Abstract
Methadone is a mu-opioid receptor agonist used for treating opiate dependence. The range of effective methadone doses is broad. Part of the large inter-individual variability in efficacy may be accounted for by genetic factors. Methadone is a substrate of the transporter P-glycoprotein (P-gp) 170 that is encoded by the ABCB1 (MDR1) gene. Thus, P-gp variants may play a role in methadone absorption and distribution. We assessed the association between ABCB1 polymorphisms and methadone dose requirements in 98 methadone-maintained patients. The stabilizing methadone doses were normally distributed with a mean and median dose of 160 mg/day (range 30–280 mg/day). Statistical analysis showed significant difference in genotype frequencies between the ‘higher’ (>150 mg/day) and ‘lower’ (≤150 mg/day) methadone dose groups for single nucleotide polymorphism (SNP) 1236C>T (rs1128503) (experiment-wise P = 0.0325). Furthermore, individuals bearing the 3-locus genotype pattern TT-TT-TT (rs1045642, rs2032582 and rs1128503) have an approximately 5-fold chance of requiring the ‘higher’ methadone dose, while individuals heterozygous for these three SNPs have an approximately 3-fold chance of stabilizing at the ‘lower’ methadone dose (point-wise P-value = 0.026). These data suggest that specific ABCB1 variants may have clinical relevance by influencing the methadone dose required to prevent withdrawal symptoms and relapse in this population.
INTRODUCTION
Opiate addiction is a chronic relapsing disorder that is treated world-wide with methadone (1). Successful treatment relies, in part, on individual methadone dose optimization to block heroin effect, reduce drug craving, prevent relapse and adverse reaction. Part of the large inter-individual variability in drug response may be accounted for by genetic factors that can determine drug absorption, distribution, metabolism and action. There are several factors involved in drug response, including metabolizing enzymes, drug targets, regulatory factors and drug transporters. Although, initially, genetics variations in metabolizing enzymes were thought to play the major role, it is now clear that genetic variations in drug transporters also influence drug response (2).
Methadone is a synthetic opioid that is generally administered as a racemic mixture of (R)- and (S)-methadone enantiomers, yet the (R)-methadone accounts for the opioid effects (3). Methadone is a mu-opioid receptor agonist and a weak N-methyl-d-aspartic acid (NMDA) receptor antagonist. There is some inter-individual variation in the pharmacokinetics of methadone that may partly be explained by variability in cytochrome P450 activity. Methadone is rapidly absorbed with peak plasma concentrations 2–4 h after oral administration (4). The methadone bioavailability ranges from 70 to 90%. Methadone is metabolized primarily in the liver and is slowly released back into plasma or bile and is excreted by fecal or urinary route (5). About 10–15% of methadone is unbound and the rest is bound to alpha 1-acid glycoprotein, albumin and globulin. Biotransformation of methadone includes N-demethylation to pyrrolidine, an inactive major metabolite, and a second N-demethylation to two inactive pyrroline metabolites (6). The major methadone-metabolizing enzymes are cytochrome P450 CYP3A4, CYP2D6 and CYP2B6. The elimination half-life for the racemic mixture in humans ranges from 16 to 28 h (4).
Methadone is a substrate of P-glycoprotein (P-gp) 170 as studies in rodents and humans have shown (7–9). P-gp is a member of the subfamily B of the ATP-binding cassette (ABC) superfamily. It is a trans-membrane protein of 1280 amino acids that is composed of two homologous sequences, each containing six trans-membrane domains and an ATP-binding domain (10). P-gp functions as an efflux pump transporting drugs from the intra-cellular to the extra-cellular domain and has a significant role in drug pharmacokinetics. P-gp is expressed in tissues with barrier function, including the epithelia of the liver, kidney, intestine and the endothelial cells lining the brain capillaries (11). It has a broad range of substrates that are also often substrates of the cytochrome P450 drug-metabolizing enzymes.
P-gp is encoded by the ABCB1 (MDR1) gene, spanning approximately 200 kb on chromosome 7p21, with 28 exons and 4.5 kb transcript. The gene is highly polymorphic with at least 38 single nucleotide polymorphisms (SNPs) in the coding region (NCBI). A significant variation in allele frequencies among different populations has been reported (12). A few common ABCB1 variants have been shown to be associated with P-gp expression, drug response and disease susceptibility (2,10,12–14). There is considerable controversy about the functional significance of various ABCB1 polymorphisms, which could be owing to emphasis on single SNPs rather than haplotypes, ethnic-specific allele frequencies (10,12), tissue-specific expression (15), the number of haplotype combinations (16,17) and the effect of other transporters or metabolizing enzymes. Most studies focused on the synonymous SNP 3435C>T (rs1045642, exon 26). Homozygosity to the T allele (TT) showed lower in vivo duodenal P-gp expression compared with CT or CC (13). Using allele-specific mRNA expression, Wang et al. (18) performed quantitative analysis of allelic expression (SNaPshot) in human liver samples, and found that the 3435C allele expresses significantly higher mRNA levels than the 3435T allele. Altered substrate specificity, as a result of distorted conformation, but similar mRNA and protein levels, was found to be caused by this variant in human and monkey cell lines (19).
The SNPs 1236C>T (rs1128503), 2677G>T (rs2032582) and 3435C>T (rs1045642) are the most common variants in the coding sequence. These SNPs are reported to be in strong linkage disequilibrium (LD) accounting for two major haplotypes, TTT and CGC (12,16). Variants 1236T, 2677T and 3435T were reported to minimize P-gp activity in vitro in a substrate-specific manner (20). A difference in the ABCB1 haplotype profile was found between different ethnic groups and low haplotype diversity was observed in Caucasians (21). A haplotype containing the haplotype subset 1236T-2677T-3435T is highly represented among non-African populations, while a haplotype containing the sub-haplotype 1236C-2677G-3435C is highly represented in the African-American population (21). Two studies reported the frequencies of ABCB1 SNPs in different Jewish ethnic groups. One study (22) demonstrated that the Ashkenazi Jewish (e.g. Eastern European) population is comparable with Caucasians in the frequency of the three common SNPs. The second study (23) reported that the frequency of the 3435T allele in four Jewish populations: Ashkenazi, Yemenite, North African and Mediterranean (from Bulgaria, Greece and Turkey) is quite similar, and is intermediate between that of the previously reported white and West African populations (24), while the frequency of the 3435T allele was higher in near Eastern Jews (from Iraq, Iran and Buchara).
