Abstract
Objectives
The aims of this study were to determine the relative influence of genetic and environmental contributions to inflammatory biomarkers, and to what extent correlations among these markers are due to genetic or environmental factors.
Methods
We performed univariate and multivariate genetic analyses of four inflammatory markers: interleukin-6 (IL-6), soluble IL-6 receptor (sIL-6R), C-reactive protein (CRP), and fibrinogen, in 166 (88 monozygotic and 78 dizygotic) middle-aged male twin pairs.
Results
The mean age (±SD) of the twins was 54 (±2.93) years. Heritability was substantial for CRP (0.61, 95% CI: 0.47–0.72) and moderate to fair for IL-6 (0.31, 0.13–0.46), sIL-6R (0.49, 0.30–0.76) and fibrinogen (0.52, 0.34–0.65). IL-6, CRP and fibrinogen showed significant correlations, but not with sIL-6R. Multivariate genetic analysis found that these correlations could be best explained by a common pathway model, where the common factor explained 27%, 73% and 25% of the variance of IL-6, CRP and fibrinogen, respectively. About 46% (95% CI: 21–64%) of the correlations among the three inflammatory markers could be explained by the genetic factors. After adjusting for covariates known to influence inflammation levels, heritability estimates were slightly decreased but the overall results remained similar.
Conclusions
A significant part of the variation in inflammatory marker levels is due to genetic influences. Furthermore, almost 50% of the shared variance among these biomarkers is due to a common genetic factor which likely plays a key role in the regulation of inflammation.
Keywords: twin study, middle-aged males, inflammatory markers, heritability, common genes
1. Introduction
The role of the immune system and inflammatory pathways in the development of atherosclerotic disease is well established [1]. C-reactive protein (CRP), an acute phase reactant, is an independent coronary heart disease (CHD) risk factor [2–4]. Other established inflammatory risk markers for CHD include fibrinogen [2] and interleukin-6 (IL-6) [5]. The pathophysiological effects of IL-6 depend strongly on a soluble form of the receptor (sIL-6R) [6, 7]. IL-6 and sIL-6R, acting in concert in atherosclerotic plaques, contribute to inflammation and myocardial injury during the acute phase of myocardial infarction [8, 9].
As reported by several twin and family studies, a considerable part of the variation in inflammatory biomarkers can be explained by genetic factors [10–13]; however, most of these studies focused on a single inflammatory marker. Substantial relationships among these markers exist, [5] and some authors have even constructed an “inflammation factor” combining IL-6 and CRP by using factor analysis or other methods [14]. Taking into account the complexity of inflammatory pathways and the inter-relatedness of cytokine signaling and acute phase proteins, it is possible that common genetic factors account for the observed association among inflammatory markers. Such shared genetic pathways may be important in the complex orchestration of the inflammatory response. To answer this question, the most efficient design is a classic twin study incorporating a multivariate analysis [15]. To date, only one study explored the genetic correlations among these inflammatory variables; however, only bivariate correlations were reported [11]. In addition, this study was limited to elderly twins who survived to 73 years old, which might have introduced selection bias [11].
In the present study, we first estimated the magnitude of genetic influences on plasma levels of IL-6, sIL-6R, CRP and fibrinogen in middle-aged male twins. Next, we determined to what extent the observed correlations among these inflammatory markers could be explained by the same genetic factors.
2. Methods
2.1 Subjects
The Twins Heart Study (THS) is an investigation of psychological, behavioral and biological risk factors for subclinical cardiovascular disease in twins. Participants were members of the Vietnam Era Twin (VET) Registry, which consists of 7,369 male-male twin pairs born between 1939 and 1955 in which both siblings served on active military duty during the Vietnam era (1965–1975). The majority (93%) was white and 6.5% was African-American. The development and characteristics of this registry were reported elsewhere [16].
THS involved a random sample of twin pairs discordant for lifetime history of major depressive disorder and a random sample of age-, race- and zygosity-matched twin pairs in which both twins were unaffected by major depression. Twins were selected for participation if they did not report a history of CHD in previous VET Registry surveys, the latest of which was in 1990 [17]. They were examined at the Emory University General Clinical Research Center between March 2002 and March 2006, where their medical history was updated. A few twins (n=35) had developed CHD between 1990 and 2002. These were not excluded, but CHD history was adjusted for in the analysis (see below). A total of 180 twin pairs were enrolled, of which 68 twin pairs were discordant for history of major depression. In the present study, we focused on subacute, low-grade systemic inflammation. Therefore, subjects and their co-twins (14 twin pairs) with plasma levels of CRP > 10 mg/L or levels of IL-6 > 10 pg/mL, which is generally considered to reflect acute and clinically significant inflammation, were excluded from the analyses. Although part of the sample was selected based on the presence of lifetime major depression, adjusting for major depression did not change the heritability of these inflammatory markers (see below). Furthermore, when discordant twin pairs were taken out, the results were virtually identical. To avoid loss of power, the entire sample was then included in the present analysis. Informed consent was obtained from each subject.
