting conformational changes that occur in the P-loop (green) on RNA binding, in the absence of a bound nucleotide (see text).
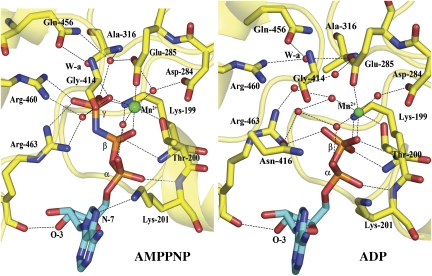
The AMPPNP–Mn2+ and ADP–Mn2+ binary complexes
In the apo-NS3h structure, the NTPase active site is filled with solvent molecules and the P-loop adopts a relaxed state with the side chain of Lys-199 pointing outwards (Figure 2C). This conformation of the P-loop is also observed for the apo DENV4 NS3 protease–helicase (Luo et al, 2008) and is likely to correspond to its resting state in solution, in the absence of either a nucleotide or an RNA ligand. In the 5′-adenylyl-β, γ-imidodiphosphate (AMPPNP)–Mn2+-bound state (Figure 3, left), the P-loop (residues 192–202) has moved towards the protein core and residues 198–202 have reordered to form an extra turn at the N-terminal end of helix α1. This disorder-to-order transition is accompanied by an inward reorientation of Lys-199 and the resulting conformation of the P-loop in the AMPPNP–Mn2+ binary complex closely resembles that shown in Figure 2C for the NS3h–RNA complex or in Figure 1A for the ternary complex formed among NS3h, AMPPNP–Mn2+ and ssRNA (see below). The triphosphate moiety of AMPPNP adopts an extended conformation stabilized by nitrogen amides and the dipolar moment of helix α1, as well as by contacts with residues from the P-loop, Arg-460 and Arg-463 (motif VI), Gly-414 and Asn-416, either through direct or water-bridged H-bonds. The 3′-OH group of the ribose (C2′ endo ring pucker) is hydrogen bonded with the main chain carbonyl oxygen of Arg-463 and the side chain amide group of Asn-329. The N-7 atom of the adenine base (in an anti-conformation) forms a weak hydrogen bond to the ɛ-amino group of Lys-201. However, the large solvent exposure of the base is consistent with a lack of discrimination between various NTP substrates by NS3 (Figure 1A). The ADP–Mn2+ binary complex, shown in Figure 3 (right) reveals a very similar set of interactions as the ADP–Mn2+moiety in the AMPPNP–Mn2+ complex, except for a slight movement of Lys-201 leading to the disruption of one hydrogen bond.
Figure 3.
The AMPPNP–Mn2+ (left) and ADP–Mn2+ binary complex (right). Carbon atoms from the adenine base and the ribose were coloured in cyan, and the phosphorus atoms in orange. The manganese ion is represented as a large green sphere and water molecules as red spheres. Conserved residues are shown as sticks and labelled, water molecules: red spheres, W-a: attacking water (see text), hydrogen bonds: dashed lines.
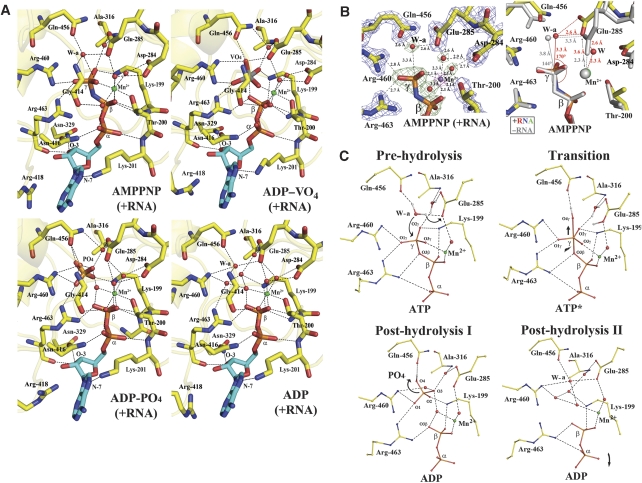
The ATP hydrolysis cycle
To shed light on the ATP hydrolytic cycle, we captured four ternary complexes of RNA-bound NS3h with AMPPNP–Mn2+, ADP–vanadate–Mn2+, ADP–Pi–Mn2+ and ADP–Mn2+, respectively (Figure 4A). Several of these complexes could be obtained either by soaking the nucleotide in preformed RNA-bound NS3h crystals or by cocrystallization of the mixture, yielding similar results. The non-hydrolysable ATP analogue AMPPNP was used to trap an enzyme–substrate ternary complex. In this complex, one water molecule hydrogen-bonded by the side chain atoms of Glu-285 (motif II) and Gln-456 (motif VI) is positioned at right distance for an in-line nucleophilic attack of the γ-phosphate (Figure 4A, top left). Interestingly, the geometry of the catalytic water (W-a) relative to the