Abstract
Using ameba coculture, we grew a Naegleria endosymbiont. Phenotypic, genetic, and phylogenetic analyses supported its affiliation as Protochlamydia naegleriophila sp. nov. We then developed a specific diagnostic PCR for Protochlamydia spp. When applied to bronchoalveolar lavages, results of this PCR were positive for 1 patient with pneumonia. Further studies are needed to assess the role of Protochlamydia spp. in pneumonia.
Keywords: Obligate intracellular bacteria, chlamydia-like organisms, pathogenicity, pneumonia, TaqMan PCR, chlamydiae, parachlamydiaceae, protochlamydia, quantitative PCR, dispatch
Recently, a Naegleria endosymbiont (KNic) was observed but remained uncultivable, precluding precise identification (1). We grew a large amount of strain KNic by using Acanthamoeba castellanii, which enabled phenotypic, genetic and phylogenetic analyses that supported its affiliation as Protochlamydia naegleriophila. This new ameba-resistant intracellular bacteria might represent a new etiologic agent of pneumonia because it is likely also resistant to human alveolar macrophages (2,3). Because other Parachlamydiaceae were associated with lung infection (4–6), we assessed the role of Pr. naegleriophila in pneumonia by developing a diagnostic PCR and applying it to bronchoalveolar lavages.
The Study
KNic growth in A. castellanii was assessed by immunofluorescence with in-house mouse anti-KNic and Alexa488-coupled anti-immunoglobulin antibodies (Invitrogen, Eugene, OR, USA). Confocal microscopy (LSM510; Zeiss, Feldbach, Switzerland) confirmed the intracellular location of KNic and demonstrated its rapid growth within A. castellanii. To precisely assess the growth rate, we performed PCR on A. castellanii/KNic coculture by using PrF/PrR primers and PrS probe. After 60 hours, we observed an increased number of bacteria per microliter of 4 logarithms (Appendix Figure).
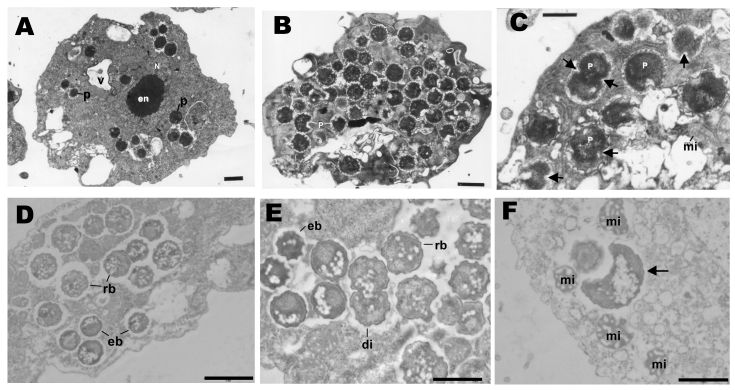
A. castellanii/KNic and N. lovaniensis/KNic cocultures were processed for electron microscopy as described. Ameba filled with bacteria exhibiting 3 developmental stages already described in other Parachlamydiaceae (8) were observed (Figure 1).
Figure 1.
Transmission electron microscopy of Protochlamydia naegleriophila. A) Naegleria lovaniensis trophozoite after transfer of endocytobionts; strain KNic (p) from the original host strain showing 15 coccoid bacteria distributed randomly within the cytoplasm of the host ameba. N, nucleus; en, endosome (karyosome) within the nucleus; v, empty food vacuoles. Magnification ×10,500; bar = 1 μm. B) N. lovaniensis trophozoite jammed with numerous endoparasitic stages of Pr. naegleriophila. Magnification ×16,800; bar = 1 μm. C) Enlarged detail of N. lovaniensis trophozoite with intracytoplasmic stages of Pr. naegleriophila. Some stages show binary fission indicated by the fission furrow (arrows). The endoparasites have a wrinkled gram-negative outer membrane rendering a spiny appearance to the endoparasites. Signs of damage are obvious within the cytoplasm of the host ameba. mi, mitochondria. Magnification ×43,500; bar = 0.5 μm. D) Pr. naegleriophila within vacuoles of Acanthamoebae castellanii ameba 2 days postinfection. Elementary bodies (eb) and reticulate bodies (rb) are visible. Elementary bodies harbor a smooth membrane compared with the reticulate bodies, which have a spiny shape. Magnification ×10,000; bar = 2 μm. E) Enlarged detail of A. castellanii trophozoite with intracytoplasmic stages of Pr. naegleriophila 3 days postinfection. Binary fission is observed (di). Magnification ×20,000; bar = 1 μm. F) Crescent body (arrow) within A. castellanii observed 3 days postinfection. Magnification ×20,000; bar = 1 μm.