2.2 Measures
All measurements were performed in the morning after an overnight fast, and both twins in a pair were tested at the same time. A medical history and a physical exam were obtained from all subjects. Height and weight were measured and used to calculate body mass index (BMI). Systolic blood pressure (SBP) and diastolic blood pressure (DBP) were measured by mercury sphygmomanometer on the right arm with the subject in sitting position after 10 minutes of rest. The average of two measurements 5 minutes apart was taken. Venous blood samples were drawn for the measurements of glucose and lipid profile after an overnight fast. Cigarette smoking was classified into current, past and never smoking. Physical activity was assessed by a modified version of the Baecke Questionnaire of Habitual Physical Activity used in the Atherosclerosis Risk in Communities (ARIC) Study [18]; this is a 16-question instrument documenting level of physical activity at work, during sports and non-sports activities. The total physical activity score (work, sports, and non-sports leisure activity) was used in the analysis. Diabetes was defined as a fasting glucose ≥ 126 mg/dl, or current pharmacological treatment for diabetes. Hypertension was defined as SBP ≥ 140 mmHg and/or DBP ≥ 90 mmHg, or current pharmacological treatment for hypertension. History of CHD was defined as previous myocardial infarction, angina pectoris or coronary revascularization procedures.
2.3 Markers of Inflammation
IL-6 and sIL-6R were assessed using commercially available ELISA kits obtained from R and D Systems. Assays were performed according to manufacturer’s specifications, and all samples were run in duplicate. Inter- and intra-assay variability of this assay is reliably <10%. CRP was measured with the Beckman Coulter High Sensitivity C-Reactive Protein assay on the Synchron LX-20 analyzer. Fibrinogen was measured by using the Dade Behring BCS coagulation analyzer.
2.4 Statistical Analyses
Initial descriptive analysis examined means and percents of the inflammatory markers and other study variables. Correlations between inflammatory markers and other cardiovascular risk factors were assessed using Pearson correlations for continuous risk factors and Spearman correlations for categorical risk factors. To improve the distributional properties of the inflammatory markers, data were transformed to the logarithmic scale. Analyses were performed using the statistical software SAS, version 9.0 (SAS, Inc., Cary, NC).
Structural equation modeling (SEM) was used to obtain estimates of the genetic and environmental influences on inflammatory factors. The assumptions under these models are that MZ twins share 100% of their genes whereas DZ twins share, on average, 50% of their genes. Thus, a greater phenotypic similarity in MZ twins as compared with DZ twins suggests genetic influences on that trait. Shared environmental factors are assumed to be 100% for both MZ and DZ twins if siblings were reared together, as in our sample, while unique environment is not shared between the siblings for either MZ or DZ twins [19]. Model fitting is based on the comparison of the variance-covariance matrices in MZ and DZ twin pairs and allows separation of the observed phenotypic variance into additive (A) genetic components and shared (C) or unique (E) environmental components. Dividing each of these components by the total variance yields standardized components of variance. One of these is the heritability (h2), which can be defined as the ratio of additive genetic variance to total phenotypic variance. Extension of univariate to multivariate models allows exploration of the question of whether the origin of the covariance between the different variables is genetically and/or environmentally determined.
In the present study, we fit both univariate and multivariate models. The intraclass correlations (ICC) were calculated to assess the similarity in MZ and DZ twins for inflammatory variables, and the heritability was estimated before and after adjusting for other factors, including age, BMI, smoking status, physical activity, history of CHD, hypertension, diabetes, and major depressive disorder. Phenotypic correlations between inflammatory markers were calculated and all variables showing significant correlations (P < 0.05) were entered into the multivariate analysis, which tested to what extent these correlations can be explained by common genes or common environment.
