Abstract
Protein sumoylation regulates a variety of nuclear functions and has been postulated to be involved in meiotic chromosome dynamics as well as other processes of spermatogenesis. Here, the expression and distribution of sumoylation pathway genes and proteins was determined in mouse male germ cells, with a particular emphasis on prophase I of meiosis. Immunofluorescence microscopy revealed that SUMO1, SUMO2/3 and UBE2I (also known as UBC9) were localized to the XY body in pachytene and diplotene spermatocytes, while only SUMO2/3 and UBE2I were detected near centromeres in metaphase I spermatocytes. Quantitative RT-PCR and western blotting were used to examine the expression of sumoylation pathway genes and proteins in enriched preparations of leptotene/ zygotene spermatocytes, prepubertal and adult pachytene spermatocytes, as well as round spermatids. Two general expression profiles emerged from these data. The first profile, where expression was more prominent during meiosis, identified sumoylation pathway participants that could be involved in meiotic chromosome dynamics. The second profile, elevated expression in post-meiotic spermatids, suggested proteins that could be involved in spermiogenesis-related sumoylation events. In addition to revealing differential expression of protein sumoylation mediators, which suggests differential functioning, these data demonstrate the dynamic nature of SUMO metabolism during spermatogenesis.
Keywords: SUMO, sumoylation, spermatogenesis, UBE2I, SUMO ligases, SUMO proteases, XY body, meiosis, spermatocytes, male germ cells
INTRODUCTION
Creation of haploid spermatozoa (spermatogenesis) requires proliferation of spermatogonia, meiotic division of spermatocytes and post-meiotic maturation of spermatids (spermiogenesis) in a coordinated and timely manner. Events taking place during prophase of meiosis I, in particular pairing, synapsis and reciprocal recombination of homologous chromosomes, are critical in establishing the haploid genome of the gamete (Morelli and Cohen, 2005). A key event is formation of the synaptonemal complex (SC), the proteinaceous structure postulated to facilitate both synapsis and recombination of homologous chromosomes. In contrast to autosomal chromosomes, the X and Y sex chromosomes are largely non-homologous and form a heterochromatic and transcriptionally inactive sub-nuclear domain known as the XY body in spermatocytes, where chromatin is uniquely modified with specific histone variants and other proteins of unknown function (Handel, 2004; Turner, 2007). All of these meiotic prophase events are characterized by recruitment of specialized proteins to meiotic nuclei and active chromatin remodeling.
Recruitment and sub-nuclear localization of proteins is frequently regulated by post-translational modifications. Prominent among these is sumoylation, or tagging of lysine residues on target proteins with SUMOs. SUMOs (small ubiquitin-related modifiers) are ~100 amino acid ubiquitin-like proteins that function as post-translational regulators of protein function and localization (Gill, 2004; Johnson, 2004). Unlike ubiquitination, which is often but not exclusively associated with protein degradation, sumoylation of target proteins is involved in modulating protein-protein interactions as well as nuclear-cytoplasmic transport. SUMO modification plays important roles in formation of specific nuclear heterochromatic territories, regulation of transcription, and DNA replication, recombination and repair (Gill, 2004; Johnson, 2004). Lower eukaryotes express only one SUMO protein (SUMO/ Smt3), while there are three SUMO paralogues in higher organisms: SUMO1, and the closely related SUMO2 and SUMO3 (usually referred to as SUMO2/3 as they are ~95% homologous to each other) (Johnson, 2004). SUMO1 and a variety of SUMO pathway proteins are expressed during meiotic prophase in yeast, where dynamic SUMO modification of proteins may be involved in formation of the SC (Cheng et al., 2006; Cheng et al., 2007; de Carvalho and Colaiacovo, 2006; Hooker and Roeder, 2006). In yeast, SUMO1 localizes to synapsed (but not asynapsed) chromosome regions, and a key component of the yeast SC, the Zip3 protein, is a SUMO E3 ligase that is crucial to initiating SC formation (Cheng et al., 2006; Cheng et al., 2007; de Carvalho and Colaiacovo, 2006; Hooker and Roeder, 2006). SUMO-mediated regulation of SC formation may be a conserved function across sexually reproducing organisms, and indeed, we, as well as others, have shown that SUMO1 accumulates in mammalian spermatocyte chromatin. Most notably, SUMO1 is found within the entire XY body chromatin in mouse pachytene spermatocytes (Rogers et al., 2004; Vigodner and Morris, 2005), while in human male germ cells, SUMO1 localization is more spatially restricted, associated primarily with the XY chromosome axes (Vigodner et al., 2006). These observations, coupled with the known functional properties of protein sumoylation, suggest sumoylation could function in XY body formation and/or maintenance. During human meiotic prophase, SUMO1 is also found in centromeric and pericentromeric heterochromatin in addition to sex chromosomes, and localization of SUMO1 becomes exclusively centromeric by the end of prophase I, a pattern not observed in mouse spermatocytes (Vigodner et al., 2006). Additionally, SUMO1 is detected in the centrosome and manchette of spermatids, as well as in specific domains of human, mouse and rat testicular somatic cells (Vigodner and Morris, 2005; Vigodner et al., 2006). These observations suggest that SUMO modification of protein substrates and SUMO turnover are complex, highly regulated events during spermatogenesis.
SUMO1 and SUMO2/3 are reversibly linked to substrate proteins following a three-step enzymatic process similar to, but distinct from, the ubiquitination pathway (Gill, 2004). Upon proteolytic maturation, SUMO is first activated by the SUMO E1-specific activating complex SAE1/SAE2 before being transferred to the SUMO E2-conjugating enzyme UBE2I (also known as UBC9); a SUMO E3 ligase then promotes the transfer of SUMO from the E2-conjugating enzyme to the substrate. Although UBE2I ultimately catalyzes the link between SUMO and the target protein, SUMO E3 ligases play important roles in enhancing substrate identification and specificity. The PIAS (protein inhibitor of activated STAT) proteins and RANBP2 are amongst the proteins that possess E3 ligase activity (Schmidt and Muller, 2003; Gill, 2004; Johnson, 2004). An essential component of the control mechanisms that regulate the availability of “ready-to-conjugate” SUMO and the modification status of individual protein substrates are SUMO/ sentrin specific peptidases (SENPs). A number of SENP enzymes have been characterized and shown to function in proteolytic processing of SUMO precursors and in deconjugation of SUMO from modified proteins (Hay, 2007; Mukhopadhyay and Dasso, 2007). Various studies have reported on the expression of genes encoding protein sumoylation factors in the testis (Kim et al., 2000; Kovalenko et al., 1996; Matsuura et al., 2005; Moilanen et al., 1999; Santti et al., 2003; Tan et al., 2002; Yamaguchi et al., 2006; Yan et al., 2003), but a detailed developmental study analyzing the expression of sumoylation pathway genes in male germ cells has not yet been conducted.
The importance of identifying the meiotic players of protein sumoylation in mammalian cells is underscored by the role of protein sumoylation in chromosome synapsis, synaptonemal complex formation and meiotic progress in yeast (Cheng et al., 2006; Hooker and Roeder, 2006). We determined the expression and distribution of sumoylation pathway genes and proteins during mouse male germ cell development, with a particular emphasis on prophase of meiosis I. Both SUMO1 and SUMO2/3 were present not only in the XY body, but also in heterochromatic chromocenters of both pachytene and diplotene spermatocytes; however, only SUMO2/3 localized to the pericentromeric regions of metaphase I (MI) spermatocytes. UBE2I localized to both the XY body in pachytene and diplotene spermatocytes, and to centromeres in MI spermatocytes. To identify sumoylation pathway genes expressed during spermatogenesis, real-time RT-PCR and immunoblot expression analyses were conducted on enriched populations of leptotene/ zygotene spermatocytes, prepubertal and adult pachytene spermatocytes, as well as round spermatids. Two general expression profiles emerged from these data. The first profile, where expression was more prominent during meiosis, identified sumoylation pathway components that could be related to meiotic chromosome dynamics. The second profile, elevated expression in post-meiotic spermatids, suggested proteins that could be involved in spermiogenesis-related sumoylation events. Taken together, our findings support intricate expression patterns for SUMO pathway genes, reflecting dynamic regulation of protein sumoylation in meiotic progression and spermatogenesis.
MATERIALS AND METHODS
Mice
All experiments were conducted using B6SJL F1 male mice obtained from a colony maintained by the investigators at The Jackson Laboratory (Bar Harbor, ME); the day of birth was designated as postnatal day (dpp) 0. Mice were maintained under normal conditions in accordance with the National Institutes of Health and U.S. Department of Agriculture standards; all procedures conducted were approved by The Jackson Laboratory Animal Care and Use Committee.
Cytological methods
Mixed germ cell preparations were obtained by enzymatic digestion of 21 dpp testes. Briefly, testes were detunicated, digested in 0.5 mg/ml collagenase (Sigma, St. Louis, MO) in Krebs-Ringer bicarbonate (KRB) media at 32°C for 20 min, then digested with 0.5 mg/ml of trypsin (Sigma) and 1 µg/ml DNaseI (USB, Cleveland, OH) in KRB at 32°C for 13 min; germ cells were subsequently released by pipetting for 3 min. The germ cell suspension was filtered through a Nitex mesh and washed three times in 0.5% bovine serum albumin (BSA; Sigma) in KRB. After the final wash, cells were counted and resuspended at a concentration of 2.5 × 106 cells /ml in KRB media; the cells were then centrifuged at 5000 rpm for 5 min. All solutions used for the preparation of surface-spread chromatin were made with distilled water. Cell pellets were resuspended in equal volumes of 2% paraformaldehyde (PFA)/ 0.03% SDS, pH 8.2 and 0.4% Kodak Professional Photo-Flo 200 solution, pH 8.2 (Electron Microscopy Sciences (EMS), Washington, PA); 2-µl aliquots were gently spread onto the wells of 12-well Shandon slides (Fisher Scientific, Pittsburg, PA). Cells were air-dried 5–10 min and then processed through 2% PFA/ 0.03% SDS, 2% PFA, and finally 0.4% Photo-Flo. Slides were allowed to completely air-dry at room temperature and were stored at −20°C before being processed for immunofluorescence as described below.