A haplotype containing the promoter SNP 129T>C (rs3213619) was found to be associated with high level of P-gp expression and an increase in transport activity, independent of 3435C>T (15). A significant association was shown between common ABCB1 haplotypes and ulcerative colitis in Caucasians. This association was dependent on the intronic variant rs3789243 and independent of 3435C>T (14).
In this study, we examined the association of alleles, genotypes and multi-locus genotype patterns of nine SNPs spanning the ABCB1 gene with stabilizing dose of methadone in former severe heroin-dependent Jewish patients from Israel.
RESULTS
Ninety-eight former severe heroin-dependent patients from an Israeli methadone maintenance treatment (MMT) clinic participated in this study. All subjects met strict criteria for opiate dependence and were in a stable drug-free phase of methadone treatment (see Materials and Methods). The stabilizing methadone doses were normally distributed with a mean and median dose of 160 mg/day (range 30–280 mg/day).
ABCB1, single nucleotide polymorphism alleles, genotypes
Nine ABCB1 common SNPs (rs1045642, rs6949448, rs2235067, rs2032583, rs2032582, rs1922242, rs1128503, rs2520464 and rs3789243) were genotyped (Table 1). These SNPs were selected for stage I of the study that was designed as an initial effort. SNP rs2032582 (2677G/T/A) is non-synonymous (Ser893Ala/Thr), SNPs rs1045642 (3435C>T) and rs1128503 (1236C>T) are synonymous and the other six SNPs are intronic. rs2032582 (2677G/T/A) is tri-allelic and was genotyped by direct sequencing of a flanking PCR product. The allele frequencies for each SNP are listed in Table 1. Observed genotype distributions were consistent with Hardy–Weinberg equilibrium (HWE) (P > 0.3, Table 1).
Table 1.
ABCB1 single nucleotide polymorphisms (SNPs) analyzed in this study
| No. | SNP | Alleles | Positiona | Amino acid change | Exon/intron (i) | MAFb | Map positionc | HWE P-valued |
|---|---|---|---|---|---|---|---|---|
| 1 | rs1045642 | C/T | 3435 | Ile1145Ile | 26 | 0.45 | 86976581 | 0.8 |
| 2 | rs6949448 | A/G | i25 | 0.43 | 86979750 | 0.7 | ||
| 3 | rs2235067 | G/A | i22 | 0.07 | 86987858 | 0.8 | ||
| 4 | rs2032583 | C/T | i21 | 0.08 | 86998497 | 0.8 | ||
| 5 | rs2032582 | G/T (A) | 2677 | Ser893Ala (Thr) | 21 | 0.43 (0.02) | 86998554 | 0.7 |
| 6 | rs1922242 | A/T | i16 | 0.50 | 87011603 | 0.3 | ||
| 7 | rs1128503 | C/T | 1236 | Gly412Gly | 12 | 0.42 | 87017537 | 0.6 |
| 8 | rs2520464 | A/G | i4 | 0.45 | 87039022 | 0.6 | ||
| 9 | rs3789243 | C/T | i3 | 0.49 | 87058822 | 0.4 |
HWE, Hardy–Weinberg equilibrium.
aReference sequence NM_000927.
bMAF, minor allele frequencies in this sample.
cNCBI Build 36.2.
dDeviation from HWE.
Linkage disequilibrium
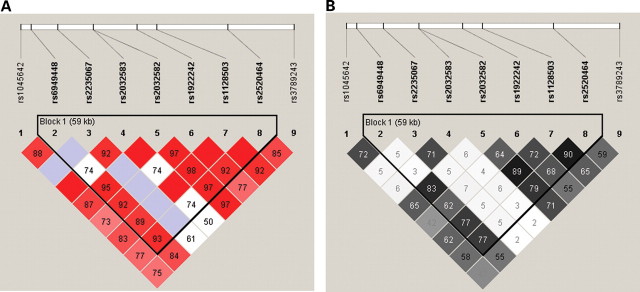
Linkage disequilibrium analysis revealed that all nine SNPs are correlated in this population and SNP nos 2–8 form a haplotype block of 59 kb from intron 4 to intron 25 (Fig. 1) (25). The highest LD value (r2 = 0.90; D′ = 1.00) corresponds to SNPs 1236 (rs1128503)–rs2520464. Also, strong association is found between SNPs 1236 (rs1128503) and 2677 (rs2032582) (r2 = 0.89; D′ = 0.98). Lower LD value was found between the SNPs 2677 (rs2032582) and 3435 (rs1045642) (r2 = 0.65; D′ = 0.87) and between SNPs 1236 (rs1128503) and 3435 (rs1045642) (r2 = 0.62; D′ = 0.83).
Figure 1.
Linkage disequilibrium plots of nine ABCB1 single nucleotide polymorphisms. (A) D′. (B) r2. Values (100×) corresponding to each single nucleotide polymorphism pair are displayed inside the squares. (A) The degree of red corresponds with the D′ score (0<D′<1); blue represents high D′ value with low significance. (B) The degree of shade of gray corresponds with the r2 score (0<r2<1).