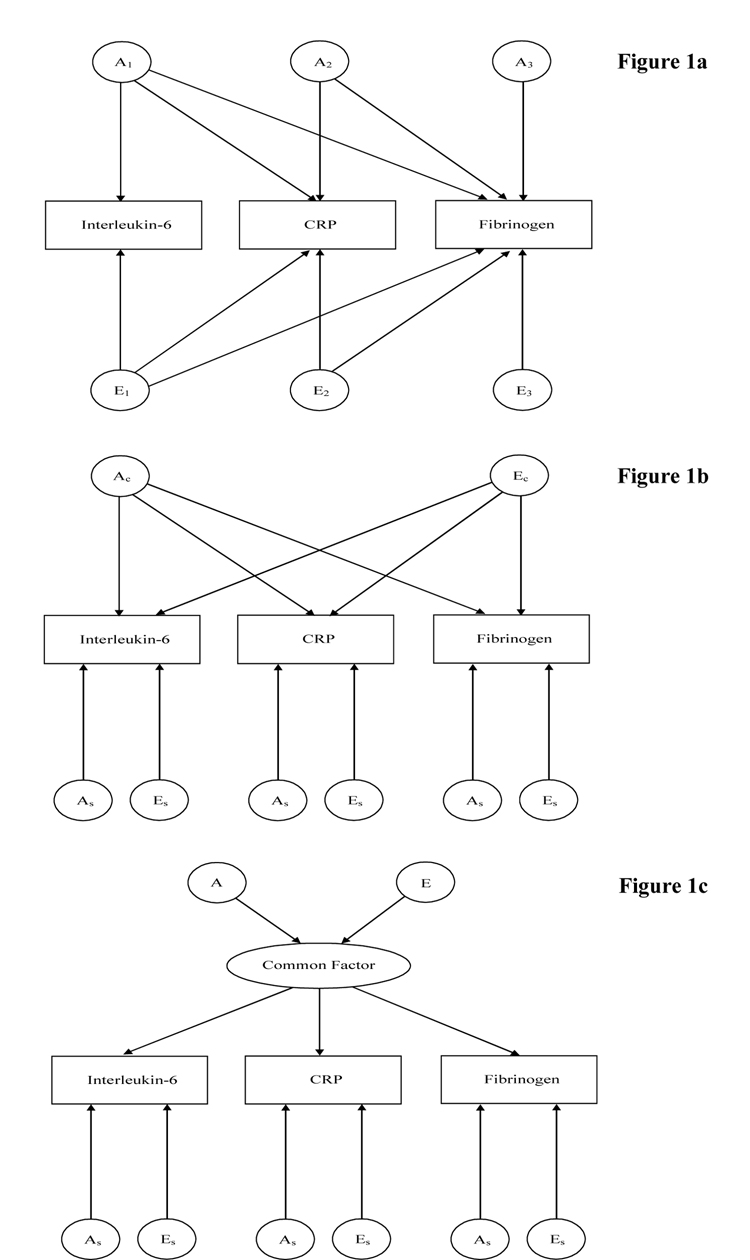
Three types of multivariate models were used: the Cholesky decomposition, the independent pathway model and the common pathway model [19]. Although all these models decompose the variance into the three components of variation, A, C and E, each represents different ways in which genes and the environment may affect the observed covariation between the measures. Briefly, the Cholesky model represents the most general model, without specific hypotheses regarding the variance-covariance structure being tested. The number of latent factors equals the number of variables: the first factor loads on all traits, and the second factor loads on all traits except the first one, and so on (Figure 1a). The independent pathway model, a submodel of the Cholesky decomposition, assumes that one common genetic or environmental factor can explain the covariance structure of the data. Besides these common factors, each outcome measure is also influenced by specific genetic and environmental factors (Figure 1b). Finally, the common pathway model is a further simplification of the independent pathway model. Both genes and environment contribute to one latent (unmeasured) variable, which is responsible for the observed covariance between the outcome measures. Genetic and environmental factors specific to each measure are also incorporated in the model (Figure 1c).
Figure 1.
Figure 1a. Cholesky decomposition of the genetic and environmental factors for inflammatory markers. An represents the additive genetic effect on the subset of measures, for example A1 represents the genetic factor loading on IL-6 and the other two markers. En represents the unique environmental factors. For clarity only genetic and unique environmental factors are illustrated. Similar models will include the shared environmental component (C)
Figure 1b. Independent pathway, where one common genetic factor (Ac) and environmental factor (Ec) explain the relationship among inflammatory markers. Apart from the common genetic and environmental factors, variation is explained by specific genetic (As) and environmental (Es) factors. For clarity only genetic and unique environmental factors are illustrated. Similar models will include the shared environmental component (C)
Figure 1c. Common pathway model where both genes and environment are assumed to contribute to one latent (unmeasured) variable which is responsible for the correlation among inflammatory markers. Specific genetic (As) and unique environmental (Es) influences are also incorporated in the model. For clarity only genetic and unique environmental factors are illustrated. Similar models will include the shared environmental component (C).
A series of models were fitted to multivariate covariance matrices. The significance of A, C and E was tested by removing them sequentially in specific submodels and comparing these with the full model. Standard likelihood-ratio tests between models were used to assess the importance of each variance component (A, C or E) on the fit of the model, leading to a model in which the pattern of variance-covariance is explained by as few parameters as possible. Another statistic, the Akaike’s Information Criterion (AIC = χ2 − 2*df), was also used to determine the optimal model fitting, where a lower AIC indicates a more parsimonious and thus a better fitting model. All genetic model fitting was carried out with the Mx statistical program [20]. To avoid the potential influences of race/ethnicity and of a previous history of CHD, we repeated all the analyses after exclusion of Africa-Americans, and after exclusion of the subjects who developed CHD between 1990 and 2001. The results were virtually identical and are not reported.
3. Results
Table 1 shows the distribution of cardiovascular risk factors and the mean concentrations of inflammatory biomarkers. For all variables, there were no significant differences between MZ and DZ twins. BMI, physical activity and smoking were highly correlated with IL-6, CRP and fibrinogen (Table 2). Individuals with a history of CHD, hypertension or diabetes had higher levels of some inflammatory markers, but not others.