Solutions used for immunolabeling of surface-spread chromatin were prepared in phosphate buffered saline (PBS), pH 7.4, and procedures were carried out at room temperature. Slides were washed 3X in 0.3% BSA, 1% goat serum and 0.005% Triton X-100 (Sigma); the second wash was supplemented with 0.05% Triton X-100. Prior to the addition of primary antibodies, the space between the wells was carefully dried. Primary antibodies were diluted in 3% BSA, 10% goat serum and 0.05% Triton X-100 (antibody dilution buffer, ADB), applied to the appropriate wells and incubated overnight at room temperature in a humidified chamber; at least one well per slide received only ADB to serve as negative control for primary antibody specificity. Antibodies used to label surface-spread chromatin preparations were as follows: rabbit polyclonal anti-SYCP3 (NB300-231, 1:1000; Novus Biologicals, Littleton, CO); mouse monoclonal anti-SYCP3 (NB100-2065, 1:100; Novus Biologicals); mouse monoclonal anti-SUMO1, clone 21C7 (1:250; Matunis et al., 1996); mouse monoclonal anti-SUMO2/3, clone 8A2 (1:250; Zhang et al., 2008); and rabbit polyclonal anti-UBE2I (1:250; Zhang et al., 2002). Following primary antibody incubation, slides were washed as described above and incubated with the appropriate secondary antibodies tagged with Alexa Fluor 488 or 594 (Molecular Probes/ Invitrogen, Eugene, OR) for 1 hr in a humidified dark chamber. Slides were then passed through a series of washes: 0.2% Photo-Flo diluted in PBS and supplemented with 4′, 6-diamidino-2-phenylindole (DAPI; Sigma), 0.05% Triton X-100 in PBS, and finally 0.2% Photo-Flo in water, and mounted with Prolong Antifade (Molecular Probes/ Invitrogen). Slides were analyzed using a Leica DMRE epifluorescence microscope (Leica Microsystems Inc., Bannockburn, IL) equipped with a MicroMax CCD camera (Princeton Instrument, Trenton, NJ); captured images were analyzed using Metamorph version 6.3r5 (Molecular Devices Corporation, Downingtown, PA). All experiments were repeated at least two times using different sets of samples.
Isolation of enriched populations of male germ cells
Enriched populations of male germ cells were obtained from the testes of 17- and 70-dpp mice according to the sedimentation velocity cell separation method (Bellvé, 1993). Mixed germ cells suspensions were prepared as described above from males of each age group, but after the three 0.5% BSA/KRB washes, cells were allowed to separate by cellular sedimentation at unit gravity in a 2–4% BSA gradient generated over 2.5 hr in a STA-PUT apparatus (ProScience Inc., GlassShop, Toronto, ON, Canada). Following sedimentation, 10-ml fractions were collected and examined using light microscopy and differential interference contrast Nomarski optics. Cells were identified on the basis of morphological criteria and size as described by Bellvé (1993). Populations of leptotene/zygotene spermatocytes (average purity = 81%) and prepubertal pachytene spermatocytes (average purity = 85%) were obtained from the testes of 17 dpp mice (n = 3 cell separations); adult pachytene spermatocytes (average purity = 84%) and round spermatids (average purity = 90%) were obtained from 70 dpp mice (n = 3 cell separations). For every cell separation, and for each population of cells collected, an aliquot of cells was snap-frozen for subsequent RNA extraction, while the rest of the cells were processed immediately for protein extraction as described below.
RNA extraction and real-time (quantitative) RT-PCR
Total RNA was extracted from snap-frozen pellets of enriched populations of male germ cells using the RNeasy extraction kit with DNaseI treatment as described by the manufacturer (Qiagen Inc. - USA, Valencia, CA). Samples were diluted to a concentration of 10 ng/µl, dispensed in single-use aliquots, and stored at −80°C. Quantitative RT-PCR (qRT-PCR) was performed on the Applied Biosystems 7500 Real-Time PCR System (Foster City, CA) using the QuantiTect SYBR Green® RT-PCR kit (Qiagen). Gene-specific primers were used to determine the overall relative expression levels of all sumoylation pathway genes tested according to the standard curve method (Bustin, 2002). Primers were designed to span introns and amplify all known transcript variants of the various genes, with the exception of Senp6, for which primers amplifying each of the two transcript variants were designed (see Table 1 for primer sequences and annealing temperatures; 18S primer sequences are from La Salle et al, 2004). SYBR Green® was used to detect the double-stranded DNA produced during the amplification reaction. Reactions were performed using approximately 10 ng or 100 pg of total RNA for the sumoylation pathway genes and 18S, respectively. One-step RT-PCR reactions were performed in 25-µl volumes as directed by the manufacturer for 40 cycles. For each gene tested, a specific standard curve was established using single-use aliquots of the same stock of RNA (total RNA extracted simultaneously from multiple 17dpp testes). Because the same stock of RNA was used to prepare all standard curves, the relative quantities determined for a given gene using this method could be compared across individual experiments. All reactions were performed in triplicate on the same three independent sets of germ cells and specificity was assessed by melting curve analysis. qRT-PCR results were normalized to their corresponding 18S rRNA content and are presented as the mean normalized expression in 10 ng of RNA. Data are reported as mean ± SEM.
TABLE 1.
Details of primers used for real-time RT-PCR
| Gene | Forward (top) and reverse (bottom) primers (5’- … -3’) | Amplicon size (bp) | Annealing temperature (°C) |
|---|---|---|---|
| Sumo1 | GCGGTGTTGTGCTGTAGAGA TCAGACATGGTGACGTGGA |
114 | 58 |
| Sumo2 | CAGCCAATCAACGAAACAGA ATGTGGTGGGACCAAATTGT |
184 | 56 |
| Sumo3 | CCGAAGGATTCTCCTTTTCC GGATGGGAGGGAACAAAAAT |
165 | 56 |
| Sae1 | CCCAGTATGTGCTGTGGTTG AAGGCACTCCACAATTCCAC |
130 | 56 |
| Sae2 | AAGCCTACCCAGAGGACGTT GCTTCAGCCTCTGTTGGTTC |
182 | 53 |
| Ube2i | TCTCCCTGCCTGTTAGCTGT GAATGCAGGTCAGGAGGTGT |
173 | 56 |
| Pias1 | ATCAGGTAGCGTCCCACAAC CGAGGCTTGATGAGGAAGAC |
185 | 54 |
| Pias2 | CCACCTGGATCTGTCCTGTT AGGTCTCATGGGACACCAAG |
146 | 54 |
| Pias3 | AGTTTCGATGCTGCCCTTTA TCATCACAATCCGAACAGGA |
146 | 56 |
| Pias4 | GCAGTGCTTTGATGCTGTGT CGCCAGGAACTCAATCTCAT |
166 | 54 |
| Senp1 | CGAAGTCTTTGCCTCGAAAC GCAGGCTTAATGGGAAATGA |
119 | 53 |
| Senp2 | AGCAGTTGCTGTGACAATGC CATTGTGGAGAGGCCTTCAT |
163 | 54 |
| Senp3 | CCGGCACCACTTACTCAAAT AGGCGTCTAGTTGGCACTGT |
130 | 56 |
| Senp5 | TGCTGACTGAAGCCAGAGAA CACGAGGCATGTCTTCTTGA |
180 | 56 |
| Senp6 – variant 1 | CGTCCACATGGACAGTCG GCTCACACGCCTTCTACGTC |
181 | N/D |
| Senp6 – variant 2 | CCTGGGCTAATTCTGGTCAA CAAGGGCATTAGCACAACAA |
155 | 56 |
| Senp7 | TTATGTGCTCCCAGCAAGGT CTGGGTGTCCTTTCGGATAA |
122 | 54 |
| Senp8 | CATGCAAAGCAGGTAGCAGA ACACAAGGCCTCGGTGTTAC |
144 | 54 |
| AF366264 | GCCTGTGACCTTTTCTCAGC GAACAGGGATAGGGGACCAT |
121 | 54 |
| EG408191 | CAGCGTGTGATGGAGAAGAA GCCAGGCACCTTAAATACCA |
156 | 54 |
| 18S | GCCCTGTAATTGGAATGAGTCCACTT GTCCCCAAGATCCAACTACGAGCTTT |
149 | 50–65 |
N/D: Could not be amplified under the conditions tested.
Protein extraction and immunoblotting analysis
Protein lysates were prepared from freshly isolated male germ cells using RIPA buffer supplemented with protease inhibitors according to the manufacturer’s recommendations (Santa Cruz Biotechnology Inc., Santa Cruz, CA). Protein concentration was determined using the BCA protein assay following the manufacturer’s instructions (Pierce/ Thermo Scientific, Rockford, IL). Total protein aliquots of 10 µg were heated 60°C for 10 min in reducing sample buffer, electrophoresed on 10% SDS-polyacrylamide gels, and transferred to Immobilon-P PVDF membranes (Millipore, Billerica, MA). Membranes were blocked in 5% non-fat dried milk diluted in PBS supplement with 0.1% Tween 20 (PBST) and were incubated with one of the following primary antibody diluted in blocking buffer: rabbit polyclonal anti-SAE2 (IMG-5111A, 1:5000; Imgenex, San Diego, CA); mouse monoclonal anti-UBE2I (610749, 1:10000; BD Transduction Laboratories, San Jose, CA); goat polyclonal anti-PIAS2 (sc-30879, 1:1000; Santa Cruz Biotechnology Inc.); rabbit polyclonal anti-PIAS4 (IMG-290, 1: 5000; Imgenex); rabbit polyclonal anti-SENP1 (IMG-521, 1:2500; Imgenex); and rabbit polyclonal anti-SENP2 (AP1232c, 1:2500; Abgent, San Diego, CA). Membranes were washed with PBST according to the instructions provided with the ECL Plus Western Blotting Detection Reagent kit (Amersham, GE Healthcare, Piscataway, NJ), followed by incubation with one of the following horseradish peroxidase (HRP)-conjugated secondary antibody: goat anti-rabbit IgG HRP-conjugated antibody (1:50 000; Molecular Probes/Invitrogen); rabbit anti-goat IgG HRP-conjugated antibody (1:30 000; Molecular Probes/Invitrogen); and rabbit anti-mouse IgG HRP-conjugated antibody (1:60 000; Molecular Probes/Invitrogen). Membranes were washed with PBST and exposed to the ECL Plus Western Blotting detection solution ensuing the manufacturer’s directions (Amersham). Chemiluminescence was revealed on Kodak X-Omatic film (Kodak, Rochester, NY). To confirm loading of equal amounts of proteins on gel, gels electrophoresed under identical conditions were stained with GelCode Blue Stain Reagent (Pierce/ Thermo Scientific). Additionally, membranes were stained with India ink following immunoblotting to confirm equal loading and consistency of transfer.