Association of specific single nucleotide polymorphisms and stabilizing methadone dose
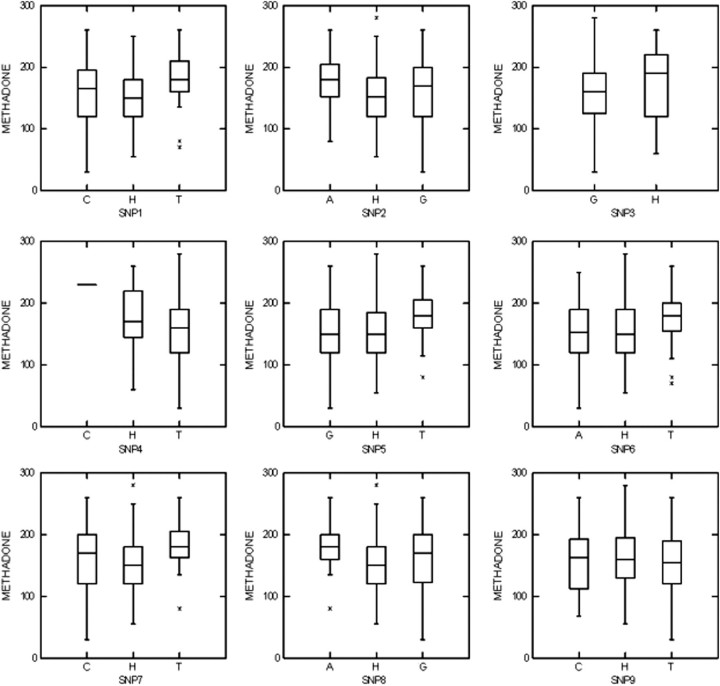
Density plots of genotypes versus methadone doses suggest that subjects with genotype ‘TT’ in SNPs rs1045642, rs2032582 and rs1128503 (nos 1, 5 and 7) and genotype ‘AA’ in SNP rs2520464 (no. 8) require relatively higher methadone doses (Fig. 2). One-way analysis of variance (ANOVA) for the three genotypes of each SNP, using methadone doses as a dependent quantitative trait, was not significant (P > 0.1).
Figure 2.
Density plots of methadone dose (mg/day) by ABCB1 genotypes. ‘A’ represents homozygous genotype AA; ‘C’ represents homozygous genotype CC; ‘G’ represents homozygous genotype GG; ‘T’ represents homozygous genotype TT; ‘H’ represents heterozygous genotype; asterisk (*) represents outliers. No homozygotes for the variant ‘A’ allele of SNP no. 3 were found.
Subjects were then dichotomized into ‘higher’ (>150 mg) and ‘lower’ (≤150 mg) groups, according to their stabilized daily dose of methadone. Comparison of allele frequencies revealed no significant difference between the two groups for each of the SNPs analyzed (P > 0.13; Table 2). Statistical analysis using contingency tables showed significant differences in genotype frequencies between the two groups, for SNPs rs1128503 and rs2520464, and a nearly significant difference for SNP rs1045642 (point-wise P = 0.007, 0.026 and 0.054, respectively; Table 3). The significance levels were adjusted for multiple testing via 10,000 permutation samples. The only genotype frequencies difference that remains significant after this correction was for SNP rs1128503 (experiment-wise P = 0.0325; Table 3). Homozygosity for the ‘T’ allele in SNP rs1128503 confers a 6.66-fold chance of stabilizing at a higher methadone dose than heterozygosity or homozygosity for the reference ‘C’ allele [odds ratio (OR) = 6.66, 95% confidence interval (CI) (1.41–31.36)].
Table 2.
Tests for association between allele frequencies of ABCB1 single nucleotide polymorphisms (SNPs) and methadone dose
| No. | SNP | Alleles | ‘Lower’ dose na | ‘Higher’ dose n | Point-wise P-value | ||
|---|---|---|---|---|---|---|---|
| Allele 1b | Allele 2b | Allele 1 | Allele 2 | ||||
| 1 | rs1045642 | C/T | 52 | 36 | 55 | 51 | 0.38 |
| 2 | rs6949448 | A/G | 34 | 54 | 50 | 58 | 0.31 |
| 3 | rs2235067 | G/A | 4 | 84 | 9 | 99 | 0.39 |
| 4 | rs2032583 | C/T | 4 | 84 | 11 | 97 | 0.18 |
| 5 | rs2032582 | G/T | 56 | 32 | 52 | 48 | 0.14 |
| 6 | rs1922242 | A/T | 49 | 39 | 48 | 60 | 0.15 |
| 7 | rs1128503 | C/T | 54 | 34 | 59 | 49 | 0.38 |
| 8 | rs2520464 | A/G | 35 | 51 | 52 | 56 | 0.31 |
| 9 | rs3789243 | C/T | 41 | 47 | 55 | 53 | 0.57 |
aNumber of alleles.
b‘1’ represents the first allele and ‘2’ represents the second allele.
Table 3.
Tests for association between genotype frequencies of ABCB1 single nucleotide polymorphisms (SNPs) and methadone dose
| No. | SNPs | Alleles | ‘Lower dose’ na | ‘Higher dose’ n | Point-wise P-value 11/12/22 | χ2 (2 degree of freedom) | Experiment-wise P-value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 11b | 12b | 22b | 11 | 12 | 22 | ||||||
| 1 | rs1045642 | C/T | 12 | 28 | 4 | 16 | 23 | 14 | 0.054 | 6.1 | 0.1977 |
| 2 | rs6949448 | A/G | 4 | 14 | 26 | 12 | 16 | 26 | 0.212 | nac | na |
| 3 | rs2235067 | G/A | 40 | 4 | 0 | 45 | 9 | 0 | 0.373 | na | na |
| 4 | rs2032583 | C/T | 0 | 4 | 40 | 1 | 9 | 44 | 0.373 | na | na |
| 5 | rs2032582 | G/T | 15 | 26 | 3 | 14 | 24 | 12 | 0.086 | na | na |
| 6 | rs1922242 | A/T | 10 | 29 | 5 | 10 | 28 | 16 | 0.094 | na | na |
| 7 | rs1128503 | C/T | 12 | 30 | 2 | 18 | 23 | 13 | 0.007 | 10.1 | 0.0325 |
| 8 | rs2520464 | A/G | 3 | 11 | 29 | 14 | 16 | 24 | 0.026 | 7.9 | 0.0892 |
| 9 | rs3789243 | C/T | 8 | 25 | 11 | 12 | 31 | 11 | 0.788 | na | na |
aNumber of subjects.
b‘11’ represents homozygote for the first allele, ‘12’ represents heterozygote and ‘22’ represents homozygote for the second allele.
c‘na’ represents not applicable.
The proportions of different sub-ethnicities in the ‘lower’ and ‘higher’ methadone dose groups were comparable. The distribution of different sub-ethnic groups in each of the three genotype categories of SNP rs1128503 was not significantly different.