Table 1.
Cardiovascular risk factors and concentrations of inflammatory markers in MZ and DZ twins
| Risk Factors | Mean ± SD or Percent | |
|---|---|---|
| MZ (N=176) | DZ (N=156) | |
| Age, year | 54.0 ± 2.93 | 54.6 ± 2.91 |
| Body mass index, kg/m2 | 28.7 ± 3.70 | 29.5 ± 5.25 |
| Physical Activity | 7.6 ± 1.57 | 7.3 ± 1.58 |
| Total cholesterol, mg/dL | 190.7 ± 36.55 | 187.5 ± 38.02 |
| HDL-Cholesterol, mg/dL | 38.2 ± 9.42 | 40.4 ± 956 |
| LDL-Cholesterol, mg/dL | 128.9 ± 34.01 | 121.0 ± 32.93 |
| Glucose, mg/dL | 97.2 ± 14.99 | 103.1 ± 19.79 |
| Current smoker, n (%) | 30 (17.1) | 30 (19.2) |
| History of CHD (yes), n (%) | 12 (6.8) | 19 (12.2) |
| Hypertension (yes), n (%) | 79 (44.9) | 80 (51.3) |
| Diabetes (yes), n (%) | 13 (7.4) | 16 (10.3) |
| IL-6, pg/mL | 2.2 ± 1.60 | 2.1 ± 1.50 |
| sIL-6R | 29.4 ± 9.42 | 28.4 ± 8.19 |
| CRP | 1.9 ± 2.01 | 1.9 ± 1.79 |
| Fibrinogen | 323.7 ± 47.65 | 338.46 ± 57.07 |
MZ, monozygotic; DZ, dizygotic; SD, standard deviation; HDL, high-density lipoprotein; LDL, low-density lipoprotein; CHD, coronary heart disease; IL-6, interleukin-6; sIL-6R, soluble interleukin-6 receptor; CRP, C-reactive protein
Table 2.
Correlations between cardiovascular risk factors and the inflammatory markers
| Variables | Ln IL-6 | Ln sIL-6R | Ln CRP | Ln Fibrinogen |
|---|---|---|---|---|
| Agea | 0.007 | 0.16† | 0.05 | 0.04 |
| Body mass indexa | 0.13◊ | −0.01 | 0.33‡ | 0.17† |
| Physical activitya | −0.24‡ | 0.06 | −0.28‡ | −0.15◊ |
| Smoking statusb | 0.24‡ | −0.06 | 0.12◊ | 0.20‡ |
| History of CHDb | 0.21‡ | −0.13◊ | 0.10 | −0.07 |
| Hypertensionb | 0.08 | 0.13◊ | 0.12◊ | −0.02 |
| Diabetesb | 0.11◊ | −0.10 | 0.09 | 0.03 |
P < 0.05
P < 0.01
P < 0.001
Pearson correlation coefficients for continuous variables
Spearman correlation coefficients for categorical variables
Ln, natural logarithm; CHD, coronary heart disease; IL-6, interleukin-6; sIL-6R, soluble interleukin-6 receptor; CRP, C-reactive protein
3.1 Heritability Estimation and Univariate Model Fitting
The intraclass correlations in MZ twins were consistently higher than those in DZ twins for all inflammatory markers, indicating genetic influence (Table 3). This was confirmed by univariate analysis. The best fitting models for IL-6, CRP and fibrinogen included only genetic and unique environmental contributions. Only for sIL-6R did the best fitting model include both genetic and shared environment as well as the unique environment (Table 3). The heritability of IL-6 was estimated at 31% (95% CI: 13–46%). Genes accounted for 61% (95% CI: 47–72%) of the variance for CRP, and 52% (95% CI: 34–65%) of the variance for fibrinogen. For sIL-6R, the heritability was 49% (95% CI: 30–76%) and the common environmental effect was 44% (95% CI: 17–63%). After adjusting for age, BMI, smoking status, physical activity, history of CHD, hypertension, diabetes, and major depressive disorder, the heritability estimations for all inflammation variables were slightly decreased but the overall results remained similar (data not shown).
Table 3.