Statistical analyses
JMP 7.0 software (SAS Institute, Cary, NC) was used for statistical analysis of the qRT-PCR data obtained. Prior to performing statistical testing, the data were analyzed to determine whether they conformed to a normal distribution. For each gene, comparison of the expression in individual germ cell types was performed using a univariate analysis of variance (one-way Anova), and then means comparisons for all pairs were made using the Tukey-Kramer HSD post hoc test. A p-value of < 0.05 was taken to be significant.
RESULTS
Spatial and temporal localization of SUMO1, SUMO2/3 and the E2 conjugating enzyme, UBE2I, in mouse spermatocytes
Previous studies reported on the prominent localization of SUMO1 to sex chromosomes in both mouse and rat pachytene spermatocytes (Rogers et al., 2004; Vigodner and Morris, 2005), but the developmental expression pattern of SUMO2/3 had not been previously reported in male germ cells. SUMO2/3 share only ~50% sequence identity with SUMO1 (Johnson, 2004), suggesting that these modifiers could have different roles in the germ line. We further characterized the relationship between SUMO1 and SUMO2/3 expression during meiosis by immunolabeling surface-spread chromatin with antibodies directed against either SUMO1 or SUMO2/3. Substages of meiotic prophase were identified by patterns of labeling with anti-SYCP3, which labels the axial and lateral axes of the synaptonemal complex (Cobb and Handel, 1998; Cohen and Pollard, 2001; Inselman et al., 2003). Briefly, in leptonema, axial axes appear discontinuous; in zygonema, homologous axes initiate pairing and lateral axes of the synaptonemal complex form; in pachynema, chromosome synapsis is completed; and in diplonema, homologous chromosomes begin to separate and are held together by chiasmata. Metaphase I (MI) spermatocytes were also identified via their characteristic SYCP3 distribution pattern; SYCP3 is found in association with sister centromeres at this stage (Dobson et al., 1994; Hodges et al., 2001).
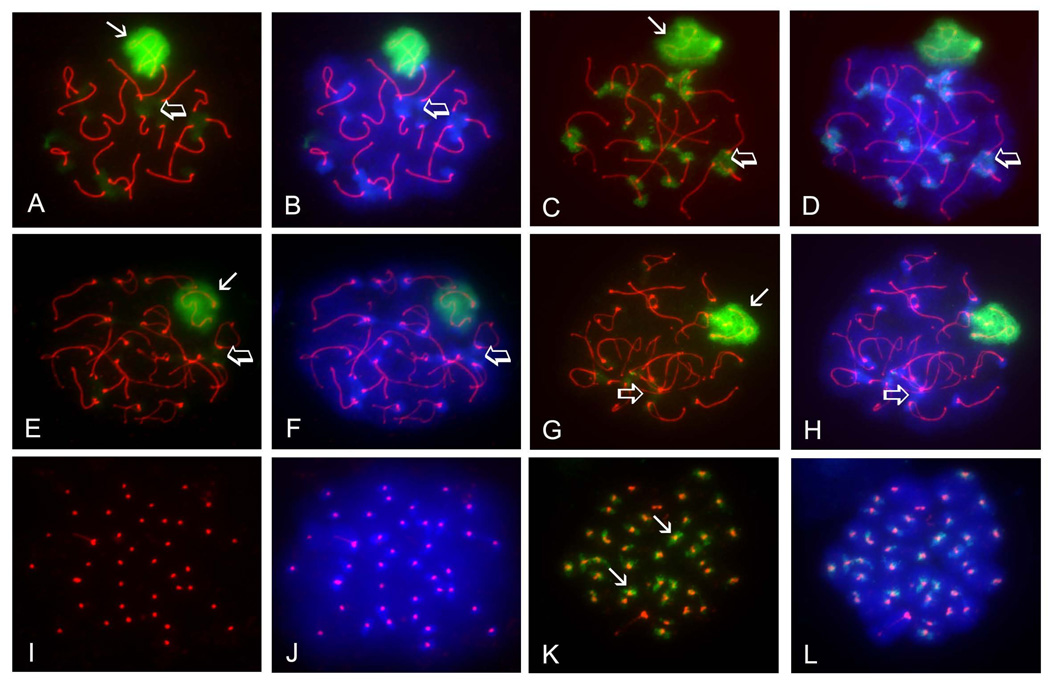
As previously reported (Rogers et al., 2004; Vigodner and Morris, 2005), SUMO1 localized to the XY body in pachytene spermatocytes (Fig. 1A, arrow). SUMO1 could also be detected in heterochromatic chromocenters in pachytene spermatocytes, as the signal obtained with the anti-SUMO1 antibody overlapped the bright patches of heterochromatin detected using the fluorescent DNA-binding dye 4’, 6-diamidino-2-phenylindole (DAPI) (Fig. 1A and B, open arrows). A very similar localization pattern was observed for SUMO2/3 in pachytene spermatocytes. The XY body (Fig. 1C, arrow) and chromocenters (Fig. 1 C and D, open arrows) were both marked by the antibody directed against SUMO2/3. In diplotene spermatocytes, the SUMO1 signal was diminished, but persisted over the XY body (Fig. 1E, arrow), while staining at chromocenters could be observed but was generally fainter than in pachytene spermatocytes (Fig. 1E and F, open arrows). The same was observed for SUMO2/3, although there was an even greater loss of signal at chromocenters in diplotene spermatocytes (Fig. 1G and H, open arrows). Interestingly, a SUMO1-positive signal was never detected in MI spermatocytes (Fig. 1I and J), but a SUMO2/3-positive signal was clearly observed in MI spermatocytes in the vicinity of SYCP3-positive sister centromeres, also co-localizing to the centromeres, as depicted by the presence of a yellow signal (Fig. 1K and L, arrows). Overall, nuclear localization of SUMO2/3 was very similar to that of SUMO1, but appeared to be exclusive in MI spermatocytes, as SUMO1 was not detected in these cells.
FIGURE 1. Both SUMO1 and SUMO2/3 accumulate in the XY body and heterochromatic chromocenters of pachytene and diplotene spermatocytes, but only SUMO2/3 localize to pericentromeric chromatin in metaphase I spermatocytes.
Surface-spread meiotic chromatin was immunolabeled with a rat polyclonal antibody specific for the synaptonemal complex marker SYCP3 (red), a mouse monoclonal antibody specific for SUMO1 (A, B, E, F, I, J) or a mouse monoclonal antibody specific to SUMO2/3 (C, D, G, H, K, L). Cells were counterstained with the fluorescent dye 4’, 6-diamidino-2-phenylindole (DAPI; blue) to visualize chromatin (B, D, F, H, L). A–B) Pachytene spermatocyte immunolabeled with the anti-SUMO1 antibody (green) showed localization of SUMO1 to the XY body (arrow) and chromocenters (open arrows). C–D) SUMO2/3 (green) also localize to the XY body (arrow) and chromocenters (open arrows) in pachytene spermatocytes. E–F) The SUMO1 signal (green) persists over the XY body (arrow) in diplotene spermatocytes, but diminishes at the chromocenters (open arrows). G–H) The SUMO2/3 signal (green) also persists in the XY body (arrow) in diplotene spermatocytes, but also diminishes at the chromocenters (open arrows). I–J) SUMO1 is not detected in meiosis I spermatocytes, as seen by the absence of green signal in these cells. Centromeres are identified by anti-SYCP3 labeling (red). K–L) SUMO2/3 (green) accumulate in the vicinity of sister centromeres (arrows) in MI spermatocytes. Magnification = 1000X.