γ-phosphate appears more favourable in the ternary complex (with bound RNA) compared with the binary AMPPNP–Mn2+ complex (Figure 4B, right panel): the distance to the γ-phosphate is 3.3 Å and the angle subtended by W-a, the γ-phosphorus and the nitrogen atom of AMPPNP is 170° in the ternary complex, whereas the corresponding values are 3.8 Å and 144°, respectively for the binary complex. Activation of W-a presumably occurs through proton transfer to Glu-285 and/or polarization by Gln-456 leading to an in-line attack of the γ-phosphate of ATP. As a result, a trigonal bipyramidal pentavalent intermediate is formed. We obtained a model for the structure of this transition state by solving the structure of an ADP–vanadate–Mn2+ ternary complex (Smith and Rayment, 1996; Chen et al, 2006) (Figure 4A, top right). In this structure determined at 2.2-Å resolution, three oxygen atoms and the vanadium atom approximately lie in a plane forming a trigonal planar VO3 group. The fourth oxygen of vanadate, which is in an apical position relative to the VO3 plane, now occupies the position of the attacking water molecule (W-a) and is stabilized by the side chain of Gln-456. Directly opposite, the other axial position is occupied by an oxygen atom from the ADP β-phosphate, hence completing the pentacoordination shell of the vanadium atom. The distance between the vanadium atom and the phosphorus atom of the β-phosphate is 2.4 Å (Supplementary Figure 2). By comparison, the distance between the phosphorus atom of the phosphate group and the β-phosphate is 3.7 Å in the ADP–Pi–Mn2+ ternary complex (Figure 4A; Supplementary Figure 2).
Figure 4.
ATP hydrolysis. (A) Snapshots of the ATP-binding site during hydrolysis. Upper panel (left to right): the AMPPNP–Mn2+ and ADP–VO4–Mn2+ ternary complexes (with bound RNA). Lower panel: the ADP–Pi–Mn2+ and ADP–Mn2+ ternary complexes. The manganese ion and water molecules are pictured as green and red spheres, respectively, Hydrogen bonds as dashed lines (see text). (B) Close-up view of the AMPPNP-bound structures. Left: the AMPPNP–Mn2+ ternary complex. An anomalous Fourier map (magenta) was calculated and displayed at a level of 10σ, confirming the presence of the manganese ion displayed as a purple sphere. Also displayed: in green, the difference Fourier map showing water molecules (that were omitted for phase calculation) in the manganese coordination shell (at a level of 3σ), and the 2Fo−Fc map at a level of 2.0σ in cyan. The attacking water molecule that is activated through interactions with side chain atoms of Glu-285 and Gln-456 is labelled W-a. Hydrogen bonds are represented by dashed lines and the corresponding interatomic distances are indicated. Right: differences in the geometry of the catalytic water W-a relative to the γ-phosphate between the AMPPNP–Mn2+ binary complex (‘−RNA' with atoms coloured in grey) and the AMPPNP–Mn2+ ternary complex (with bound RNA, in colours; oxygen: red, nitrogen: blue, carbon: yellow). (C) The proposed catalytic mechanism for ATP hydrolysis. The attacking water molecule, labelled W-a, gets activated through proton transfer to Glu-285 that functions as a base. The scissile bond is indicated by an arrow and possible exit routes of the hydrolysis products are indicated by curved arrows. The hydrogen atoms were modelled based on chemical grounds (see text for details).