Multi-locus genotype patterns
Multi-locus genotype patterns (MLGPs), were constructed from the three common SNPs (nos 1, 5 and 7—rs1045642, rs2032582 and rs1128503; respectively; Table 4) and the set of nine SNPs that includes these three common SNPs (Table 5). There were five 3-locus patterns with frequency higher than 5%. The three major patterns were ‘HHH’ (e.g. heterozygous for the three SNPs, CT-GT-CT, 40.8%), ‘CGC’ (e.g. CC-GG-CC, homozygosity for the reference allele; 20.4%) and ‘TTT’ (TT-TT-TT, homozygosity for the variant allele; 13.3%). The three major 9-locus patterns were ‘HHGTHHHHH’ (31.6%), ‘CGGTGACGT’ (9.2%) and ‘TAGTTTTAC’ (13.3%), (SNP order nos 1–9; Table 5), corresponding to the three major 3-locus patterns, respectively. Association analyses of the 3-locus MLGPs showed evidence of association with point-wise overall P-value of 0.026 (Table 4). Comparison of genotype pattern distributions revealed that individuals heterozygous for the three SNPs (‘HHH’) have an approximately 3-fold chance of stabilizing at a relatively ‘lower’ methadone dose (≤150 mg/day) than individuals with any other genotype pattern [OR = 2.85, 95% CI (1.15, 7.16), point-wise P = 0.0144; Table 4]. Individuals homozygous for the variant allele (‘TTT’) have an approximately 5-fold (1/0.19) chance of requiring a relatively ‘higher’ methadone dose (>150 mg/day) than individuals with any other genotype pattern [OR = 0.19, 95% CI (0.03, 0.98, point-wise P = 0.0335; Table 4]. All individuals with the coding sequence 3-locus pattern (‘TTT’, SNPs nos 1, 5 and 7) also have the genomic 9-locus pattern ‘TAGTTTTAC’ (SNP nos 1–9). Individuals with the 9-locus MLGP ‘TAGTTTTAC’ might require a relatively ‘higher’ methadone dose [OR = 0.19, 95% CI (0.03, 0.98), P = 0.0336, Table 5]. After correction for multiple testing, the P-values obtained in the MLGP analysis does not reach the significance level of 0.05.
Table 4.
Tests for association between ABCB1 3-locus genotype patterns (MLGPs) and methadone dose
| MLGPs SNP nos 1, 5, 7 | Code | ‘Lower dose’ | ‘Higher dose’ | OR (95% CI) | Point-wise P-value | ||
|---|---|---|---|---|---|---|---|
| na | % | n | % | ||||
| CT-GT-CT | ‘HHH’b | 24 | 54.5 | 16 | 29.6 | 2.85 (1.15, 7.16) | 0.014 |
| TT-TT-TT | ‘TTT’ | 2 | 4.5 | 11 | 20.4 | 0.19 (0.03, 0.98) | 0.034 |
| CC-GG-CC | ‘CGC’ | 10 | 22.7 | 10 | 18.5 | 1.29 (0.44, 3.85) | 0.624 |
| CT-GG-CC | ‘HGC’ | 2 | 4.5 | 4 | 7.4 | 0.60 (0.07, 4.06) | 0.688 |
| TT-GT-CT | ‘THH’ | 2 | 4.5 | 3 | 5.5 | 0.81 (0.09, 6.36) | 1.000 |
| Othersc | 3 | 6.8 | 11 | 20.4 | 0.29 (0.06, 1.22) | 0.081 | |
| Total | 44 | 100 | 54 | 100 | |||
χ2 = 12.74; df = 5; overall P = 0.026; experiment-wise P = 0.13. SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
an denotes number of individuals with the pattern.
b‘H’ denotes heterozygote.
cMLGPs with frequency <0.05 were combined under ‘Others’.
Table 5.
Test for association between ABCB1 9-locus genotype patterns and methadone dose
| MLGPs (order nos 1–9) | ‘Lower dose’ | ‘Higher dose’ | OR (95% CI) | Point-wise P-value | ||
|---|---|---|---|---|---|---|
| n | % | n | % | |||
| HHGTHHHHH | 17 | 38.6 | 14 | 25.9 | 1.80 (0.70, 4.65) | 0.1968 |
| CGGTGACGT | 5 | 11.4 | 4 | 7.4 | 1.60 (0.34, 7.73) | 0.7272 |
| TAGTTTTAC | 2 | 4.5 | 11 | 20.4 | 0.19 (0.03, 0.98) | 0.0336 |
| Others | 20 | 45.5 | 25 | 46.3 | ||
| Total | 44 | 100 | 54 | 100 | ||
χ2 = 6.80; df = 3; overall P = 0.0787. MLGP, multi-locus genotype pattern; OR, odds ratio; CI, confidence interval.
DISCUSSION
Successful methadone treatment for opiate dependence relies to a certain extent on dosage optimization. P-gp acts as a gatekeeper at the blood–brain barrier and the gastrointestinal tract among other organs, and may contribute to the regulation of methadone distribution. Genetic variability in the ABCB1 gene may influence methadone distribution by altering P-gp expression and function. Numerous studies have been conducted to evaluate the clinical relevance of ABCB1 variants, but presented contradicting results. The present study was designed to examine the influence of specific polymorphisms of the ABCB1 gene on the methadone dose required for stabilization.
The nine SNPs analyzed (Table 1) were selected based on reported evidence for functionality, relatively high frequency, location in the gene and LD data. MLGP analysis revealed the presence of five major 3-locus genotype patterns (frequency >5%) and also three major 9-locus genotype patterns, indicating a strong LD and low diversity in this group. This phenomenon has been observed in other populations and might be a sign of positive selection when variants connected with better adaptation are expected to increase rapidly in frequency (21,26). We showed that, in this population, homozygosity for the ‘T’ allele of the common SNP 1236C>T (rs1128503) confers an approximately 7-fold chance of requiring more than 150 mg of methadone, daily, for stabilization (experiment-wise P = 0.0325). In addition, a 3-locus genotype pattern, ‘TT-TT-TT’ (rs1045642, rs2032582 and rs1128503), that occurs in 13.3% of the sample, is possibly associated with a ‘higher’ methadone dose requirement.