To measure the serologic differentiation index (SDI) between strain KNic and other Chlamydia-like organisms, we immunized Balb/c mice to produce anti-KNic antibodies. Purified Pr. amoebophila (ATCC PRA-7), Simkania negevensis (ATCC VR-1471), Parachlamydia acanthamoebae strain Seine, Waddlia chondrophila (ATCC 1470), Neochlamydia hartmannellae (ATCC 50802), Criblamydia sequanensis (CRIB 18), and Rhabdochlamydia crassificans (CRIB 01) antigens were tested by micro-immunofluorescence against mouse anti-KNic antibodies, whereas KNic antigen was tested with serum against all these different Chlamydia-like organisms. SDIs were calculated as described. Serum from mice immunized with KNic showed strong reactivity against autologous antigen (titers of 4,096). Significant cross-reactivity between KNic and Pr. amoebophila (SDI = 7) and P. acanthamoebae (SDI = 10) was observed. Mouse anti-KNic serum did not react with other Chlamydia-like organisms (Table 1). Because cross-reactivity between members of the order Chlamydiales was proportional to the relatedness between each species, the strong cross-reactivity between KNic and Pr. amoebophila supports the affiliation of KNic in the genus Protochlamydia.
Table 1. Antibody titers and serologic differentiation index (SDI) obtained from reciprocal cross-reactions of mouse antiserum with different Chlamydia-like organisms, as determined by immunofluorescence* www.cdc.gov/EID/content/14/1/168-T1.htm.
| Strains | Antigen titers (SDI) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Pr. naegleriophila | Pr. amoebophila | Simkania negevensis | P. a. strain Seine | Waddlia chondrophila | Neoclamydia hartmannellae | Criblamydia sequanensis | R. crassificans | |
| Pr. naegleriophila | 4,096 (0) | 256 (7) | <4 (14) | 512 (10) | 4 (20) | 16 (14) | <4 (20) | <4 (13) |
| Pr. amoebophila | 32 (7) | 256 (0) | <32 (5) | 1,024 (3) | 64 (10) | 32 (0) | 128 (9) | 64 (3) |
| S. negevensis | 32 (14) | 128 (5) | 512 (0) | 64 (12) | 128 (12) | <32 (12) | 512 (8) | 256 (3) |
| P. strain Seine | 128 (10) | 512 (3) | 32 (12) | 16,384 (0) | 64 (19) | 64 (12) | <32 (19) | <32 (12) |
| W. chondrophila | 32 (20) | 128 (10) | 32 (12) | <32 (19) | 32,768 (0) | <32 (18) | 512 (13) | 128 (10) |
| N. hartmannellae | 32 (14) | 128 (0) | <32 (12) | 128 (12) | <32 (18) | 2,048 (0) | <32 (18) | <32 (11) |
| C. sequanensis | 32 (20) | 128 (9) | 128 (8) | 64 (19) | 256 (13) | <32 (18) | 32,768 (0) | 128 (8) |
| R. crassificans | 32 (13) | 128 (3) | 64 (3) | 64 (12) | 64 (10) | <32 (11) | 256 (8) | 256 (0) |
*Pr., Protochlamydia; P. a., Parachlamydia acanthamoebae; R., Rhabdochlamydia. Titers highlighted in gray, previously published in (9), are provided here because these data were used to calculate SDI between Pr. naegleriophila strain KNic and the other Chlamydia-related bacteria.