Intraclass correlations of MZ and DZ twins and univariate genetic model fitting for the inflammatory markers
| Inflammatory Markers | ICC MZ | ICC DZ | Univariate Model Fitting | a2 (95% CI) |
c2 (95% CI) |
e2 (95% CI) |
−2LL | AIC |
|---|---|---|---|---|---|---|---|---|
| Ln IL-6 | 0.31 | 0.22 | ||||||
| ACE | 0.17 (0-0.46) |
0.12 (0-0.39) |
0.71 (0.54-0.89) |
581.2 | −54.8 | |||
| AE |
0.31 (0.13-0.46) |
- |
0.69 (0.54-0.87) |
581.4 | −56.6 | |||
| CE | - | 0.26 (0.11-0.39) |
0.74 (0.61-0.89) |
501.5 | −56.4 | |||
| Ln sIL-6R | 0.94 | 0.64 | ||||||
| ACE |
0.49 (0.30-0.76) |
0.44 (0.17-0.63) |
0.07 (0.05-0.10) |
−97.4 | −737.4 | |||
| AE | 0.93 (0.90-0.95) |
- | 0.07 (0.05-0.10) |
−89.1 | −731.1 | |||
| CE | - | 0.82 (0.77-0.87) |
0.18 (0.13-0.23) |
−54.6 | −696.6 | |||
| Ln CRP | 0.65 | 0.25 | ||||||
| ACE | 0.61 (0.23-0.72) |
0 (0-0.33) |
0.39 (0.28-0.53) |
782.9 | 212.9 | |||
| AE |
0.61 (0.47-0.72) |
- |
0.39 (0.28-0.53) |
782.9 | 210.9 | |||
| CE | - | 0.48 (0.34-0.59) |
0.52 (0.41-0.66) |
791.7 | 219.7 | |||
| Ln Fibrinogen | 0.50 | 0.17 | ||||||
| ACE | 0.52 (0.16-0.65) |
0 (0-0.28) |
0.48 (0.35-0.66) |
−248.8 | −820.8 | |||
| AE |
0.52 (0.34-0.65) |
- |
0.48 (0.35-0.66) |
−248.8 | −822.8 | |||
| CE | - | 0.35 (0.21-0.49) |
0.65 (0.51-0.80) |
−242.2 | −816.2 |
a2 variance explained by additive genetics (heritability)
c2 variance explained by shared environment
e2 variance explained by unique environment
Best fitting model is in bold.
ICC, intraclass correlation; MZ, monozygotic; DZ, dizygotic; CI, confidence interval; Ln, natural logarithm; IL-6, interleukin-6; sIL-6R, soluble interleukin-6 receptor; CRP, Creactive protein; -2LL, -2 times log-likelihood; AIC, Akaike's information criterion
3.2 Phenotypic Correlations
The sIL-6R showed no relation with the other three inflammatory markers (r=−0.03, 0.04 and 0.02 for IL-6, CRP and fibrinogen respectively, P>0.5), and was removed from subsequent multivariate analyses. In contrast, strong correlations were observed between CRP and IL-6 (r=0.44, p<0.001), CRP and fibrinogen (r=0.42, P<0.001), and IL-6 and fibrinogen (r=0.26, P<0.001).
3.3 Multivariate Model Fitting
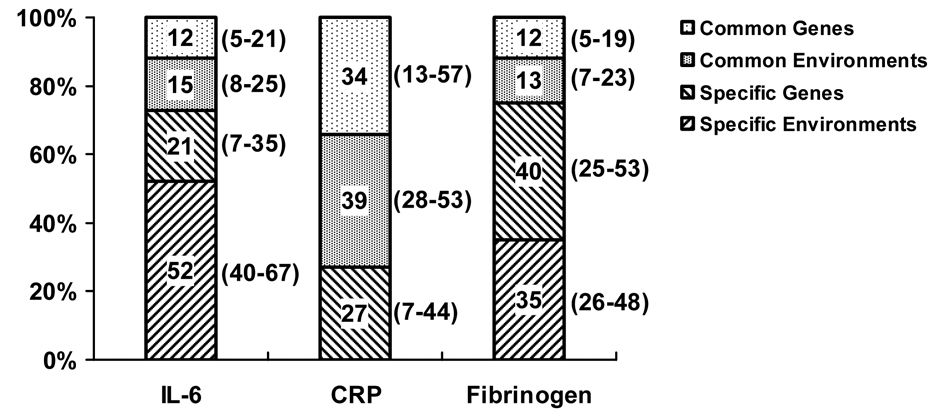
Consistent with the univariate genetic models, the multivariate genetic models consisted of additive genetic factors and unique environmental factors. Compared with the Cholesky model (AIC=−815) and independent pathway model (AIC=-816), the common pathway model (AIC=−817) was the best fitting multivariate model explaining the relationship between IL-6, CRP and fibrinogen, which consisting of one latent common factor influenced by genetics and unique environment. This latent common factor was characterized by high loading of CRP, moderate loading of IL-6, and small loading of fibrinogen. Apart from this common factor, there were also specific genetic and unique environmental influences on IL-6 and fibrinogen, except for CRP, for which no specific unique environmental influences were detected. The variance of each marker that can be explained by the common factor and specific factor is shown in Figure 2. As noted, most of the variance of IL-6 and fibrinogen was explained by the specific factors (73% and 75% respectively), while the common factor explained large part of the variance of CRP (73%). There was no specific unique environmental effect on CRP level. About 46% (95% CI: 21-64%) of the correlation between the inflammatory markers can be explained by genetic factors. The adjustment for covariates didn’t change the results.
Figure 2.