This finding is intriguing from various aspects; SNP 1236C>T (rs1128503) is a synonymous variant that does not result in change in the protein sequence. It is located at exon 12, in one of the intra-cellular loops of the protein, adjacent to an ATP-binding/utilization domain. 1236C>T could affect translation regulation, RNA stability or other molecular mechanisms. It may also have an indirect effect by being linked to a causal variant. A number of studies reported strong LD between 1236C>T and 2677G>T as well as 3435C>T. In this study population, a haplotype block of 59 kb (from intron 4 to intron 25) was identified, which includes 1236C>T and 2677G>T. A lower LD (r2 = 0.63; D′ = 0.83) was found between 1236C>T and 3435C>T. The allele frequencies of 1236C>T differ between various populations with major difference among Caucasians, Asians and Africans (10,12,13,16). A recent report (27) showed that individuals homozygous for the ‘T’ allele of 1236C>T (as well as 2677G>T and 3435C>T) had higher clearance of the tyrosine kinase inhibitor, imatinib, that is used to treat chronic myeloid leukemia or gastrointestinal stromal tumors. Xing et al. (28) suggested that individuals homozygous for the ‘T’ allele of 1236C>T have greater improvement after treatment with the P-gp substrate antipsychotic drug, risperidone, independent of 2677G>T and 3435C>T. An in vitro study using the P-gp substrate rhodamine 123, in a mammalian epithelial cell line (LLC-PK1), concluded that the three SNPS (1236C>T, 2677G>T and 3435C>T), individually or as a haplotype, minimize P-gp activity in a substrate-dependent manner (20).
A number of studies reported that the 3435T allele associates with lower P-gp expression and activity (13) as well as reduced mRNA stability (18). Accordingly, one would expect that homozygosity to the 3435T allele would similarly reduce P-gp expression in the blood–brain barrier, which could lead to lower efflux function, increase methadone brain entry, and hence require an overall lower methadone dose. Our finding seems to contradict this assumption, but there is no consensus about the functionality of the 3435T variant and a few studies reported opposite data (29,30). It was also shown that haplotype analysis may be more effective than the analysis of individual SNPs and the 3435T allele is positioned on at least two major distinct 3-locus haplotypes.
The various organs involved in methadone exposure, in which P-gp is expressed, should also be taken into account as the same variant may simultaneously affect different processes, such as absorption and elimination, in addition to brain distribution. It has been suggested that P-gp variants will have little impact on net methadone intestinal absorption because of the fact that even relatively low doses of methadone (80–150 mg/day), as used in methadone treatment of opiate addiction, would reach a sufficient concentration to saturate the transporters (2,31).
It is hard to predict the extent of the relative contribution of ABCB1 genetic variability to the inter-individual variation in methadone dose requirement. There are a number of confounding factors that have to be considered when interpreting this data, including genetic variation in the metabolizing enzymes, methadone clearance, increased opiate tolerance by chronic exposure, psychiatric co-morbidity, drug co-dependence, delivery methods, sex, age and ethnicity, as well as the actual target of the medication and the downstream signal transduction mechanism. For instance, studies have found that patients with chronic pain, benzodiazepines or alcohol abuse, and patients treated with medication known to alter the biotransformation of methadone, such as rifampin or phenytoin, need to be prescribed higher doses of methadone (32–36).
Our finding cannot be directly compared with those of a similar study by Coller et al. (37) in Australia. Although three out of the five SNPs in their study are similar to the SNPs we studied, it seems that about half of the patients were in MMT treatment <2 months, a time when full stabilization would not be expected. In addition, the methadone doses reported were significantly lower than those in our study (range 15–110 mg/day, mean 59.2 ± 24.8).
Many clinicians hesitate to use very high methadone doses (above 200 mg/day) because of the possible adverse effects of prolonged QTc intervals (38,39). In Israel, permission is still required for the administration of any methadone dose above 120 mg/day. In the USA, there used to be a similar dose limit, which later was increased to 150 mg/day. Now there is no federal limit on methadone doses, but many states have a limit ranging from 120 to 200 mg/day. Therefore, determining which genotypes may require higher doses could be clinically significant. Predicting individual sensitivity to methadone may help determine the most effective methadone dose for each individual patient.
The mechanism underlying the association between P-gp variants and methadone requirements is uncertain. Further studies are necessary to evaluate the function of specific variations on P-gp expression in different tissues and to replicate this study in a larger sample and with different ethnicities.
MATERIAL AND METHODS
Subjects
All patients currently treated in an MMT program (The Dr. Miriam and Sheldon G. Adelson Clinic for Drug Abuse, Treatment and Research, Tel Aviv, Israel) (40) were formerly severe heroin addicts. All subjects had (i) at least 1 year of daily multiple self-administration of heroin, which is required, by Israeli law, for entry into an MMT clinic, and (ii) at least one withdrawal or failure in a detoxification center. Subjects were further ascertained using the modified Addiction Severity Index (ASI), which addresses any current additional drug and alcohol abuse (41). The study was approved by the Helsinki Committee of the Tel Aviv Sourasky Medical Center and The Rockefeller University Hospital Institutional Review Board and all subjects signed informed consent for genetic studies. A total of 138 consecutive subjects agreed to participate in genetic studies during the period 2004–2006. Blood samples were obtained and genomic DNA was isolated. Totally 109 subjects met the following criteria for this specific study: (i) at least 6 months in MMT; (ii) Jewish ancestry; (iii) negative urine, for illicit opiates, and a stable methadone dose for at least 4 weeks prior to obtaining blood specimen; (iv) not related to another subject already in the study. Out of these, three samples failed DNA isolation. Eight samples were later excluded because of genotype failure; ending up with a total of 98 individuals (64 males; 34 females). The mean age of the subjects was 45 years (range 20–67). Nine subjects had positive urine for cocaine over the 4 weeks prior to obtaining blood specimen. This study dataset includes five major sub-groups: Sephardic (Western Europe, Balkans and Morocco) (39%), Ashkenazi (Central and Eastern Europe) (22%), Oriental (Iraq, Iran, Yemen and Syria) (16%), mixed (13%) and unknown (10%). The stabilizing methadone dose ranged between 30 and 280 mg/day with a mean and median dose of 160 mg/day.