The NS3h protein appears to be active within the crystal environment because soaking with ATP leads to the observation of an ADP-Pi–Mn2+ complex, which results from ATP hydrolysis (Figure 4A, bottom left). Following ATP hydrolysis, both the phosphate and ADP moieties must vacate the nucleotide-binding site and we have no direct evidence regarding the exact sequence of these post-hydrolysis events. A small tunnel lined by Pro-195, Ala-316, Thr-317, Pro-326, Ala-455 and Gln-456 could serve as an exit route for the phosphate product of the hydrolytic reaction. In the ADP-Pi–Mn2+ complex, the phosphate has moved away from the β-phosphate of ADP and a water molecule is now intercalated between the two species, presumably screening the electrostatic repulsion between their negative charges. In the ADP–Mn2+ ternary complex (Figure 4A, bottom right), several water molecules occupy the location of the phosphate, including the position formerly occupied by W-a. A summary of the catalytic mechanism proposed for ATP hydrolysis by the flavivirus helicase is displayed in Figure 4C.
Coordination of the divalent ion
Interestingly, during the ATPase catalytic cycle, the coordination of the Mn2+ ion appears affected by the presence of ssRNA (Figure 4B; compare also Figures 3 and 4A). In the absence of ssRNA, Mn2+ is octahedrally coordinated by oxygen atoms from Thr-200, the γ-phosphate and two water molecules in the equatorial plane and by two oxygen atoms emanating from the β-phosphate and Glu-285 in the apical position. Conversely, in the presence of ssRNA, Glu-285 no longer directly interacts with the Mn2+ ion (the distance increases from 2.3 to 3.5 Å) but instead through an additional water molecule, which completes the octahedral coordination shell (Figure 4B). This change in the coordination shell of the divalent metal ion is likely to affect catalytic activity by shifting the negative charge of the carboxyl group of Glu-285 that functions as the base, making the catalytic water a better nucleophile. Moreover, the alteration of the metal hydration state could also facilitate the release of the ADP–Mn2+ leaving group as compared with a leaving group directly coordinated by Glu-285.
Discussion
An evolutionarily conserved ssRNA recognition module
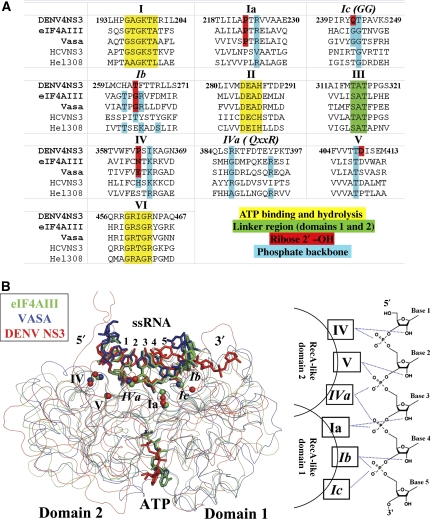
Two high-resolution crystallographic structures of DEAD box eukaryotic proteins involved in mRNA processing were recently reported: eIF4AIII, a member of the Exon Junction Complex and the Vasa protein, both as complexes with polyU (Andersen et al, 2006; Bono et al, 2006; Sengoku et al, 2006). Here, we report the first structure of a flaviviridae helicase in complex with ssRNA. A comparison of these three structures (Figure 5) reveals a striking similarity in the molecular details of ssRNA recognition and suggests the existence of an ancestral ssRNA recognition module that was borrowed by the flaviviridae during evolution. A summary of the interactions between ssRNA and conserved residues within the SF2 helicase motifs is presented in Figure 5. The two α/β (RecA-like) domains establish extensive interactions with the 2′-OH and phosphate groups of a core of five nucleotides (numbered here from 1 to 5) in a conserved manner (Figure 5). Notably, both the conformation and location of the ssRNA with respect to the three helicases DENV NS3h, eIF4AIII and Vasa are virtually identical (Figure 5B). Besides this conserved core of 5 nt that are well ordered in the protein-binding site, both 5′ and 3′ nucleotides appear disordered, an observation consistent with their susceptibility to RNase digestion for the eiF4AIII–RNA complex (Andersen et al, 2006). Several of these sequence motifs are also present in DNA helicases (Buttner et al, 2007; Jankowsky and Fairman, 2007; Pyle, 2008). It appears that through fusion with additional domains, this ancestral single-strand nucleic acid recognition module has evolved to fulfill different molecular and cellular tasks. In the case of the DENV NS3 protein, both a protease domain and an original protein recognition module have been fused at its N- and C-terminal ends, respectively.
Figure 5.