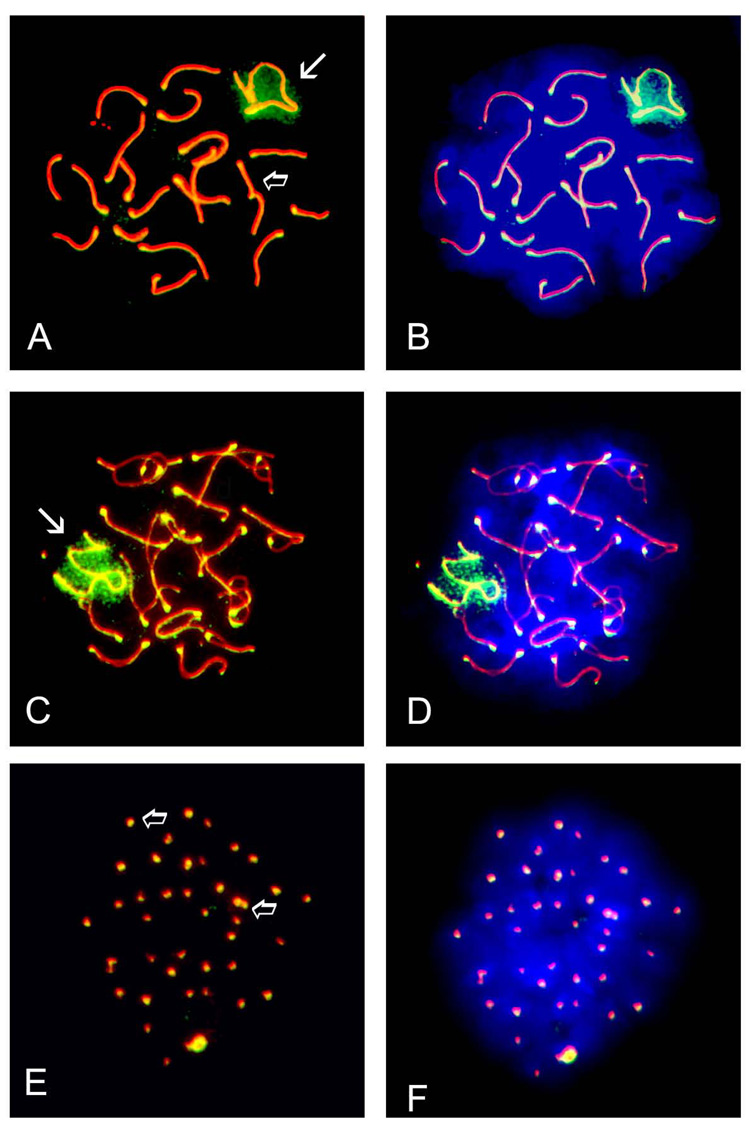
The SUMO E2 conjugating enzyme UBE2I has previously been reported to be expressed in pachytene spermatocytes and to localize to synaptonemal complexes (Kovalenko et al., 1996; Yamaguchi et al., 2006). We further investigated the cell-specific localization of UBE2I in surface-spread chromatin to determine if it correlated with either of the observed SUMO patterns; substages of germ cells were identified on the basis of SYCP3 staining pattern as described above. In pachytene spermatocytes, UBE2I localized to synaptonemal complexes (Fig. 2A, open arrow) and also to the XY body (Fig. 2A, white arrow). Immunolabeling of the XY body (white arrow) and synaptonemal complexes (open arrow) persisted in diplotene spermatocytes (Fig. 2C), while UBE2I was exclusively localized to centromeres in MI spermatocytes (Fig. 2E, open arrows). For the most part, the localization pattern obtained for UBE2I recapitulated that of both SUMO1 and SUMO2/3.
FIGURE 2. The SUMO E2 conjugating enzyme, UBE2I, localizes to synaptonemal complexes, the XY body and centromeres in spermatocytes.
Surface-spread meiotic chromatin was immunolabeled with a mouse monoclonal antibody specific for the synaptonemal complex marker SYCP3 (red) and a rabbit polyclonal antibody specific for the SUMO E2 conjugating enzyme UBE2I (green). A) UBE2I localizes to the XY body (arrow) and associates with the synaptonemal complex in pachytene spermatocytes (open arrow). B) Visualization of chromatin using the fluorescent dye DAPI - same cell as in A. C) UBE2I also localizes to the XY body (arrow) and the synaptonemal complex (open arrow) in diplotene spermatocytes. D) Visualization of chromatin using the fluorescent dye DAPI - same cell as in C. E) Exclusive localization of UBE2I to centromeres (open arrow) as seen by the co-localization of the UBE2I signal with that of SYCP3 to give a yellowish signal in MI spermatocytes. F) Visualization of chromatin using the fluorescent dye DAPI - same cell as in E. Magnification = 1000X.
Expression dynamics of sumoylation pathway genes during spermatogenesis
To identify candidate regulatory factors that could be involved in the spermatogenic sumoylation events that we observed, quantitative RT-PCR (qRT-PCR) was used to assess the expression of 19 genes known to be involved in SUMO metabolism. Four different enriched populations of postnatal male germ cell were isolated by sedimentation at unit gravity using mice of different ages. The expression assay was designed to establish the relative expression of each gene, not to determine the absolute amount of transcript present in each germ cell type tested. Analysis of variance revealed the significant expression differences.
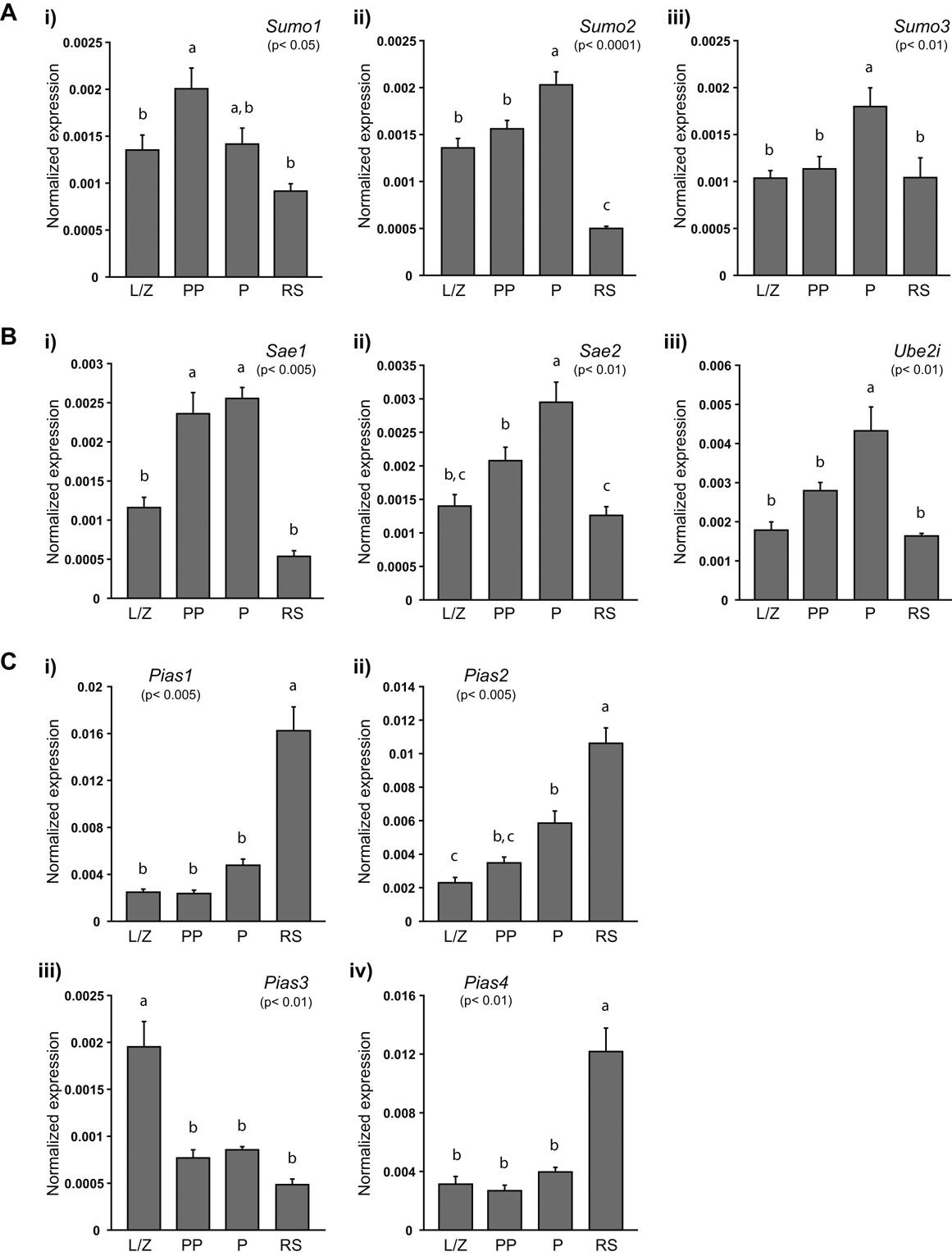
All 19 genes tested were expressed in leptotene/ zygotene spermatocytes (L/Z), prepubertal pachytene spermatocytes (PP), adult pachytene spermatocytes (P) and round spermatids (RS), albeit following different expression patterns (Fig. 3 and Fig. 4). Sumo1 expression (p< 0.05) increased between the leptotene/ zygotene and early pachytene stage, and then decreased slightly in more mature pachytene spermatocytes and reached its lowest levels in round spermatids (Fig. 3A-i). Sumo2 (p< 0.0001) expression was significantly different among all germ cells types except between leptotene/ zygotene spermatocytes and prepubertal pachytene spermatocytes (Fig. 3A-ii). Finally, for Sumo3 (p< 0.01), there were significantly higher transcript levels in adult pachytene spermatocytes then in any other cell type (Fig. 3A-iii).
FIGURE 3. Expression of sumoylation pathway genes in male germ cells.
A) Sumo1 (i), Sumo2 (ii) and Sumo3 (iii) are differentially expressed during spermatogenesis. B) Sae1 (i), Sae2 (ii) and Ube2i (iii) exhibit similar expression patterns in mouse germ cells. C) Pias1 (i), Pias2 (ii), and Pias4 (iv) expression is highest in round spermatids whereas Pias3 (iii) expression is highest in leptotene/ zygotene spermatocytes. Relative quantification of each gene was determined in leptotene/ zygotene (L/Z), prepubertal pachytene (PP) and adult pachytene (P) spermatocytes, as well as in round spermatids (RS) using real-time RT-PCR. Expression was determined in triplicate in each of the three series of germ cells and normalized to 18S expression. For each gene, data are presented as the mean expression in 10 ng of RNA normalized to 18S expression. The p-value obtained for the analysis of variance is given in brackets under each gene name; expression levels in germ cell types not identified by the same letter are considered significantly different. Data reported as mean ± SEM; n= 3 enriched germ cell preparations.