Study design
The subjects were divided into two groups according to their stabilized daily dose of methadone: the first group was comprised of subjects stabilized on a methadone dose between 30 and 150 mg/day (‘lower dose’) and the second group comprised of subjects stabilized at a methadone dose between 151 and 280 mg/day (‘higher dose’). The decision to split the groups at a methadone dose of 150 mg/day was based on the fact that, until recently, the majority of patients were maintained on doses <150 mg/day.
Single nucleotide polymorphism selection
A set of eight SNPs spanning most of the ABCB1 gene was selected (Table 1). The selection criteria were: (a) potential functionality; (b) minor allele frequency (MAF) >0.02; (c) tagging SNPs (tSNPs) for Caucasians based on HapMap (www.hapmap.org) and others (14). The set of SNPs comprised of one non-synonymous (tri-allelic rs2032582G/T/A), two synonymous (rs1045642 and rs1128503) and five intronic tSNPs. The ninth SNP, rs2032583 (intron 21), was not initially selected by the criteria described above but was included in the final analysis. This SNP is located 57 bp from SNP no. 5 (the tri-allelic 2677G>T/A, rs2032582) and was amplified by polymerase chain reaction (PCR) and sequenced with the same primers.
Genotyping
Genotypes were determined by TaqMan® technology (SNP nos 1–3, 7–9; Table 1). SNP nos 4 and 5 (rs2032582, rs2032583) were directly sequenced. The TaqMan® Pre-Designed SNP Genotyping Assays (Applied Biosystems, Foster City, CA) are listed in Supplementary Material, Table S1a. PCR was performed in replicates, on a GeneAmp® PCR system 9700, using TaqMan® universal PCR master mix with uracil-N-glycosylate (AmpErase UNG) (Applied Biosystems) according to the manufacturer's protocol. Briefly, TaqMan® assay mix (20× or 40×) and universal PCR master mix (2×) were mixed, and the volume was adjusted with water to 4 µl and was then added to 10 ng of genomic DNA (1 µl) in a 384-well optical reaction plate. PCR amplification lasted for 2 min at 50°C; 10 min at 95°C followed by 40 cycles of 15 s at 92°C and 1 min at 60°C. Genotype analysis was performed on an ABI Prism® 7900 sequence detection system using SDS 2.1 software (Applied Biosystems).
Sequencing
PCR amplification of exon 21 and the overlapping intronic sequences was performed using AmpliTaq Gold® (Applied Biosystems). Primers were designed using software Primer 3 (42) and the sequences are listed in Supplementary Material, Table S1. In addition, representative samples from each TaqMan analysis were confirmed by sequencing. Primer sequences are listed in Supplementary Material, Table S1.
PCR amplification consisted of 10 min at 94°C, eight ‘touch-down’ cycles of 30 s at 94°C, 30 s at 63–56°C and 30 s at 72°C, followed by 35 cycles of 30 s at 94°C, 30 s at 56°C and 30 s at 72°C with a final step of 7 min at 72°C. PCR products were purified using Qiagen BioRobot 9600® (Venlo, The Netherlands) and sequenced using ABI 3700 Big Dye Terminators (v3)® on an ABI Prism 3700® capillary sequencer (Applied Biosystems). Electropherograms were scored twice, independently, using the Sequencer 4.5 software (Gene Codes Corporation, Ann Arbor, MI, USA).
Statistical analysis
χ2 Tests were carried out to test for deviation from HWE with a cut-off of 0.05 showing deviation from HWE (Table 1). Fisher's exact test and χ2 test were used for allelic and genotypic analyses for individual SNPs (Tables 2 and 3). The four individuals carrying the rare ‘A’ allele of rs2032582 (2677) were excluded from the analysis. ANOVA was used for the analysis of the three genotypes of each SNP, using methadone doses as a dependent quantitative trait. LD values between SNP pairs were determined using LD statistics (r2 and D′) with the Haploview program version 3.2 (Broad Institute, MIT and Harvard, MA, USA) (43) (Fig. 1). Density plots of methadone dose by genotypes were produced using the SYSTAT 11 program (Systat Software, Inc., San Jose, CA, USA) (Fig. 2). MLGPs (a set of genotypes one each at different loci) were tabulated for SNP nos 1, 5 and 7 and for SNP nos 1–9 (Tables 4 and 5). Fisher's exact test and likelihood ratio χ2 test were performed to determine individual and global P-values, respectively. MLGPs with frequency <0.05 were pooled into a single class called ‘others’. Experiment-wise significance levels were obtained by permutation testing (44). The SUMSTAT program (http://www.genemapping.cn/sumstat.html) was used to perform random permutation with 10 000 randomization samples. In each of these ‘null’ samples, a χ2 value was computed for the observed data. The proportion of randomization samples with χ2 exceeding the χ2 in the observed data is an estimate for its associated significance level, which automatically adjusts for multiple testing and allows for the dependence among SNPs. Comparing these randomization P-values with Bonferroni corrected P-values shows that the nine observed SNPs approximately correspond to five independent SNPs. Thus, experiment-wise P-values may be obtained from uncorrected (point-wise) P-values by multiplying the latter with 5 (Table 3).