(A) Structure-based sequence alignment of SF2 helicases discussed in the text. Only the regions encompassing residues involved in ATP binding and hydrolysis, interdomain communication, binding of the ribose 2′-hydroxyl of RNA or backbone phosphate are included. Residue numbers are for the DENV NS3 protein. Clusters of residues that are structurally equivalent (based on the superposition in (B)) but are not conserved are indicated in italics (e.g. motif GG or Ic). (B) ssRNA recognition by DEAD/DExH helicases. Left panel: comparison of ssRNA complexes of eIF4AIII (in green; PDB code: 2J0S), Vasa (in blue; PDB code: 2DB3) and DENV NS3h (in red, this work). The structures are overlaid based on the ssRNA sugar–phosphate backbone. Protein α-carbon atoms are displayed as smooth ribbon and ssRNA as sticks. Cα atoms of the spatially conserved RNA-binding residues (belonging to motifs I–V) are indicated by coloured spheres. Right panel: schematic illustration of the ssRNA recognition module of DEAD/DExH helicases. For subdomain 1, motif Ia interacts with 2′-OH of base 3 and phosphate group of base 4; motif Ib with 2′-OH of base 4 and phosphate group of base 5; motif Ic (also named GG) with the phosphate of base 5. For subdomain 2, motif IV interacts with 2′-OH of base 1 and phosphate of base 2; motif V with 2′-OH of base 2 (observed in DENV NS3) and phosphate of base 3; motif Iva with phosphate of bases 3 and 4.
Conformational changes during ATP hydrolysis and RNA unwinding
On the basis of our crystallographic results, RNA binding appears as the major determinant that dictates the conformation of the flavivirus NS3 helicase (Figure 2). The NS3h helicase was crystallized in three different crystal forms in the absence of RNA (Table I), all of which are closely superposable and RNA binding appears sufficient to convert the helicase to its closed conformation. However, no change in the quaternary structure of NS3h was observed in the various altered nucleotide states, either for binary or for ternary (RNA-bound) complexes. This was slightly surprising in the light of the unwinding mechanism proposed for the HCV NS3 protein, a DExH helicase, where the protein cycles between open and closed states depending on the nucleotide-binding state and where the ATP hydrolysis cycle is coupled to loose versus tight RNA binding and translocation (Pyle, 2008). This observation might simply relate to constraints imposed on the NS3h protein by forces from the crystal lattice (e.g. in its RNA-bound C2 crystal form) that would restrict conformational changes. The fact that NS3h retains ATPase activity in the C2 crystalline form (ATP can be soaked into a preformed crystal and the ATPase reaction occurs) suggests that both the NS3h–ATP and the NS3h–vanadate–ADP–Mn2+ transition-like states and ADP–Pi–Mn2+ structures represent biologically relevant snapshots. Conversely, the biological relevance of the crystal structures we obtained for the ADP–Mn2+ binary and ternary complexes is probably more questionable because the enzyme is expected both to release the reaction products and translocate along RNA, a step presumably accompanied by several conformational changes (Myong et al, 2007). Thus, at least another conformational state of NS3h probably exists that could not be trapped in our experiments and which might correspond to one of the conformations predicted for the flaviviridae helicase using normal mode analysis (Zheng et al, 2007).
Insights into the RNA-unwinding mechanism by NS3
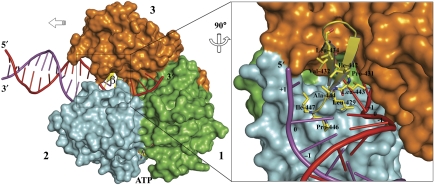
On the basis of the experimental complexes between DENV NS3h and ssRNA described above, we performed a simple modelling exercise, which consisted in extending the bound ssRNA strand into the 5′ direction and pairing it with a complementary strand to form a dsRNA substrate for NS3h (Figure 6). Modelling was aided by a crystallographic Hel308 protein–DNA structure, a SF2 helicase with a related architecture for both the single-strand nucleic acid-binding tunnel and the ATPase active site (Wu et al, 2005; Xu et al, 2005; Buttner et al, 2007). As seen in Figure 6, one β-hairpin element of DENV NS3 (joining strands β4A′ and β4B′, see Xu et al (2005)) is likely to have a crucial function for separating the two strands by disrupting base stacking and stabilizing the unwound duplex (Buttner et al, 2007). In this model, one RNA strand would migrate through the tracking ssRNA tunnel, whereas the other strand is forced towards the back of the protein by the β-hairpin. We note a relative variability of the residues exposed at the surface of the β-hairpin scaffold among several SF2 helicases: in the Hel308 DNA helicase, it consists of a stretch of charged or aromatic residues, of charged residues in HCV NS3, whereas the β-hairpin from dengue NS3 