Variance of inflammatory markers explained by common factors and specific factors. For IL-6 and fibrinogen, about 25% of the variance is explained by common factors, and about 75% by specific, not shared factors. For CRP, however, most of the variance (75%) is due to common factors, i.e., factors that are shared with the other biomarkers. There is no specific unique environmental effect on CRP level. Values in brackets are 95% confidence intervals. IL-6: interleukin-6; CRP: C-reactive protein.
4. Discussion
By using a classic twin study and multivariate structural equation modeling, the present study provides evidence, for the first time, that a latent common genetic factor accounts for a substantial portion of the observed variability in a number of plasma inflammation biomarker levels, including IL-6, CRP and fibrinogen.
The 31% heritability for IL-6 is higher than a previously reported estimate of 17% in elderly twins [11], but lower than that reported for younger twins (58%) [21]. It is possible that the relative contribution of genetic and non-genetic influences on inflammation may change with age [21]. Another explanation is that the prevalence of conditions which may trigger inflammation, such as CHD and diabetes, increases with advancing age, which may contribute to a larger proportion of unshared unique environment in elderly twins. Our estimate of 61% heritability for CRP is somewhat higher than in two other twin studies [11, 12]; however, one of the studies was limited, again, to elderly twins [11] while the other only involved female twins [12]. Family studies report heritabilities for CRP between 0.25 and 0.40 [10, 13]. The 52% heritability estimate that we found for fibrinogen was comparable to or higher than the estimates from previous studies in twins and families (0.34–0.51) [22, 23]. To date, no information has been published on the heritability of the sIL-6R levels, but the 49% estimate in our study makes this an interesting candidate trait for future studies to identify quantitative trait loci (QTL) for inflammation.
We found a substantial correlation among IL-6, CRP and fibrinogen, a finding that was previously observed [5]. By using factor analysis, Koukkunen et al. extracted an “inflammation factor” combining IL-6, CRP and fibrinogen, and found that it was a strong predictor of future coronary events in patients with unstable angina [14]. Our finding that much of the inflammatory factor (46%) is due to common genetic factors is consistent with the one previous twin study [11]. In this earlier study of elderly twins the authors found a high genetic correlation between IL-6 and CRP, as well as between IL-6 and fibrinogen, but a small genetic correlation between CRP and fibrinogen.
The multivariate genetic model in our study showed that the environmental variance of CRP plasma level is caused by the common factor (Figure 2). In addition, most of the variance of CRP (73%) was explained by unique environmental or genetic factors common to all three inflammatory markers. The role of CRP in development of atherosclerosis has been well documented, and elevated CRP levels have been associated with the risk of future cardiovascular events [3, 4]. However, whether CRP plays a direct role in cardiovascular risk or is merely a marker of increased risk is still controversial [24, 25]. Our results suggest that CRP may not have a large unique role in atherosclerosis, but strongly covaries with other biomarkers; however, additional research is required.
What genes may underlie the genetic correlation among inflammatory biomarkers? Genetic polymorphisms in or around previously investigated candidate genes for inflammation only explain a small, insignificant part of the variance of inflammatory biomarker plasma levels.[11] The heritability of these biomarkers may therefore depend mostly upon enhancer elements further away from the genes or upon other genes with regulatory function. Recent interest has focused on heat shock protein transcription factors (HSFs),[26, 27] which control the level of inflammatory cytokines directly via regulation of cytokine genes [26]. For example, HSF1 suppresses the expression of proinflammatory cytokines TNF-α and IL-1B in vitro and in vivo in mice [26]. HSFs can also affect the inflammatory response indirectly through regulation of the transcription of heat shock proteins (HSPs) [28]. HSPs are an important family of endogenous proteins induced in response to environmental stresses [29]. Growing evidence has demonstrated that HSPs are involved in activation of the innate immune system and stimulate the expression of inflammatory markers through the CD14 receptor, Toll-like receptors 2 and 4, or calcium-dependent signaling [30–33]. Because of their physiological and immunological role, as well as their intercellular signaling capacity, HSF-HSP factors and HSP receptors should be considered as candidate genes responsible for the plasma level of inflammatory markers.
Our study has several limitations. First, the sample is derived from a twin registry of military veterans, where the members may be healthier than, or in other ways different from the general population of similarly aged Americans [34]. Second, our analyses included only male twins, and it is difficult to generalize to women. However, the heritabilities of inflammatory markers in our study are comparable to previous estimates in female twins [12]. In addition, the correlation among the inflammatory markers in our study was similar to that seen in both men and women in the general population [5].
In conclusion, a significant part of the variation in inflammatory marker levels is due to genetic influences. Furthermore, almost half of the shared variance among inflammatory markers is due to a common genetic factor. This factor likely plays a key role in the regulation of inflammation. The specific genetic mechanisms involved in controlling inflammatory response are unknown but our findings can be used to help guide gene discovery.