SUPPLEMENTARY MATERIAL
FUNDING
National Institute of Health (DA-P60-05130 and DA-K05-00049) to M.J.K., (MH-R01-44292) to J.O., National Natural Science Foundation of China (30730057) from the Chinese Government to J.O. and National Institute of Health/NCRR-CTSA (UL1-RR024143) The Rockefeller University Center for Clinical and Translational Science.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge the contribution of Drs Sara Hamon, David Nielsen, Ann Ho, Dmitri Proudnikov, Vadim Yuferov, Ms Susan Russo and Mr Matthew Randesi.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Kreek M.J., Vocci F.J. History and current status of opioid maintenance treatments: blending conference session. J. Subst. Abuse. Treat. 2002;23:93–105. doi: 10.1016/s0740-5472(02)00259-3. [DOI] [PubMed] [Google Scholar]
- 2.Kerb R. Implications of genetic polymorphisms in drug transporters for pharmacotherapy. Cancer Lett. 2006;234:4–33. doi: 10.1016/j.canlet.2005.06.051. [DOI] [PubMed] [Google Scholar]
- 3.Kreek M.J. Introduction to addictive disorders: implications for pharmacotherapies. In: Sibley D.R., Hanin I., Kuhar M., Skolnick P., editors. Handbook of Contemporary Neuropharmacology. Vol. 1. Hoboken, NJ, USA: Wiley; 2007. pp. 451–463. [Google Scholar]
- 4.Kreek M.J. Plasma and urine levels of methadone. Comparison following four medication forms used in chronic maintenance treatment. NY. State J. Med. 1973;73:2773–2777. [PubMed] [Google Scholar]
- 5.Kreek M.J., Oratz M., Rothschild M.A. Hepatic extraction of long- and short-acting narcotics in the isolated perfused rabbit liver. Gastroenterology. 1978;75:88–94. [PubMed] [Google Scholar]
- 6.Kreek M.J., Gutjahr C.L., Garfield J.W., Bowen D.V., Field F.H. Drug interactions with methadone. Ann. NY. Acad. Sci. 1976;281:350–371. doi: 10.1111/j.1749-6632.1976.tb27945.x. [DOI] [PubMed] [Google Scholar]
- 7.Crettol S., Digon P., Golay K.P., Brawand M., Eap C.B. In vitro P-glycoprotein-mediated transport of (R)-, (S)-, (R,S)-methadone, LAAM and their main metabolites. Pharmacology. 2007;80:304–311. doi: 10.1159/000107104. [DOI] [PubMed] [Google Scholar]
- 8.Somogyi A.A., Barratt D.T., Coller J.K. Pharmacogenetics of opioids. Clin. Pharmacol. Ther. 2007;81:429–444. doi: 10.1038/sj.clpt.6100095. [DOI] [PubMed] [Google Scholar]
- 9.Dagenais C., Graff C.L., Pollack G.M. Variable modulation of opioid brain uptake by P-glycoprotein in mice. Biochem. Pharmacol. 2004;67:269–276. doi: 10.1016/j.bcp.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 10.Marzolini C., Paus E., Buclin T., Kim R.B. Polymorphisms in human MDR1 (P-glycoprotein): recent advances and clinical relevance. Clin. Pharmacol. Ther. 2004;75:13–33. doi: 10.1016/j.clpt.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Fojo A.T., Ueda K., Slamon D.J., Poplack D.G., Gottesman M.M., Pastan I. Expression of a multidrug-resistance gene in human tumors and tissues. Proc. Natl Acad. Sci. USA. 1987;84:265–269. doi: 10.1073/pnas.84.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim R.B., Leake B.F., Choo E.F., Dresser G.K., Kubba S.V., Schwarz U.I., Taylor A., Xie H.G., McKinsey J., Zhou S., et al. Identification of functionally variant MDR1 alleles among European Americans and African Americans. Clin. Pharmacol. Ther. 2001;70:189–199. doi: 10.1067/mcp.2001.117412. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmeyer S., Burk O., von Richter O., Arnold H.P., Brockmoller J., Johne A., Cascorbi I., Gerloff T., Roots I., Eichelbaum M., et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc. Natl Acad. Sci. USA. 2000;97:3473–3478. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho G.T., Soranzo N., Nimmo E.R., Tenesa A., Goldstein D.B., Satsangi J. ABCB1/MDR1 gene determines susceptibility and phenotype in ulcerative colitis: discrimination of critical variants using a gene-wide haplotype tagging approach. Hum. Mol. Genet. 2006;15:797–805. doi: 10.1093/hmg/ddi494. [DOI] [PubMed] [Google Scholar]
- 15.Takane H., Kobayashi D., Hirota T., Kigawa J., Terakawa N., Otsubo K., Ieiri I. Haplotype-oriented genetic analysis and functional assessment of promoter variants in the MDR1 (ABCB1) gene. J. Pharmacol. Exp. Ther. 2004;311:1179–1187. doi: 10.1124/jpet.104.069724. [DOI] [PubMed] [Google Scholar]
- 16.Tang K., Ngoi S.M., Gwee P.C., Chua J.M., Lee E.J., Chong S.S., Lee C.G. Distinct haplotype profiles and strong linkage disequilibrium at the MDR1 multidrug transporter gene locus in three ethnic Asian populations. Pharmacogenetics. 2002;12:437–450. doi: 10.1097/00008571-200208000-00004. [DOI] [PubMed] [Google Scholar]
- 17.Kroetz D.L., Pauli-Magnus C., Hodges L.M., Huang C.C., Kawamoto M., Johns S.J., Stryke D., Ferrin T.E., DeYoung J., Taylor T., et al. Sequence diversity and haplotype structure in the human ABCB1 (MDR1, multidrug resistance transporter) gene. Pharmacogenetics. 2003;13:481–494. doi: 10.1097/00008571-200308000-00006. [DOI] [PubMed] [Google Scholar]
- 18.Wang D., Johnson A.D., Papp A.C., Kroetz D.L., Sadee W. Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharmacogenet. Genom. 2005;15:693–704. [PubMed] [Google Scholar]
- 19.Kimchi-Sarfaty C., Oh J.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M. A ‘silent’ polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 20.Salama N.N., Yang Z., Bui T., Ho R.J. MDR1 haplotypes significantly minimize intracellular uptake and transcellular P-gp substrate transport in recombinant LLC-PK1 cells. J. Pharm. Sci. 2006;95:2293–2308. doi: 10.1002/jps.20717. [DOI] [PubMed] [Google Scholar]