appears more hydrophobic (Figure 6). Interestingly, a topologically equivalent hairpin (christened ‘separation pin') was also proposed to have an active function in DNA unwinding by UvrD, a bacterial SF1 helicase (Lee and Yang, 2006). Thus, this important functional feature is not limited to SF2 helicases but appears to be shared between helicases of these two superfamilies. How residues from the β-hairpin precisely interact with the fork and how this affect RNA unwinding and processivity remain to be determined for a DExH helicase. Moreover, it is not known with certainty whether subdomain 3 of the DExH viral helicase establishes any interaction with dsRNA during the unwinding reaction, an attractive hypothesis considering that the topologically equivalent subdomains 1B and 2B from SF1 helicase UvrD do interact with the DNA duplex (Lee and Yang, 2006). Recent single molecule studies would favour a second point of contact of the flaviviridae helicase ahead of the fork, presumably by contacting dsRNA (Cheng et al, 2007). The driving force for strand separation itself remains elusive (Betterton and Julicher, 2005): in an active mechanism, the β-hairpin could function as a ‘helix opener' by actively disrupting base pairs hydrogen bonds. Quaternary movements in the three NS3h subdomains gradually accumulating tension on the protein–nucleic acid complex might also assist this process by exerting a force or torque on the duplex, as was suggested by single molecule studies using the HCV helicase (Myong et al, 2007; Pyle, 2008). In a passive mechanism, the β-hairpin would merely exert an effect as an ‘insulator' by taking advantage of the spontaneous opening of the duplex at the fork, through thermal motion, and by stacking with the melted bases. In our model, base +1 would interact with the conserved Leu-443 that projects from the β-hairpin disrupting base stacking and thus splaying apart dsRNA at the fork region (Figure 6). We also noticed departure from A-form RNA between bases 5 and 6 at the 3′ end of the RNA12 oligomer bound to DENV NS3. A distorted RNA conformation was indeed observed in the Vasa–polyU complex and was proposed to be important for the mechanism of RNA unwinding (Sengoku et al, 2006). Another possibility is that this protein-induced strain is not directly related to RNA duplex unwinding but rather to prevent NS3 from sliding backwards along the ssRNA substrate.
Figure 6.
RNA unwinding by the NS3 helicase. Left panel: proposed model for the NS3 helicase complexed with dsRNA and ATP. Right panel: close-up view of the proposed ‘helix opener' the β-hairpin from domain 2. Conserved hydrophobic residues are shown as sticks and labelled. Partially melted dsRNA is modelled based on comparison between NS3–ssRNA and the archaeal Hel308-dsDNA (PDB code: 2P6R) structure. colour code: subdomain 1, green; subdomain 2, cyan; subdomain 3, orange; β-hairpin, yellow ribbon and sticks; dsRNA strands are in red (tracking strand) and purple. At the dsRNA fork, three hydrophobic patches protrude from the β-hairpin: patch 1, formed by Pro-431 and Leu-443 stack with base +1 of the tracking strand bound to NS3; patch 2, Leu-429 and Ala-444, stack with base 0, possibly disrupting base stacking between bases 0 and base +1. Likewise, patch 3 formed by Pro-446 and Ile-447 located opposite to patches 2 and 4 Val-432, Leu-434 and Ile-442 stack with bases 0 and +1 of the other strand.
Concluding remarks
For several positive-strand RNA viruses, including poliovirus and flaviviruses, RNA replication, translation and encapsidation occur in a concerted manner, presumably to minimize amplification and packaging of defective RNA (Novak and Kirkegaard, 1994). Moreover, a role for NS3 in virus assembly, independent of its known enzymatic functions and in association with NS2A, was proposed (Khromykh et al, 2001; Patkar and Kuhn, 2008). Given the strict conservation of the two bases AG at the 5′ ends of both the + and − strands genomic flaviviral RNA, it is tempting to propose that having translocated towards the 5′ end, the bound NS3 helicase would exert an effect as an ‘RNA-anchoring device' by both preventing strand reannealing and helping redirect NS5 RdRp to the 3′ end of the genome (Filomatori et al, 2006), thereby ensuring the complete replication of RNA. Reasons for the asymmetry in genome replication and encapsidation might derive in part from specific recognition of capped viral RNA by the NS3 protease–helicase, presumably in association with NS2A (Patkar and Kuhn, 2008) and capsid proteins. Finally, the development of a reliable high-throughput unwinding assay for inhibitors screening has been elusive for DENV NS3h. The availability of small RNA ligands should now stimulate the search for inhibitors that can compete with RNA binding.