Taxonomic position of KNic was further defined by sequencing 16Sr RNA (rrs, DQ635609) and ADP/ATP translocase (nnt, EU056171) encoding genes. The rrs was amplified/sequenced using 16SIGF/RP2Chlam primers. The nnt was amplified/sequenced using nntF2p (5′-TGT(AT)GAT(CG)CATGGCAA(AG)TTTC-3′) and nntR1p (5′-GATTT(AG)CTCAT(AG)AT(AG)TTTTG-3′) primers. Genetic and phylogenetic analyses were conducted by using MEGA software (12). The 1,467-bp rrs sequence showed 97.6% similarity with Pr. amoebophila, 91.8%–93.2% with other Parachlamydiaceae, and 85.7%–88.6% with other Chlamydiales. Based on the Everett genetic criteria (13), KNic corresponds to a new species within the Protochlamydia genus because its sequence similarity with Pr. amoebophila is >95% (same genus) and <98.5% (different species). Phylogenetic analyses of rrs gene sequences showed that KNic clustered with Pr. amoebophila, with bootstraps of 98% and 95% in neighbor-joining and minimum-evolution trees, respectively. The 569-bp nnt sequence exhibited 91.1% similarity with Pr. amoebophila, 65.5%–72.6% with other Parachlamydiaceae, and 55.4%–72.6% with other Chlamydiales. Phylogenetic analyses of nnt sequences showed that KNic clustered with Pr. amoebophila. On the basis of these analyses, we propose to name strain KNic “Protochlamydia naegleriophila.”
We then developed a specific diagnostic PCR for Protochlamydia spp. Primers PrF (5′-CGGTAATACGGAGGGTGCAAG-3′) and PrR (5′-TGTTCCGAGGTTGAGCCTC-3′) as well as probe PrS (5′-TCTGACTGACACCCCCGCCTACG-3′) were selected. The 5′-Yakima-Yellow probe (Eurogentec, Seraing, Belgium) contained locked nucleic acids (underlined in sequence above). The reactions were performed with 0.2 μM each primer, 0.1 μM probe, and iTaqSupermix (Bio-Rad, Rheinach, Switzerland). Cycling conditions were as described, and PCR products were detected with ABIPrism7000 (Applied Biosystems, Rotkreuz, Switzerland). Each sample was amplified in duplicate. Inhibition, negative PCR mixture, and extraction controls were systematically tested.
To allow quantification, a plasmid containing the target gene was constructed by cloning PCR products into pCR2.1-TOPO vector (Invitrogen, Basel, Switzerland). Recombinant plasmid DNA quantified using Nanodrop ND-1000 (Witech, Littau, Switzerland) was 10-fold diluted and used as positive controls.
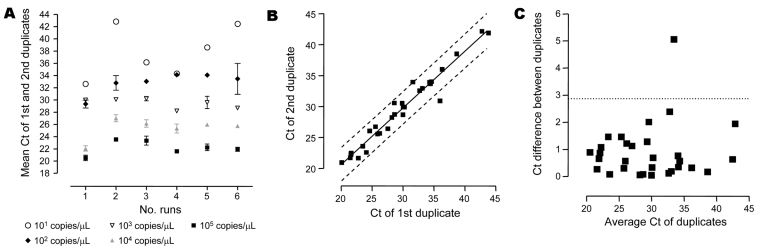
The analytical sensitivity was 10 copy/μL (Figure 2, panel A). Intra-run variability was good (Figure 2 panel B) with a Bland-Altman bias of 0.99 and a limit of agreement of 2.87 (Figure 2, panel A). Inter-run variability was low at high concentration, 1.12, 1.71, 0.82, 1.77 cycles for 105, 104, 103, 102 copies/μL, respectively. Inter-run variability was higher at low concentration, 4.22 cycles for 101 copies/μL (Figure 2, panel A). Analytical specificity was tested with bacterial and eukaryotic DNA (Table 2). The PCR slightly amplified DNA from R. crassificans, another Chlamydia-like organism. No cross-amplification was observed with any other bacteria or with human cells. The absence of cross-amplification of P. acanthamoebae is important because this Chlamydia-related bacteria is considered an emerging agent of pneumonia (4–6).