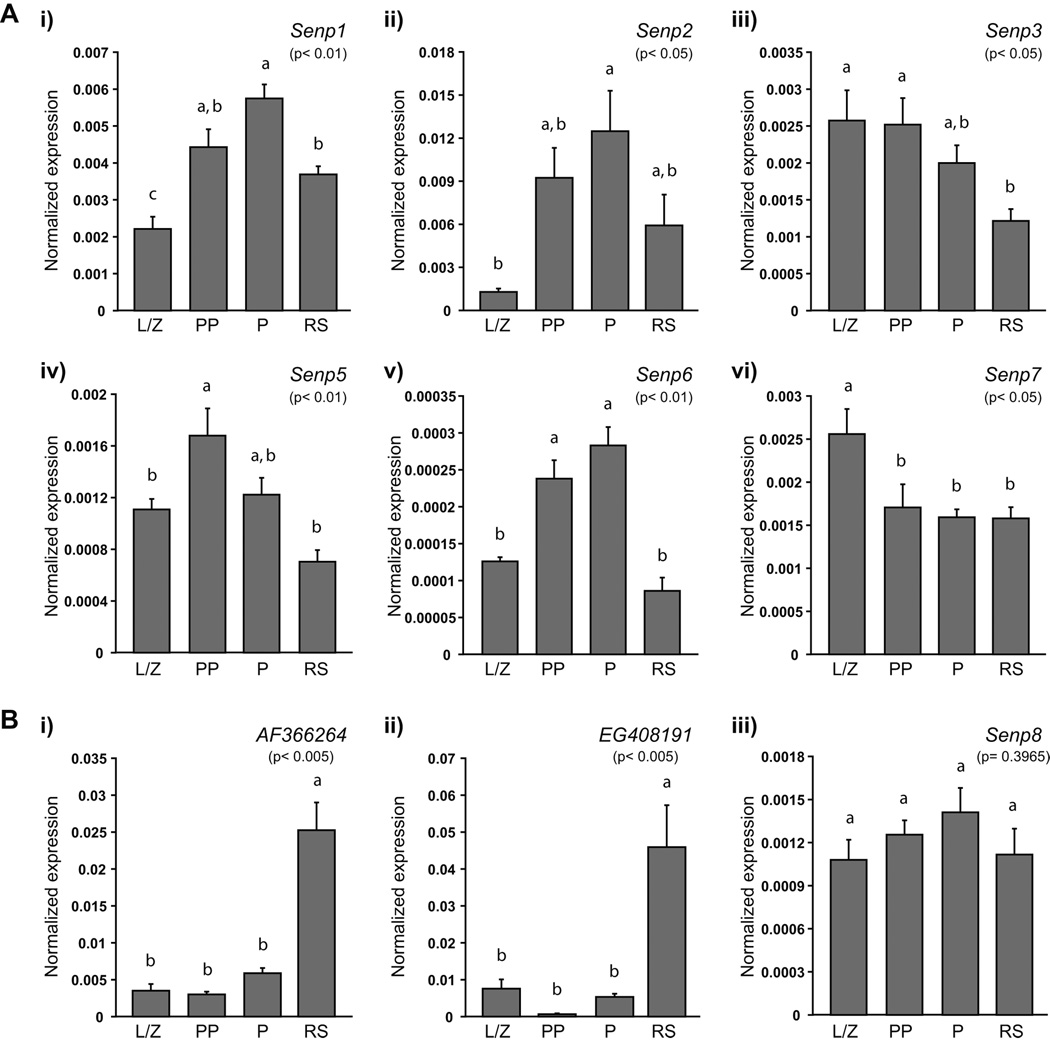
FIGURE 4. Expression of the genes encoding for the SUMO-specific proteases during spermatogenesis.
A) Individual SUMO-specific protease genes present distinct expression profiles. B) Two genes predicted to encode novel SUMO-specific proteases, as well as Senp8, another Senp gene family member, are expressed in male germ cells. AF366264 (i) and EG408191 (ii) are expressed similarly to Pias1, Pias2 and Pias4, while Senp8 (iii) expression does not significantly vary in the germ cells analyzed. Relative quantification of each gene was determined in leptotene/ zygotene (L/Z), prepubertal pachytene (PP) and adult pachytene (P) spermatocytes, as well as in round spermatids (RS) using real-time RT-PCR. Expression was determined in triplicate in each of the three series of germ cells and normalized to 18S expression. For each gene, data are presented as the mean expression in 10 ng of RNA normalized to 18S expression. The p-value obtained for the analysis of variance is given in brackets under each gene name; expression levels in germ cell types not identified by the same letter are considered significantly different. Data reported as mean ± SEM; n= 3 enriched germ cell preparations.
Analysis of the expression of the genes coding for the SUMO E1 activating heterodimeric complex, Sae1 and Sae2, as well as the gene encoding the SUMO E2 conjugating enzyme, Ube2i, showed that all three genes were expressed in a pattern whereby expression of these transcripts increased steadily from the leptotene/zygotene stage to the pachytene stage and then decreased in round spermatids (Fig. 3B). The levels of Sae1 (p< 0.005) transcripts in prepubertal and adult pachytene spermatocytes were higher then in leptotene/ zygotene spermatocytes or round spermatids, whereas they did not vary significantly between these two cell types (Fig. 3B-i). In the case of Sae2 (p< 0.01), expression in adult pachytene spermatocytes differed significantly from that found in all other cell types. Additionally, expression in prepubertal pachytene spermatocytes was also different from that detected in round spermatids (Fig. 3B-ii). Similarly to Sumo3, Ube2i transcript levels were higher in adult pachytene spermatocytes than in any other cell types (p< 0.01; Fig. 3B-iii).
Determination of the expression of the PIAS SUMO E3-protein ligase encoding genes revealed two opposing expression profiles (Fig. 3C). In the case of Pias1 (p< 0.005) and Pias4 (p< 0.01), expression was significantly higher in round spermatids than in any other cell type (Fig. 3C-i and -iv), whereas for Pias2 (p< 0.005), expression in adult pachytene spermatocytes was also higher than in leptotene/ zygotene spermatocytes (Fig. 3C-ii). For Pias3 (p< 0.01), transcript levels were significantly elevated in leptotene/ zygotene spermatocytes compared to levels observed in the other germ cells tested (Fig. 3C-iii).
Assessing the expression of the six genes coding for SUMO-specific proteases (Senp1, Senp2, Senp3, Senp5, Senp6 and Senp7) revealed distinct expression patterns (Fig. 4A). Senp1, Senp2 and Senp6 presented the same expression pattern as Sumo2, Sumo3, Sae1, Sae2 and Ube2i: expression increased from the leptotene/ zygotene stage until the pachytene stage and decreased in round spermatids (Fig. 4A-i, -ii and -v). As observed for Sumo1 (Fig. 3A-i), the transcript levels of Senp5 (p< 0.01) were elevated in prepubertal pachytene spermatocytes compared to leptotene/ zygotene spermatocytes or round spermatids, but they were not significantly different from the levels observed in adult pachytene spermatocytes (Fig. 4A-iv). Senp3 and Senp7 exhibited no specific expression pattern (Fig. 4A-iii and -vi). Senp3 transcript levels did not vary greatly in spermatocytes and decreased slightly in round spermatids (Fig. 4A-iii). Senp7 transcripts were most abundant in leptotene/ zygotene spermatocytes, similar to Pias 3 (Fig. 3C-iii), decreased by about 0.5 fold in prepubertal pachytene spermatocytes and remained so in pachytene spermatocytes and round spermatids (Fig. 4A-vi).
Expression of two predicted genes, AF366264 (also known as Susp4 (Lee et al., 2006); MGI ID: 2667157) and EG408191 (MGI ID: 3644887), as well as that of Senp8, encoding another member of the SENP family that acts on NEDD8, a different ubiquitin-like protein, was also evaluated (Fig. 4B). A very distinct expression profile, reminiscent of the one obtained for Pias1, Pias2 and Pias4, was observed for AF366264 and EG408191 (Fig. 4B-i and -ii); expression was low in spermatocytes and increased by more than 4- and 8- fold, respectively, in round spermatids. Senp8 expression levels were similar in all germ cell populations tested (Fig. 4B-iii). Interestingly, only two of three expression profiles reached statistical significance, the differences in Senp8 expression never being significantly different (p= 0.3965). However, for both AF366264 (p< 0.005) and EG408191 (p< 0.005), expression in round spermatids was significantly higher than in any of the other cell types.