Materials and methods
Cloning and expression
The catalytic domain of the NS3 helicase domain from dengue virus (DENV) serotype 4 (residues 172–618), hereafter named NS3h, was amplified by PCR using forward primer (5′-ATTGGTGAGCCAGATTATGAAGTGG-3′) and the reverse primer (5′-GTGGTGCTCGAGTTACTTTCTTCCACTGGCAAA-3′). The XhoI site is underlined. The fragment was digested and cloned into a modified pET32b plasmid using T4 ligase (Roche), where the S tag and enterokinase cleavage sequence are absent. The expressed construct thus comprises the thioredoxin (Trx) protein followed by a hexa-histidine tag and a thrombin cleavage site fused to the N terminus of NS3h. Transformed Escherichia coli BL21-CodonPlus cells (Stratagene) were grown at 37°C in LB medium supplemented with 100 μg ml−1 ampicillin and 50 μg ml−1 chloramphenicol to an OD600 nm of 0.6–0.8. Protein expression was induced at 16°C by adding isopropyl-β-D-thiogalactopyranoside with a final concentration of 0.4 mM. After overnight growth, cells were harvested by centrifugation at 8000 g for 10 min at 4°C and stored at −20°C.
Protein purification
Cells resuspended in buffer A (20 mM Na3PO4, pH 7.4, 0.5 M NaCl and 40 mM imidazole) were lysed by sonication and the lysate was clarified by centrifugation at 30 000 g for 60 min at 4°C. The supernatant was purified by metal affinity using a HisTrap HP column (Amersham Bioscience) equilibrated with buffer A. Proteins were eluted using a linear gradient of imidazole from 40 to 500 mM. The fraction containing Trx-(His)6-D4NS3hel was dialysed against buffer B (20 mM Na3PO4, pH 7.4 and 0.2 M NaCl), with concomitant cleavage of the thioredoxin tag by thrombin (10 enzyme units per mg of protein) at 4°C for approximately 18 h. The cleavage mixture was loaded onto a HisTrap HP column equilibrated with buffer B to remove the Trx-(His)6 protein from the mixture. Concentrated NS3h proteins were subjected to a final gel-filtration purification step through a HiPrep 16/60 Superdex 75 column (Amersham Bioscience) in buffer C (20 mM Tris–HCl, pH 7.4, 150 mM NaCl, 5 mM dithiothreitol and 5% glycerol) or buffer C′ (20 mM Tris–HCl, pH 7.4, 50 mM NaCl, 5 mM β-mercaptoethanol and 5% glycerol). Fractions containing NS3h were pooled and concentrated to 20 mg ml−1.
Crystallization and data collection
Crystals of native NS3h (stored in buffer C) were grown at 13°C by the hanging drop vapour diffusion method over wells containing 0.1 M 2-(N-morpholino)ethanesulphonic acid (MES), pH 6.5 and 20% polyethylene glycol 3350. A volume of 2 μl of precipitating solution was mixed with an equal volume of NS3h at a concentration of 5 mg ml−1. The drop was equilibrated against a reservoir containing 1 ml of the precipitating solution. Crystals for the AMPPNP (Roche) complex were obtained by cocrystallization of NS3h at a concentration of 5 mg ml−1 (∼0.1 mM), with 5 mM MnCl2 and 5 mM AMPPNP using a slightly different precipitating solution (0.1 M MES, pH 6.5 and 10% polyethylene glycol 3350) at 13°C. Crystals with ADP (Roche) were obtained by cocrystallization at a concentration of 2.5 mg ml−1 with 5 mM MnCl2 and 5 mM ADP in 0.1 M Tris–HCl, pH 7.0 and 7.5% polyethylene glycol 3350 at 23°C. The binary complex with RNA12 was also obtained through cocrystallization: a final solution containing NS3h (storage buffer: buffer C) at 2.5 mg ml−1 and RNA12 (Sigma) at 0.1 mM (∼2-fold molar excess) was incubated at 25°C for 1 h. Subsequently, a volume of 3 μl of the NS3h–RNA12 solution was mixed with 2 μl of precipitating solution (0.1 M MES, pH 6.0 and 15% polyethylene glycol 8000) and the drop was equilibrated against a reservoir containing 1 ml of the precipitating solution. Interestingly, using identical cocrystallization conditions, isomorphous crystals can be obtained either with a AGU10 12-mer or with the 5′ untranslated region (AGUUGUUAGUCU) of the viral genome, but not with a polyU 12-mer, suggesting that a guanine at position 2 is needed for crystallization with a 12-mer.