Figure 2.
A) Intra and inter-run reproducibility of the real-time PCR assessed on duplicate of plasmidic positive controls performed at 10-fold dilutions from 105 to 101 plasmid/μL during 6 successive runs. Standard deviations show the intra-run reproducibility of the real-time PCR. B) Plots of the cycle threshold (Ct) of first and second duplicates, showing intra-run and inter-run variability of the real-time PCR between duplicates of positive control. 95% confidence interval is shown by the dashed lines. C) Bland-Altman graph showing the difference of Ct of both duplicates according to the mean of the Ct of duplicates. The dashed line shows the 95% confidence interval (i.e., limit of agreement).
Table 2. Bacterial and eukaryotic DNA used to determine the specificity of the real-time PCR.
| Bacterial DNA | Source/strain |
|---|---|
| Bordetella pertussis | Clinical specimen |
| Chlamydia trachomatis | Clinical specimen |
| Chlamydophila pneumoniae | ATCC VR-1310 |
| Criblamydia sequanensis | CRIB-18 |
| Enterococcus faecalis | ATCC 29212 |
| Escherichia coli | ATCC 35218 |
| Gardnerella vaginalis | Clinical specimen |
| Haemophilus influenzae | ATCC 49247 |
| Klebsiella pneumoniae | ATCC 27736 |
| Lactobacillus spp. | Clinical specimen |
| Legionella pneumophila | Clinical specimen |
| Listeria monocytogenes | Clinical specimen |
| Moraxella catharralis | Clinical specimen |
| Mycobacterium tuberculosis | Clinical specimen |
| Neisseria lactamica | Clinical specimen |
| Neisseria weaveri | Clinical specimen |
| Neochlamydia hartmanella | ATCC 50802 |
| Parachlamydia acanthamoebae strain BN9 | ATCC VR-1476 |
| Parachlamydia acanthamoebae strain Hall's coccus | ATCC VR-1476 |
| Protochlamydia amoebophila strain UWE25 | ATCC PRA-7 |
| Pseudomonas aeruginosa | ATCC 27853 |
| Rhabdochlamydia crassificans | CRIB-01 |
| Simkania negevensis | ATCC VR-1471 |
| Staphylococcus epidermidis | Clinical specimen |
| Streptococcus agalactiae | ATCC 13813 |
| Streptococcus mutans | Clinical specimen |
| Streptococcus pneumoniae | Clinical specimen |
| Streptococcus pyogenes | ATCC 19615 |
| Waddlia chondrophila | ATCC VR-1470 |
| Eukaryotic DNA | Source/strain |
| Acanthamoebae castellanii | ATCC 30010 |
|
Candida albicans
|
ATCC 10231 |
| Human cells | ATCC CCL-185 |
We tested 134 bronchoalveolar lavage samples from patients with (n = 65) and without (n = 69) pneumonia and extracted DNA by using a Bio-Rad Tissue Kit. One sample was positive, with 543 and 480 copies/μL. This positive result was confirmed using the 16sigF/16sigR PCR (13), which targets another DNA segment. This sequence exhibited 99.6% (284/285) similarity with Pr. naegleriophila strain KNic and 95.1% (269/283) with Pr. amoebophila. The presence of Protochlamydia antigen in the sample was confirmed by immunofluorescence performed using rabbit anti-KNic antibody directly on the bronchoalveolar lavage sample and by ameba coculture (Appendix Figure).