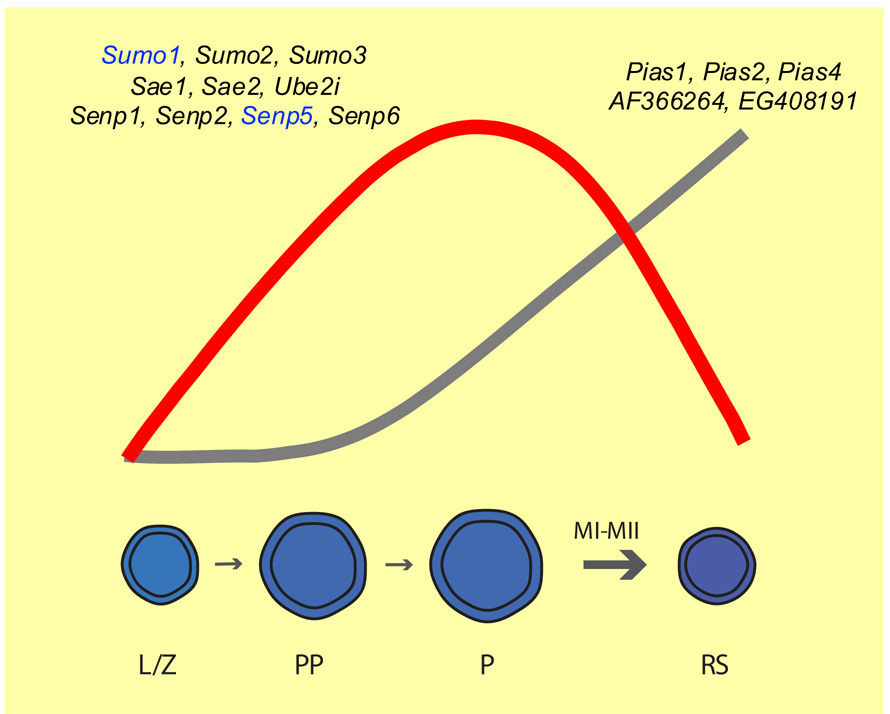
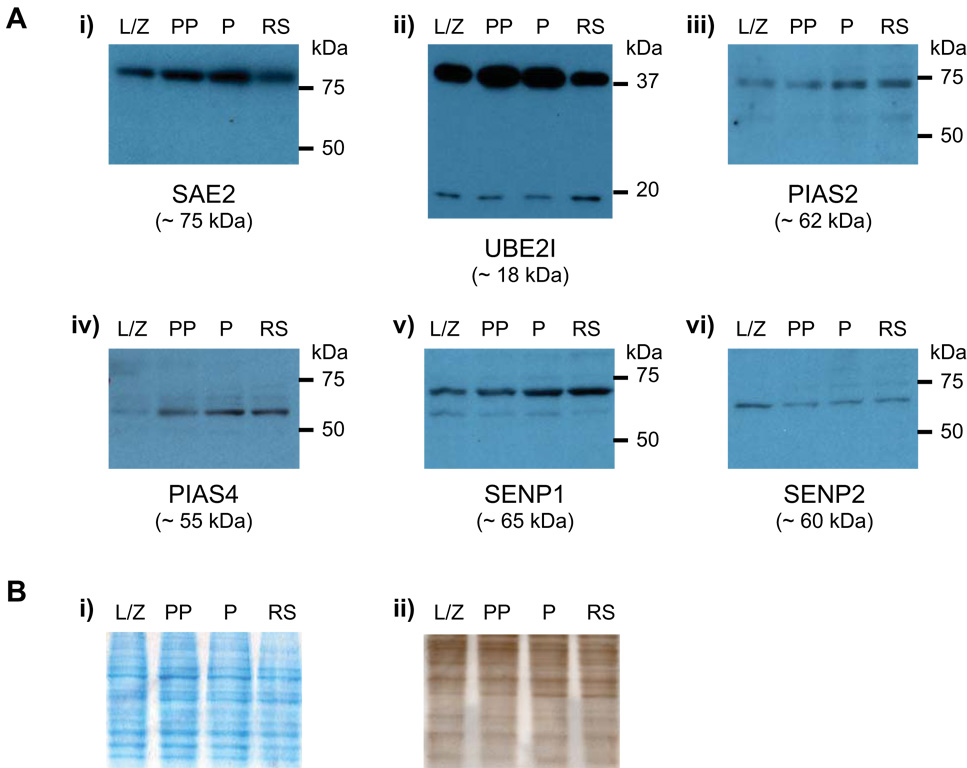
From these analyses, expression of most sumoylation pathway genes can be classified into two distinct profiles, whereby expression is either more prominent during meiosis or in post-meiotic spermatids (summarized in Fig. 6). A protein expression analysis was undertaken to confirm these patterns. Lysates of purified male germ cells prepared from an aliquot of the same cells used for the qRT-PCR studies were analyzed by immunoblotting using commercially available antibodies directed against six candidate protein selected because their mRNA expression corresponded to either one of the two major expression profiles. An antibody directed against SAE2, one of the two subunits of the heterodimeric SUMO E1-specific activating complex, detected a band slightly larger than 75 kDa in all cell types; SAE2 has a calculated molecular weight of ~75 kDa. Levels of this protein appeared highest in adult pachytene spermatocytes, just as established by real-time RT-PCR (Fig. 5A-i). The pattern obtained with the anti-UBE2I antibody confirmed the expression pattern obtained at the mRNA level, and also revealed that most UBE2I in germ cells is in a modified form, as two bands were detected: a smaller band of ~18 kDa, consistent with the calculated molecular weight of UBE2I, and a larger band of ~38 kDa (Fig. 5A-ii).
FIGURE 6. Schematic representation of sumoylation pathway gene dynamics during spermatogenesis.
Expression of sumoylation pathway genes follows two main patterns. The first one, elevated expression during meiosis (red line), identifies sumoylation pathway genes that could be involved in XY body-related sumoylation functions. The second one, elevated expression in post-meiotic spermatids (grey line), suggests genes that could be involved in spermiogenesis-related sumoylation events. Genes expressed according to either one pattern are depicted above each profile. Sumo1 and Senp5 are represented in blue as their expression is higher in prepubertal pachytene spermatocytes than in more mature pachytene spermatocytes. L/Z = leptotene/ zygotene spermatocytes; PP = prepubertal pachytene spermatocytes; P = pachytene spermatocytes; RS = round spermatids; MI, MII= meiotic divisions I & II.
FIGURE 5. Immunoblot detection of SUMO metabolism proteins in male germ cells.
A) Expression of SAE2 (i), UBE2I (ii), PIAS2 (iii), PIAS4 (iv), SENP1 (v) and SENP2 (vi) in leptotene/ zygotene (L/Z), prepubertal pachytene (PP) and adult pachytene (P) spermatocytes, as well as in round spermatids (RS). i) Expression of SAE2 increases from leptonema/ zygonema to pachynema and decreases in round spermatids. ii) Two bands are detected using an antibody against UBE2I, a smaller band of ~ 18kDa, and a larger band of ~ 38kDa. Expression peaks in pachytene spermatocytes. iii) PIAS2 levels are highest in pachytene spermatocytes and round spermatids. iv) An antibody directed against PIAS4 shows increasing levels of the protein with male germ cell development. v) SENP1 levels increase from leptonema/ zygonema until pachynema, and remain elevated in round spermatids. vi) SENP2 is prominent in leptotene/ zygotene spermatocytes. Equal amounts of total protein (10 µg) were loaded in each lane. Apparent molecular weights are given in brackets below each protein name (in kDa), while appropriate molecular weight markers are indicated to the right of each panel (in kDa). Western blots were conducted on each of the three series of germ cells; representative results for one series are presented here. B) i) Gel electrophoresed under identical conditions but stained with GelCode Blue Stain demonstrating equal protein loading in each lane. ii) Membrane stained with India ink following transfer illustrating equal protein loading as well as transfer consistency. Equal amounts of total protein (10 µg) were loaded in each lane. Lane identification is the same as in A).
Antibodies against PIAS2 (Fig. 5A-iii) and PIAS4 (Fig. 5A-iv) also confirmed the patterns obtained by qRT-PCR, demonstrating that expression of both of these proteins increased with male germ cell development, and was more prominent in adult pachytene spermatocytes and round spermatids. For SENP1 (Fig. 5A-v), the immunoblot revealed increasing amounts of the protein during developmental stages from leptotene/ zygotene spermatocytes to adult pachytene spermatocytes, similarly to the real-time RT-PCR results. However, the levels of SENP1 appeared to be sustained in round spermatids, whereas Senp1 transcript levels decreased in these same cells compared to adult pachytene spermatocytes (Fig. 4A-i). Finally, in the case of SENP2 a profile different than the one obtained by qRT-PCR emerged from the immunoblotting analysis (Fig. 5A-vi). Levels of SENP2 were highest in leptotene/ zygotene spermatocytes, whereas transcript levels were lowest in these same cells, suggesting that there might be accumulation of stable SENP2 protein from earlier stages in spermatogenesis, or differences in translation efficiency between cell types. In general, however, the results obtained by immunoblotting corroborated the two expression patterns obtained via real-time RT-PCR.
DISCUSSION
Sumoylation, a post-translational modification of proteins, has previously been linked to a number of spermatogenic processes, including XY body formation, microtubule nucleation, nuclear reshaping and modulation of steroid action (Rogers et al., 2004; Vigodner and Morris, 2005; Vigodner et al., 2006). In this study, we examined the expression and distribution of sumoylation pathway genes using immunocytochemistry, real-time RT-PCR and immunoblotting in order to asses the requirements for protein sumoylation in the male germ line, in particular during prophase I of meiosis. We expanded on our previous finding regarding SUMO1 localization in male germ cells (Rogers et al., 2004), and presented the first localization profile of SUMO2/3 in spread spermatocytes, revealing that SUMO1 and SUMO2/3 are distributed similarly in pachytene and diplotene spermatocytes, but that only SUMO2/3 are detectable in MI spermatocytes. We also found that localization of UBE2I, the SUMO E2 conjugating enzyme, was similar to that of SUMO1 and SUMO2/3, in addition to being exclusively present in synaptonemal complexes (SCs) of both pachytene and diplotene spermatocytes. Finally, the expression analysis conducted on a number of sumoylation pathway genes in enriched populations of spermatocytes and spermatids using qRT-PCR and immunoblotting uncovered factors that could be involved in meiotic and post-meiotic sumoylation events.
Protein sumoylation and meiotic progression in the mouse
Previous studies conducted on mouse and rat spermatocytes have shown that SUMO1 localizes to a specific nuclear heterochromatic territory, the XY body, at pachynema, suggesting that protein sumoylation plays an important role during meiosis in mammals (Rogers et al., 2004; Vigodner and Morris, 2005). Our analysis of SUMO2/3 localization during meiosis in the mouse revealed that, like SUMO1, SUMO2/3 exist in specific nuclear territories in spermatocytes (Fig. 1A–H). In fact, both SUMO1 and SUMO2/3 are detected in the XY body and in chromocenters of pachytene and diplotene spermatocytes. As previously described, the XY body is comprised of the partially paired, transcriptionally repressed heterochromatic sex chromosomes, while meiotic chromocenters contain aggregated centromeric heterochromatin of two or more autosomal bivalents. The results presented here clearly demonstrate that SUMO1 and SUMO2/3 mark these two types of heterochromatic territories in mouse germ cells. In somatic cells, the proteins that are modified by SUMO regulate a variety of functions, including chromatin structure, gene expression and formation of specific nuclear territories such as PML nuclear bodies, and most of these proteins localize to the nucleus (Gill, 2004; Johnson, 2004). Our data suggest that SUMO modification of proteins could accomplish similar functions in male germ cells, in particular in the course of heterochromatic territory formation. However, immunolocalization alone does not allow distinguishing between SUMO conjugated to proteins, and free SUMO. Although there are currently no reports of a functional role for free SUMO, this possibility cannot be excluded and further studies involving substrate identification will be required to resolve this issue. Additionally, whether SUMO modification of proteins targets them to specific nuclear territories, or whether proteins acquire sumoylation in situ remains to be clarified. Localization of UBE2I to the XY body during pachynema and diplonema argues that at least some proteins might be sumoylated directly in the XY body (Fig. 2A–D). In fact, localization of UBE2I to specific sites could be an important factor in controlling the balance of modified substrates, as most proteins appear to be modified only transiently by SUMO, and then rapidly desumoylated, further emphasizing the dynamic nature of SUMO modification.