ACKNOWLEDGMENTS
The United States Department of Veterans Affairs provides financial support for the development and maintenance of the Vietnam Era Twin (VET) Registry. Numerous organizations have provided invaluable assistance in the conduct of this study, including: the Department of Defense; the National Personnel Records Center, National Archives and Records Administration; the Internal Revenue Service; the National Institutes of Health; the National Opinion Research Center; the National Research Council, National Academy of Sciences; the Institute for Survey Research, Temple University. Most importantly, the authors gratefully acknowledge the continued cooperation and participation of the members of the VET Registry and their families. Without their contribution this research would not have been possible. This study was supported by K24HL077506, R01 HL68630 and R01 AG026255 from the National Institutes of Health; by the Emory University General Clinical Research Center MO1-RR00039 and by grant 0245115N from the American Heart Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 2.Danesh J, Collins R, Appleby P, et al. Association of fibrinogen, C-reactive protein, albumin, or leukocyte count with coronary heart disease: meta-analyses of prospective studies. Jama. 1998;279:1477–1482. doi: 10.1001/jama.279.18.1477. [DOI] [PubMed] [Google Scholar]
- 3.Lagrand WK, Visser CA, Hermens WT, et al. C-reactive protein as a cardiovascular risk factor: more than an epiphenomenon? Circulation. 1999;100:96–102. doi: 10.1161/01.cir.100.1.96. [DOI] [PubMed] [Google Scholar]
- 4.Danesh J, Wheeler JG, Hirschfield GM, et al. C-reactive protein and other circulating markers of inflammation in the prediction of coronary heart disease. N Engl J Med. 2004;350:1387–1397. doi: 10.1056/NEJMoa032804. [DOI] [PubMed] [Google Scholar]
- 5.Luc G, Bard JM, Juhan-Vague I, et al. C-reactive protein, interleukin-6, and fibrinogen as predictors of coronary heart disease: the PRIME Study. Arterioscler Thromb Vasc Biol. 2003;23:1255–1261. doi: 10.1161/01.ATV.0000079512.66448.1D. [DOI] [PubMed] [Google Scholar]
- 6.Jones SA, Rose-John S. The role of soluble receptors in cytokine biology: the agonistic properties of the sIL-6R/IL-6 complex. Biochim Biophys Acta. 2002;1592:251–263. doi: 10.1016/s0167-4889(02)00319-1. [DOI] [PubMed] [Google Scholar]
- 7.Scheller J, Rose-John S. Interleukin-6 and its receptor: from bench to bedside. Med Microbiol Immunol (Berl) 2006;195:173–183. doi: 10.1007/s00430-006-0019-9. [DOI] [PubMed] [Google Scholar]
- 8.Ueda K, Takahashi M, Ozawa K, et al. Decreased soluble interleukin-6 receptor in patients with acute myocardial infarction. Am Heart J. 1999;138:908–915. doi: 10.1016/s0002-8703(99)70016-5. [DOI] [PubMed] [Google Scholar]
- 9.Rose-John S, Scheller J, Elson G, et al. Interleukin-6 biology is coordinated by membrane-bound and soluble receptors: role in inflammation and cancer. J Leukoc Biol. 2006 doi: 10.1189/jlb.1105674. [DOI] [PubMed] [Google Scholar]
- 10.Pankow JS, Folsom AR, Cushman M, et al. Familial and genetic determinants of systemic markers of inflammation: the NHLBI family heart study. Atherosclerosis. 2001;154:681–689. doi: 10.1016/s0021-9150(00)00586-4. [DOI] [PubMed] [Google Scholar]
- 11.de Maat MP, Bladbjerg EM, Hjelmborg JB, et al. Genetic influence on inflammation variables in the elderly. Arterioscler Thromb Vasc Biol. 2004;24:2168–2173. doi: 10.1161/01.ATV.0000143856.01669.e7. [DOI] [PubMed] [Google Scholar]
- 12.MacGregor AJ, Gallimore JR, Spector TD, et al. Genetic effects on baseline values of C-reactive protein and serum amyloid a protein: a comparison of monozygotic and dizygotic twins. Clin Chem. 2004;50:130–134. doi: 10.1373/clinchem.2003.028258. [DOI] [PubMed] [Google Scholar]
- 13.Dupuis J, Larson MG, Vasan RS, et al. Genome scan of systemic biomarkers of vascular inflammation in the Framingham Heart Study: evidence for susceptibility loci on 1q. Atherosclerosis. 2005;182:307–314. doi: 10.1016/j.atherosclerosis.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Koukkunen H, Penttila K, Kemppainen A, et al. C-reactive protein, fibrinogen, interleukin-6 and tumour necrosis factor-alpha in the prognostic classification of unstable angina pectoris. Ann Med. 2001;33:37–47. doi: 10.3109/07853890109002058. [DOI] [PubMed] [Google Scholar]