The ternary complexes containing RNA12 and AMPPNP, ADP-Pi, ADP–vanadate or ADP, respectively (Tables I and II), were obtained through 20 h soak of crystals of the binary RNA complex in a cryoprotecting solution (0.1 M MES, pH 6.0, 25% PEG8000 and 25% glycerol) that contained 5 mM MnCl2 in the presence of 5 mM AMPPNP, ATP, ADP and orthovanadate (Sigma) or ADP at 13°C. Prior to data collection, native and AMPPNP crystals were briefly soaked in a cryoprotecting solution containing 0.1 M MES, pH 6.5, 20% PEG3350 and 25% glycerol; ADP complex crystals were soaked in 0.1 M Tris–HCl, pH 7.0, 20% PEG3350 and 25% glycerol; RNA12-bound crystals were soaked in 0.1 M MES, pH 6.0, 25% PEG8000 and 25% glycerol for 20 h. Following cryoprotection, crystals were rapidly cooled to 100 K in a nitrogen gas stream (Oxford Cryosystems) and diffraction intensities were recorded at the Nanyang Technological University (Singapore) on an R-AXIS IV++ imaging plate detector. For the RNA12–ADP–VO4–Mn2+ and RNA12–AMPPNP–Mn2+ ternary complexes, data at higher resolution were collected at the ESRF, Grenoble (beamline ID23-1) and at the Swiss Light Source, Villingen (beamline X10SA) respectively, using charged-coupled device detectors.
Integration, scaling and merging of the intensities were carried out using the MOSFLM and Scala programs (CCP4, 1994).
Structure solution and refinement
The native DENV4 NS3 helicase structure was solved by molecular replacement with the program Phaser (McCoy, 2007) using the DENV2 NS3 helicase (PDB code: 2BMF) (Xu et al, 2005) as a search probe. The complex with RNA12 was solved with the program Phaser, using the native refined DENV4 NS3 helicase structure as the search probe. Refinement cycles were carried out using REFMAC5 (Murshudov et al, 1997) with the TLS (translation, liberation, screw-rotation displacement) refinement option towards the end, using the four following groups for each independent molecule: subdomain 1 (residues 168–326), subdomain 2 (327–481), subdomain 3 (482–618) and the RNA ligand. Refinement cycles were interspersed with model rebuilding using the computer graphics program Coot (Emsley and Cowtan, 2004). The quality of the structures was analysed using PROCHECK (Laskowski et al, 1993). A summary of the refinement statistics and stereochemistry analysis is given in Table II. Superpositions of structures were carried out using the program LSQKAB from the CCP4 suite. Figures were prepared using the program Pymol (DeLano, 2002).
Supplementary Material
Supplementary Figures 1 and 2
Acknowledgments
This study was supported by BMRC grants 06/1/22/19/447 and 05/1/22/19/394, an ARC grant from the Singapore Ministry of Education ARC5/07 and an ATIP grant from the CNRS to the laboratory of JL.