- 21.Tang K., Wong L.P., Lee E.J., Chong S.S., Lee C.G. Genomic evidence for recent positive selection at the human MDR1 gene locus. Hum. Mol. Genet. 2004;13:783–797. doi: 10.1093/hmg/ddh099. [DOI] [PubMed] [Google Scholar]
- 22.Kimchi-Sarfaty C., Marple A.H., Shinar S., Kimchi A.M., Scavo D., Roma M.I., Kim I.W., Jones A., Arora M., Gribar J., et al. Ethnicity-related polymorphisms and haplotypes in the human ABCB1 gene. Pharmacogenomics. 2007;8:29–39. doi: 10.2217/14622416.8.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ostrovsky O., Nagler A., Korostishevsky M., Gazit E., Galski H. Genotype and allele frequencies of C3435T polymorphism of the MDR1 gene in various Jewish populations of Israel. Ther. Drug Monit. 2004;26:679–684. doi: 10.1097/00007691-200412000-00015. [DOI] [PubMed] [Google Scholar]
- 24.Ameyaw M.M., Regateiro F., Li T., Liu X., Tariq M., Mobarek A., Thornton N., Folayan G.O., Githang'a J., Indalo A., et al. MDR1 pharmacogenetics: frequency of the C3435T mutation in exon 26 is significantly influenced by ethnicity. Pharmacogenetics. 2001;11:217–221. doi: 10.1097/00008571-200104000-00005. [DOI] [PubMed] [Google Scholar]
- 25.Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z., Wang J., Tantoso E., Wang B., Tai A.Y., Ooi L.L., Chong S.S., Lee C.G. Signatures of recent positive selection at the ATP-binding cassette drug transporter superfamily gene loci. Hum. Mol. Genet. 2007;16:1367–1380. doi: 10.1093/hmg/ddm087. [DOI] [PubMed] [Google Scholar]
- 27.Gurney H., Wong M., Balleine R.L., Rivory L.P., McLachlan A.J., Hoskins J.M., Wilcken N., Clarke C.L., Mann G.J., Collins M., et al. Imatinib disposition and ABCB1 (MDR1, P-glycoprotein) genotype. Clin. Pharmacol. Ther. 2007;82:33–40. doi: 10.1038/sj.clpt.6100201. [DOI] [PubMed] [Google Scholar]
- 28.Xing Q., Gao R., Li H., Feng G., Xu M., Duan S., Meng J., Zhang A., Qin S., He L. Polymorphisms of the ABCB1 gene are associated with the therapeutic response to risperidone in Chinese schizophrenia patients. Pharmacogenomics. 2006;7:987–993. doi: 10.2217/14622416.7.7.987. [DOI] [PubMed] [Google Scholar]
- 29.Kim K.A., Park P.W., Park J.Y. Effect of ABCB1 (MDR1) haplotypes derived from G2677T/C3435T on the pharmacokinetics of amlodipine in healthy subjects. Br. J. Clin. Pharmacol. 2007;63:53–58. doi: 10.1111/j.1365-2125.2006.02733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ambudkar S.V., Kimchi-Sarfaty C., Sauna Z.E., Gottesman M.M. P-glycoprotein: from genomics to mechanism. Oncogene. 2003;22:7468–7485. doi: 10.1038/sj.onc.1206948. [DOI] [PubMed] [Google Scholar]
- 31.Sakaeda T., Nakamura T., Okumura K. Pharmacogenetics of drug transporters and its impact on the pharmacotherapy. Curr. Topics Med. Chem. 2004;4:1385–1398. doi: 10.2174/1568026043387692. [DOI] [PubMed] [Google Scholar]
- 32.Peles E., Schreiber S., Gordon J., Adelson M. Significantly higher methadone dose for methadone maintenance treatment (MMT) patients with chronic pain. Pain. 2005;113:340–346. doi: 10.1016/j.pain.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 33.Bleich A., Gelkopf M., Weizman T., Adelson M. Benzodiazepine abuse in a methadone maintenance treatment clinic in Israel: characteristics and a pharmacotherapeutic approach. Isr. J. Psychiatry Relat. Sci. 2002;39:104–112. [PubMed] [Google Scholar]
- 34.Kreek M.J., Garfield J.W., Gutjahr C.L., Giusti L.M. Rifampin-induced methadone withdrawal. N. Engl. J. Med. 1976;294:1104–1106. doi: 10.1056/NEJM197605132942008. [DOI] [PubMed] [Google Scholar]
- 35.Tong T.G., Pond S.M., Kreek M.J., Jaffery N.F., Benowitz N.L. Phenytoin-induced methadone withdrawal. Ann. Intern. Med. 1981;94:349–351. doi: 10.7326/0003-4819-94-3-349. [DOI] [PubMed] [Google Scholar]
- 36.Borg L., Ho A., Peters J.E., Kreek M.J. Availability of reliable serum methadone determination for management of symptomatic patients. J. Addict. Dis. 1995;14:83–96. doi: 10.1300/J069v14n03_06. [DOI] [PubMed] [Google Scholar]
- 37.Coller J.K., Barratt D.T., Dahlen K., Loennechen M.H., Somogyi A.A. ABCB1 genetic variability and methadone dosage requirements in opioid-dependent individuals. Clin. Pharmacol. Ther. 2006;80:682–690. doi: 10.1016/j.clpt.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 38.Pearson E.C., Woosley R.L. QT prolongation and torsades de pointes among methadone users: reports to the FDA spontaneous reporting system. Pharmacoepidemiol. Drug Saf. 2005;14:747–753. doi: 10.1002/pds.1112. [DOI] [PubMed] [Google Scholar]
- 39.Peles E., Bodner G., Kreek M.J., Rados V., Adelson M. Corrected-QT intervals as related to methadone dose and serum level in methadone maintenance treatment (MMT) patients: a cross-sectional study. Addiction. 2007;102:289–300. doi: 10.1111/j.1360-0443.2006.01668.x. [DOI] [PubMed] [Google Scholar]
- 40.Peles E., Schreiber S., Adelson M. Factors predicting retention in treatment: 10-year experience of a methadone maintenance treatment (MMT) clinic in Israel. Drug Alcohol Depend. 2006;82:211–217. doi: 10.1016/j.drugalcdep.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 41.McLellan A.T., Kushner H., Metzger D., Peters R., Smith I., Grissom G., Pettinati H., Argeriou M. The Fifth edition of the Addiction Severity Index. J. Subst. Abuse Treat. 1992;9:199–213. doi: 10.1016/0740-5472(92)90062-s. [DOI] [PubMed] [Google Scholar]
- 42.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 43.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 44.Hoh J., Wille A., Ott J. Trimming, weighting, and grouping SNPs in human case-control association studies. Genome Res. 2001;11:2115–2119. doi: 10.1101/gr.204001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.