In yeast, SUMO1-modified proteins accumulate along the length of chromosomes during prophase, and Zip3, a SC-initiating protein possessing SUMO E3 ligase activity, regulates assembly of the synaptonemal complex by promoting the association of Zip1 with Zip3-dependent SUMO conjugates (Cheng et al., 2006). Zip1, like the mammalian SYCP1 protein, is an integral component of the yeast SC, and is found throughout the entire length of the SC. We, as well as others, have not observed SUMO1 or SUMO2/3 on, or in the vicinity of, synaptonemal complexes (Fig.1; Rogers et al., 2004; Vigodner and Morris, 2005). However, we show here that in addition to localizing to the XY body, UBE2I localizes along synaptonemal complexes in both pachytene and diplotene spermatocytes (Fig. 2A–D). Recent work has demonstrated that the carboxyl terminus of UBE2I possesses activity independent of its ability to catalyze SUMO conjugation, suggesting that the C-terminus of this protein could be involved in regulating nuclear receptor and coactivator functions in a sumoylation-independent fashion (Chang et al., 2007). Although UBE2I associates with at least five different nuclear receptors, it is premature to conclude that localization of UBE2I along synaptonemal complexes reflects its sumoylation-independent functions. We cannot exclude that localization of SUMO1 and SUMO2/3 along synaptonemal complexes falls below the limits of detection of the microscopy techniques used, or that rapid SUMO turnover on its substrates makes it a difficult state to capture using immunolocalization.
Different components of the sumoylation machinery have also been linked to chromosome condensation, cohesion and segregation during both mitosis and meiosis in Saccharomyces cerevisiae (Cheng et al., 2007; Watts, 2007). Staining of centromeric chromatin by SUMO1 persists until the end of meiotic prophase in human spermatocytes, and has been implicated in sister chromatid cohesion or kinetochore formation at metaphase (Vigodner et al., 2006). We observed exclusive localization of SUMO2/3 in the vicinity of sister centromeres in MI mouse spermatocytes (Fig. 1I–L). Lack of SUMO1, as well as the restricted localization of SUMO2/3 at MI, underscore both the dynamic interplay between sumoylation and desumoylation events, and the diverse roles SUMO1 and SUMO2/3 might be playing during male germ cell development. UBE2I co-localizes to sister centromeres at MI (Fig. 2D–E), suggesting that proteins are modified in situ, and not simply relocated to sister centromeres, further stressing the dynamic nature of protein sumoylation during meiosis. Studies conducted using yeast and Xenopus egg extracts have shown that sumoylation of the centromeric protein topoisomerase II (TOP2) is necessary for proper centromere cohesion and chromosome segregation at mitosis (Azuma et al., 2003; Azuma et al., 2005; Bachant et al., 2002). In Xenopus, SUMO2 is specifically conjugated to TOP2 during mitosis by the SUMO E3 ligase PIAS4 (Azuma et al., 2005). We have previously shown that TOP2 is involved in chromosome condensation at meiosis (Cobb et al., 1997), and that TOP2A, one of the two TOP2 isoforms, specifically localizes to pericentromeric chromatin of MI spermatocytes in a pattern very reminiscent of the one observed for SUMO2/3 (Fig. 2; Cobb et al., 1999). Protein sumoylation could therefore be involved in centromere function during meiosis by regulating the function of proteins such as TOP2.
The mediators of protein sumoylation during spermatogenesis
Examining the expression of sumoylation pathway genes in four enriched populations of male germ cells revealed that the expression of these genes can be classified into two main general profiles, as summarized in Figure 6. It is important to note that these expression profiles are not mutually exclusive, and should be viewed as reflective of the dynamic state of protein sumoylation during male germ cell development. In the first profile (Fig. 6, red line), genes that could be involved in meiosis-related sumoylation functions are identified, with Sumo1, Sumo2, Sumo3, Sae1, Sae2, Ube2i, Senp1, Senp2, Senp5 and Senp6 being genes whose expression peaks in pachytene spermatocytes. The current SUMO1 and SUMO2/3 immunofluorescence studies, as well as those previously conducted (Rogers et al., 2004; Vigodner and Morris, 2005), are consistent with these Sumo1, Sumo2, and Sumo3 expression results, whereby the highest levels of SUMO1 and SUMO2/3 are found in pachytene spermatocytes (Fig. 1 and 3). Therefore, the genes identified in this expression study could be involved in the sumoylation events detected in pachytene, diplotene and metaphase I spermatocytes in Figure 1. The immunolocalization and immunoblotting results obtained for UBE2I are also strikingly consistent with the expression pattern obtained for Ube2i (Fig. 2, 3B-iii and 5A-ii). Interestingly, the immunoblotting analysis revealed that most UBE2I appears to be modified by another molecule in male germ cells, perhaps by SUMO (Fig. 5A-ii). Although SUMO1, SUMO2 and SUMO3 have calculated molecular weights close to 10 kDa, their intrinsic charge gives them apparent molecular weights closer to 20 kDa. The apparent molecular weight of ~38 kDa of the larger band suggests that UBE2I could be conjugated to SUMO, but further studies will be required to verify this. Of note, the expression profiles of Sae1, Sae2 and Ube2i, as well as the immunoblots for SAE2 and UBE2I, present a striking degree of correspondence, not only amongst themselves, but also to those obtained for Sumo1, Sumo2 and Sumo3 (Fig. 3A and B). As all sumoylation reactions have three common factors, the SUMO E1 activating heterodimer composed of SAE1 and SAE2, and the SUMO E2 conjugating enzyme, UBE2I, Sae1, Sae2 and Ube2i would be predicted to be expressed similarly, and we observed exactly this at both the mRNA and protein levels.
Candidates that could be implicated in spermiogenesis-associated events are uncovered in the second profile (Fig. 6, grey line). Previous work by Vigodner and Morris (2005) showed that SUMO1 is present in chromocenters of round spermatids, and in the perinuclear ring and centrosome of elongating spermatids. Prominent expression of Pias1, Pias2, Pias4, AF366264 and EG408191 in round spermatids could be related to those sumoylation events. In situ hybridization conducted on adult mouse testis sections has revealed that Pias1, Pias2 (also referred to as Piasx) and Pias4 (also known as Piasy) are expressed in a stage-specific manner, and that Pias1 transcripts are especially abundant in round spermatids, whereas Pias2 and Pias4 transcripts are detected in both spermatocytes and spermatids (Tan et al., 2002). Transgenic mice harboring the LacZ reporter gene under the control of a germ cell-specific Pias2 promoter exhibit β-galactosidase activity in pachytene spermatocytes, round spermatids and elongating spermatids (Santti et al., 2003). The Pias1, Pias2 and Pias4 gene expression profiles, as well as the immunoblotting results for PIAS2 and PIAS4, are consistent with these data (Fig. 3C and 5A). Matsuura and colleagues (2005) have shown by immunohistochemistry that PIAS4 localizes to the XY body of pachytene spermatocytes. Their results, as well as ours, suggest that both differential expression and localization of SUMO E3 ligases are involved in controlling the activity of these proteins. Finally, SUMO-specific proteases process the various SUMO moieties with different affinities, and differentiate strongly between processing and deconjugating substrates, in addition to presenting different subcellular localizations and expression patterns, which argues that these enzymes hold well-defined, non-redundant roles in mitotic cells (Mukhopadhyay and Dasso, 2007). Differential expression of the various Senp genes (Fig. 4 and Fig. 5A) suggests that certain SENPs, the ones expressed earlier in meiosis especially, might be involved in SUMO processing, whereas other SENPs would work in SUMO deconjugation. Our immunoblotting results for SENP1 and SENP2 suggest control of protein stability, further emphasizing that specificity and activity of these proteins is complex and likely involves restricting their localization to specific subcellular territories in germ cells. Immunolocalization studies will be instrumental in determining the distribution of all PIAS and SENP proteins in developing male germ cells, and functional analyses will be required to determine the exact role each SUMO-specific ligase and protease plays in male germ cells.
In summary, there are both similarities and striking differences in the localization patterns of both SUMO1 and SUMO2/3 in mouse spermatocytes. Genes encoding proteins that could be involved in meiotic sumoylation events are differentially expressed in male germ cells. Our results clearly illustrate the dynamic nature of SUMO metabolism during spermatogenesis, and suggest that SUMO-modification of proteins may be specifically involved in formation or function of heterochromatic nuclear territories during meiosis.
ACKNOWLEDGEMENTS
We are grateful to Drs. John Eppig and Laura Reinholdt for their thoughtful comments on this manuscript, and to Antonio Planchart for discussions. We also wish to thank Heather Lothrop for her help with animal work, Bobbi-Jo Shirley for her excellent technical assistance in microscopy, and Shuqin Xing for her expertise in statistical analysis. This work was supported by a grant from the NIH to MAH (HD48998) and a Cancer Center Core Grant to The Jackson Laboratory (CA34196). SL is the recipient of fellowships from Les Fonds de la Recherche en Santé du Québec (FRSQ) and the Canadian Institutes of Health Research (CIHR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This study was supported by NIH grant HD48998 to MAH.