References
- Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR (2006) Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science 313: 1968–1972 [DOI] [PubMed] [Google Scholar]
- Betterton MD, Julicher F (2005) Opening of nucleic-acid double strands by helicases: active versus passive opening. Phys Rev E Stat Nonlin Soft Matter Phys 71 (1 Part 1): 011904. [DOI] [PubMed] [Google Scholar]
- Bleichert F, Baserga SJ (2007) The long unwinding road of RNA helicases. Mol Cell 27: 339–352 [DOI] [PubMed] [Google Scholar]
- Bono F, Ebert J, Lorentzen E, Conti E (2006) The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 126: 713–725 [DOI] [PubMed] [Google Scholar]
- Buttner K, Nehring S, Hopfner KP (2007) Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol 14: 647–652 [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50 (Part 5): 760–763 [DOI] [PubMed] [Google Scholar]
- Chen C, Saxena AK, Simcoke WN, Garboczi DN, Pedersen PL, Ko YH (2006) Mitochondrial ATP synthase. Crystal structure of the catalytic F1 unit in a vanadate-induced transition-like state and implications for mechanism. J Biol Chem 281: 13777–13783 [DOI] [PubMed] [Google Scholar]
- Cheng W, Dumont S, Tinoco I Jr, Bustamante C (2007) NS3 helicase actively separates RNA strands and senses sequence barriers ahead of the opening fork. Proc Natl Acad Sci USA 104: 13954–13959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL User's Manual. Palo Alto, CA, USA: DeLano Scientific [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60 (Part 12): 2126–2132 [DOI] [PubMed] [Google Scholar]
- Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV (2006) A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev 20: 2238–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E, Fairman ME (2007) RNA helicases—one fold for many functions. Curr Opin Struct Biol 17: 316–324 [DOI] [PubMed] [Google Scholar]
- Khromykh AA, Meka H, Guyatt KJ, Westaway EG (2001) Essential role of cyclization sequences in flavivirus RNA replication. J Virol 75: 6719–6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, Murcko MA, Lin C, Caron PR (1998) Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 6: 89–100 [DOI] [PubMed] [Google Scholar]
- Koonin EV (1993) A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J Mol Biol 229: 1165–1174 [DOI] [PubMed] [Google Scholar]
- Kuo MD, Chin C, Hsu SL, Shiao JY, Wang TM, Lin JH (1996) Characterization of the NTPase activity of Japanese encephalitis virus NS3 protein. J Gen Virol 77 (Part 9): 2077–2084 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26: 283–291 [Google Scholar]
- Lee JY, Yang W (2006) UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 127: 1349–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Clum S, You S, Ebner KE, Padmanabhan R (1999) The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J Virol 73: 3108–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Thiel HJ, Rice CM (2007) Flaviviridae: the Viruses and their Replication. Philadelphia: Lippincott-Raven Publishers [Google Scholar]
- Luo D, Xu T, Hunke C, Gruber G, Vasudevan SG, Lescar J (2008) Crystal structure of the NS3 protease–helicase from dengue virus. J Virol 82: 173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 63 (Part 1): 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53 (Part 3): 240–255 [DOI] [PubMed] [Google Scholar]
- Myong S, Bruno MM, Pyle AM, Ha T (2007) Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science 317: 513–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak JE, Kirkegaard K (1994) Coupling between genome translation and replication in an RNA virus. Genes Dev 8: 1726–1737 [DOI] [PubMed] [Google Scholar]
- Pang PS, Jankowsky E, Planet PJ, Pyle AM (2002) The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J 21: 1168–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patkar CG, Kuhn RJ (2008) Yellow fever virus NS3 plays an essential role in virus assembly independent of its known enzymatic functions. J Virol 82: 3342–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM (2008) Translocation and unwinding mechanisms of RNA and DNA helicases. Ann Rev Biophys 37: 317–336 [DOI] [PubMed] [Google Scholar]
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S (2006) Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 125: 287–300 [DOI] [PubMed] [Google Scholar]
- Smith CA, Rayment I (1996) X-ray structure of the magnesium(II).ADP.vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 Å resolution. Biochemistry 35: 5404–5417 [DOI] [PubMed] [Google Scholar]
- Story RM, Steitz TA (1992) Structure of the recA protein–ADP complex. Nature 355: 374–376 [DOI] [PubMed] [Google Scholar]
- Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB (1999) Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97: 75–84 [DOI] [PubMed] [Google Scholar]
- Warrener P, Tamura JK, Collett MS (1993) RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J Virol 67: 989–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Bera AK, Kuhn RJ, Smith JL (2005) Structure of the Flavivirus helicase: implications for catalytic activity, protein interactions, and proteolytic processing. J Virol 79: 10268–10277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Sampath A, Chao A, Wen D, Nanao M, Chene P, Vasudevan SG, Lescar J (2005) Structure of the dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 Å. J Virol 79: 10278–10288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap TL, Xu T, Chen YL, Malet H, Egloff MP, Canard B, Vasudevan SG, Lescar J (2007) Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J Virol 81: 4753–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Liao JC, Brooks BR, Doniach S (2007) Toward the mechanism of dynamical couplings and translocation in hepatitis C virus NS3 helicase using elastic network model. Proteins 67: 886–896 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1 and 2