- 15.Boomsma D, Busjahn A, Peltonen L. Classical twin studies and beyond. Nat Rev Genet. 2002;3:872–882. doi: 10.1038/nrg932. [DOI] [PubMed] [Google Scholar]
- 16.Goldberg J, Curran B, Vitek ME, et al. The Vietnam Era Twin Registry. Twin Res. 2002;5:476–481. doi: 10.1375/136905202320906318. [DOI] [PubMed] [Google Scholar]
- 17.Scherrer JF, Xianx H, Bucholz KK, et al. A twin study of depression symptoms, hypertension, and heart disease in middle-aged men. Psychosom Med. 2003;65:548–557. doi: 10.1097/01.psy.0000077507.29863.cb. [DOI] [PubMed] [Google Scholar]
- 18.Richardson MT, Ainsworth BE, Wu HC, et al. Ability of the Atherosclerosis Risk in Communities (ARIC)/Baecke Questionnaire to assess leisure-time physical activity. Int J Epidemiol. 1995;24:685–693. doi: 10.1093/ije/24.4.685. [DOI] [PubMed] [Google Scholar]
- 19.Neale MC, Cardon LR. Methodologies for genetic studies of twins and families. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1992. [Google Scholar]
- 20.Neale MC, Boker SM, Xie G, et al. Mx: Statistical Modeling. Richmond, VA: Department of Psychiatry, Virginia Commonwealth University; 1999. [Google Scholar]
- 21.Grunnet L, Poulsen P, Klarlund Pedersen B, et al. Plasma cytokine levels in young and elderly twins: genes versus environment and relation to in vivo insulin action. Diabetologia. 2006;49:343–350. doi: 10.1007/s00125-005-0080-8. [DOI] [PubMed] [Google Scholar]
- 22.Hamsten A, Iselius L, de Faire U, et al. Genetic and cultural inheritance of plasma fibrinogen concentration. Lancet. 1987;2:988–991. doi: 10.1016/s0140-6736(87)92557-8. [DOI] [PubMed] [Google Scholar]
- 23.Bladbjerg EM, de Maat MP, Christensen K, et al. Genetic influence on thrombotic risk markers in the elderly--a Danish twin study. J Thromb Haemost. 2006;4:599–607. doi: 10.1111/j.1538-7836.2005.01778.x. [DOI] [PubMed] [Google Scholar]
- 24.Scirica BM, Morrow DA. Is C-reactive protein an innocent bystander or proatherogenic culprit? The verdict is still out. Circulation. 2006;113:2128–2134. doi: 10.1161/CIRCULATIONAHA.105.611350. discussion 51. [DOI] [PubMed] [Google Scholar]
- 25.Verma S, Devaraj S, Jialal I. Is C-reactive protein an innocent bystander or proatherogenic culprit? C-reactive protein promotes atherothrombosis. Circulation. 2006;113:2135–2150. discussion 50. [PubMed] [Google Scholar]
- 26.Xiao X, Zuo X, Davis AA, et al. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. Embo J. 1999;18:5943–5952. doi: 10.1093/emboj/18.21.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wick G, Knoflach M, Xu Q. Autoimmune and inflammatory mechanisms in atherosclerosis. Annu Rev Immunol. 2004;22:361–403. doi: 10.1146/annurev.immunol.22.012703.104644. [DOI] [PubMed] [Google Scholar]
- 28.Jurivich DA, Sistonen L, Sarge KD, et al. Arachidonate is a potent modulator of human heat shock gene transcription. Proc Natl Acad Sci U S A. 1994;91:2280–2284. doi: 10.1073/pnas.91.6.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Q. Role of heat shock proteins in atherosclerosis. Arterioscler Thromb Vasc Biol. 2002;22:1547–1559. doi: 10.1161/01.atv.0000029720.59649.50. [DOI] [PubMed] [Google Scholar]
- 30.Asea A, Kraeft SK, Kurt-Jones EA, et al. HSP70 stimulates cytokine production through a CD14-dependant pathway, demonstrating its dual role as a chaperone and cytokine. Nat Med. 2000;6:435–442. doi: 10.1038/74697. [DOI] [PubMed] [Google Scholar]
- 31.Dybdahl B, Wahba A, Lien E, et al. Inflammatory response after open heart surgery: release of heat-shock protein 70 and signaling through toll-like receptor-4. Circulation. 2002;105:685–690. doi: 10.1161/hc0602.103617. [DOI] [PubMed] [Google Scholar]
- 32.Pockley AG. Heat shock proteins as regulators of the immune response. Lancet. 2003;362:469–476. doi: 10.1016/S0140-6736(03)14075-5. [DOI] [PubMed] [Google Scholar]
- 33.Satoh M, Shimoda Y, Akatsu T, et al. Elevated circulating levels of heat shock protein 70 are related to systemic inflammatory reaction through monocyte Toll signal in patients with heart failure after acute myocardial infarction. Eur J Heart Fail. 2006;8:810–815. doi: 10.1016/j.ejheart.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Henderson WG, Eisen S, Goldberg J, et al. The Vietnam Era Twin Registry: a resource for medical research. Public Health Rep. 1990;105:368–373. [PMC free article] [PubMed] [Google Scholar]