REFERENCES
- Azuma Y, Arnaoutov A, Dasso M. SUMO-2/3 regulates topoisomerase II in mitosis. J. Cell Biol. 2003;163:477–487. doi: 10.1083/jcb.200304088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y, Arnaoutov A, Anan T, Dasso M. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. EMBO J. 2005;24:2172–2182. doi: 10.1038/sj.emboj.7600700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell. 2002;9:1169–1182. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- Bellvé AR. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol. 1993;225:84–113. doi: 10.1016/0076-6879(93)25009-q. [DOI] [PubMed] [Google Scholar]
- Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- Chang YL, Huang CJ, Chan JY, Liu PY, Chang HP, Huang SM. Regulation of nuclear receptor and coactivator functions by the carboxyl terminus of ubiquitin-conjugating enzyme 9. Int. J. Biochem. Cell Biol. 2007;39:1035–1046. doi: 10.1016/j.biocel.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Cheng CH, Lo YH, Liang SS, Ti SC, Lin FM, Yeh CH, Huang HY, Wang TF. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006;20:2067–2081. doi: 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CH, Lin FM, Lo YH, Wang TF. Tying SUMO modifications to dynamic behaviors of chromosomes during meiotic prophase of Saccharomyces cerevisiae. J. Biomed. Sci. 2007;14:481–490. doi: 10.1007/s11373-007-9176-0. [DOI] [PubMed] [Google Scholar]
- Cobb J, Reddy RK, Park C, Handel MA. Analysis of expression and function of topoisomerase I and II during meiosis in male mice. Mol. Reprod. Dev. 1997;46:489–498. doi: 10.1002/(SICI)1098-2795(199704)46:4<489::AID-MRD7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Cobb J, Handel MA. Dynamics of meiotic prophase I during spermatogenesis: from pairing to division. Semin. Cell Dev. Biol. 1998;9:445–450. doi: 10.1006/scdb.1998.0202. [DOI] [PubMed] [Google Scholar]
- Cobb J, Miyaike M, Kikuchi A, Handel MA. Meiotic events at the centromeric heterochromatin: histone H3 phosphorylation, topoisomerase II alpha localization and chromosome condensation. Chromosoma. 1999;108:412–425. doi: 10.1007/s004120050393. [DOI] [PubMed] [Google Scholar]
- Cohen PE, Pollard JW. Regulation of meiotic recombination and prophase I progression in mammals. Bioessays. 2001;23:996–1009. doi: 10.1002/bies.1145. [DOI] [PubMed] [Google Scholar]
- de Carvalho CE, Colaiacovo MP. SUMO-mediated regulation of synaptonemal complex formation during meiosis. Genes Dev. 2006;20:1986–1992. doi: 10.1101/gad.1457806. [DOI] [PubMed] [Google Scholar]
- Dobson MJ, Pearlman RE, Karaiskakis A, Spyropoulos B, Moens PB. Synaptonemal complex proteins: occurrence, epitope mapping and chromosome disjunction. J. Cell Sci. 1994;107(Pt 10):2749–2760. doi: 10.1242/jcs.107.10.2749. [DOI] [PubMed] [Google Scholar]
- Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- Handel MA. The XY body: a specialized meiotic chromatin domain. Exp. Cell Res. 2004;296:57–63. doi: 10.1016/j.yexcr.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Hay RT. SUMO-specific proteases: a twist in the tail. Trends Cell Biol. 2007;17:370–376. doi: 10.1016/j.tcb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Hodges CA, LeMaire-Adkins R, Hunt PA. Coordinating the segregation of sister chromatids during the first meiotic division: evidence for sexual dimorphism. J. Cell Sci. 2001;114:2417–2426. doi: 10.1242/jcs.114.13.2417. [DOI] [PubMed] [Google Scholar]
- Hooker GW, Roeder GS. A role for SUMO in meiotic chromosome synapsis. Curr. Biol. 2006;16:1238–1243. doi: 10.1016/j.cub.2006.04.045. [DOI] [PubMed] [Google Scholar]
- Inselman A, Eaker S, Handel MA. Temporal expression of cell cycle-related proteins during spermatogenesis: establishing a timeline for onset of the meiotic divisions. Cytogenet. Genome Res. 2003;103:277–284. doi: 10.1159/000076813. [DOI] [PubMed] [Google Scholar]
- Johnson ES. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Kim KI, Baek SH, Jeon YJ, Nishimori S, Suzuki T, Uchida S, Shimbara N, Saitoh H, Tanaka K, Chung CH. A new SUMO-1-specific protease, SUSP1, that is highly expressed in reproductive organs. J. Biol. Chem. 2000;275:14102–14106. doi: 10.1074/jbc.275.19.14102. [DOI] [PubMed] [Google Scholar]
- Kovalenko OV, Plug AW, Haaf T, Gonda DK, Ashley T, Ward DC, Radding CM, Golub EI. Mammalian ubiquitin-conjugating enzyme Ubc9 interacts with Rad51 recombination protein and localizes in synaptonemal complexes. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2958–2963. doi: 10.1073/pnas.93.7.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Salle S, Mertineit C, Taketo T, Moens PB, Bestor TH, Trasler JM. Windows for sex-specific methylation marked by DNA methyltransferase expression profiles in mouse germ cells. Dev. Biol. 2004;268:403–415. doi: 10.1016/j.ydbio.2003.12.031. [DOI] [PubMed] [Google Scholar]
- Lee MH, Lee SW, Lee EJ, Choi SJ, Chung SS, Lee JI, Cho JM, Seol JH, Baek SH, Kim KI, Chiba T, Tanaka K, Bang OS, Chung CH. SUMO-specific protease SUSP4 positively regulates p53 by promoting Mdm2 self-ubiquitination. Nat. Cell Biol. 2006;8:1424–1431. doi: 10.1038/ncb1512. [DOI] [PubMed] [Google Scholar]
- Matsuura T, Shimono Y, Kawai K, Murakami H, Urano T, Niwa Y, Goto H, Takahashi M. PIAS proteins are involved in the SUMO-1 modification, intracellular translocation and transcriptional repressive activity of RET finger protein. Exp. Cell Res. 2005;308:65–77. doi: 10.1016/j.yexcr.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moilanen AM, Karvonen U, Poukka H, Yan W, Toppari J, Janne OA, Palvimo JJ. A testis-specific androgen receptor coregulator that belongs to a novel family of nuclear proteins. J. Biol. Chem. 1999;274:3700–3704. doi: 10.1074/jbc.274.6.3700. [DOI] [PubMed] [Google Scholar]
- Morelli MA, Cohen PE. Not all germ cells are created equal: aspects of sexual dimorphism in mammalian meiosis. Reproduction. 2005;130:761–781. doi: 10.1530/rep.1.00865. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Rogers RS, Inselman A, Handel MA, Matunis MJ. SUMO modified proteins localize to the XY body of pachytene spermatocytes. Chromosoma. 2004;113:233–243. doi: 10.1007/s00412-004-0311-7. [DOI] [PubMed] [Google Scholar]
- Santti H, Mikkonen L, Hirvonen-Santti S, Toppari J, Janne OA, Palvimo JJ. Identification of a short PIASx gene promoter that directs male germ cell-specific transcription in vivo. Biochem. Biophys. Res. Commun. 2003;308:139–147. doi: 10.1016/s0006-291x(03)01339-1. [DOI] [PubMed] [Google Scholar]
- Schmidt D, Muller S. PIAS/SUMO: new partners in transcriptional regulation. Cell Mol. Life Sci. 2003;60:2561–2574. doi: 10.1007/s00018-003-3129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JA, Hall SH, Hamil KG, Grossman G, Petrusz P, French FS. Protein inhibitors of activated STAT resemble scaffold attachment factors and function as interacting nuclear receptor coregulators. J. Biol. Chem. 2002;277:16993–17001. doi: 10.1074/jbc.M109217200. [DOI] [PubMed] [Google Scholar]
- Turner JM. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- Vigodner M, Morris PL. Testicular expression of small ubiquitin-related modifier-1 (SUMO-1) supports multiple roles in spermatogenesis: silencing of sex chromosomes in spermatocytes, spermatid microtubule nucleation, and nuclear reshaping. Dev. Biol. 2005;282:480–492. doi: 10.1016/j.ydbio.2005.03.034. [DOI] [PubMed] [Google Scholar]
- Vigodner M, Ishikawa T, Schlegel PN, Morris PL. SUMO-1, human male germ cell development, and the androgen receptor in the testis of men with normal and abnormal spermatogenesis. Am. J. Physiol. Endocrinol. Metab. 2006;290:E1022–E1033. doi: 10.1152/ajpendo.00527.2005. [DOI] [PubMed] [Google Scholar]
- Watts FZ. The role of SUMO in chromosome segregation. Chromosoma. 2007;116:15–20. doi: 10.1007/s00412-006-0079-z. [DOI] [PubMed] [Google Scholar]
- Yamaguchi YL, Tanaka SS, Yasuda K, Matsui Y, Tam PP. Stage-specific importin13 activity influences meiosis of germ cells in the mouse. Dev. Biol. 2006;297:350–360. doi: 10.1016/j.ydbio.2006.04.465. [DOI] [PubMed] [Google Scholar]
- Yan W, Santti H, Janne OA, Palvimo JJ, Toppari J. Expression of the E3 SUMO-1 ligases PIASx and PIAS1 during spermatogenesis in the rat. Gene Expr. Patterns. 2003;3:301–308. doi: 10.1016/s1567-133x(03)00045-0. [DOI] [PubMed] [Google Scholar]
- Zhang H, Saitoh H, Matunis MJ. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell Biol. 2002;22:6498–6508. doi: 10.1128/MCB.22.18.6498-6508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XD, Goeres J, Zhang H, Yen TJ, Porter AC, Matunis MJ. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol. Cell. 2008;29:729–741. doi: 10.1016/j.molcel.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]