The positive sample was taken from an immunocompromised patient who had cough, dyspnea, and a lung infiltrate. Bronchoscopy examination of the lower respiratory tract showed mucosal inflammation localized at the middle lung lobe. Cytology and Gram stain of the bronchoalveolar lavage showed many leucocytes with macrophages (65%) and neutrophils (23%). Although no antimicrobial treatment was administered prior to bronchoscopy, no other etiologic agent was identified despite extensive microbiologic investigations of bronchial aspirate and bronchoalveolar lavage. Results of Gram stain, auramine stain (for Mycobacterium spp.), and silver stain (for Pneumocystis carinii) tests were negative. Only physiologic oropharyngeal flora could be grown on sheep-blood and chocolate-bacitracin agars. Cell culture, as well as culture for fungi and mycobacteria, remained sterile. Moreover, results of PCRs specific for the detection of Legionella pneumophila, Chlamydophila pneumoniae, and Mycoplasma pneumoniae (15) were all negative. The patient recovered and remained free of symptoms of acute lung infection during the next 20 months.
Conclusions
Isolating new species from environmental and clinical samples is important to better define their epidemiology and potential pathogenicity. We defined the taxonomic position of a novel Naegleria endosymbiont and proposed its affiliation within the Protochlamydia genus as Pr. naegleriophila sp. nov. Moreover, we developed a new PCR targeting Protochlamydia spp., applied it to clinical samples, and identified a possible role of Pr. naegleriophila as an agent of pneumonia.
Protochlamydia naegleriophila (nae.gle.rio′.phi.la Gr. fem.n. Naegleria, name of host cell, Gr. adj. philos, -a friendly to, referring to intracellular growth of Protochlamydia naegleriophila strain KNic within Naegleria amebae). The 16Sr RNA sequence (DQ635609) of KNic is 97.6% similar to that of P. amoebophila, making this organism a member of the genus Protochlamydia. KNic does not grow on axenic media (1) but grows by 4 logarithms in 60 h within A. castellanii. KNic exhibits a Chlamydia-like developmental cycle, with reticulate, elementary, and crescent bodies. The reticulate body is about 900 nm and has a spiny appearance similar to that of P. amoebophila (Figure 2, panel B). To be classified within the Pr. naegleriophila species, a new strain should show a 16Sr RNA similarity >98.5% (13) and similar phenotypic traits.
Supplementary Material
A) Growth rate of Protochlamydia naegleriophila within Acanthamoeba castellanii assessed using a specific quantitative real-time PCR. Number of DNA copies present in culture are plotted according to time postinfection. Standard errors of the mean of duplicate experiments are shown. B) Indirect immunofluorescence preformed using rabbit anti-KNic antibody directly on the bronchoalveolar lavage showing the presence of few Pr. naegleriophila strain KNic or C) in clusters of this obligate intracellular bacteria. D) Immunofluorescence performed on amebal coculture showing the presence of Pr. naegleriophila. This strain was lost in subsequent passages.
Acknowledgments
We thank J.L. Barblan and the staff of Pôle Facultaire Médical Universitaire at the Medical Faculty of Geneva for assisting with electron microscopy analysis, staff of the Cellular Imaging Facility for assisting with confocal microscopy analysis, Gerhild Gmeiner for technical assistance in preparing Naegleria lovaniensis with Protochlamydia strain KNic for electron microscopy, Philip Tarr for reviewing the manuscript, and M. Perrenoud and S. Aeby for technical help.
This work was supported by the Swiss National Science Foundation grants FN 3200BO-105885 and FN 3200BO-116445. G.G. is supported by the Leenards Foundation through a career award entitled “Bourse Leenards pour la relève académique en médecine clinique à Lausanne.”
Biography
Ms Casson is completing a PhD thesis at the University of Lausanne. Her research is dedicated to defining the role of the obligate intracellular Chlamydia-like organisms to humans, discovering new species, and defining their role in pneumonia.
Footnotes
Suggested citation for this article: Casson N, Michel R, Müller K-D, Aubert JD, Greub G. Protochlamydia naegleriophila as etiologic agent of pneumonia. Emerg Infect Dis [serial on the Internet]. 2008 Jan [date cited]. Available from http://www.cdc.gov/EID/content/14/1/168.htm
References
- 1.Michel R, Muller KD, Hauröder B, Zöller L. A coccoid bacterial parasite of Naegleria sp. (Schizopyrenida: Vahlkampfiidae) inhibits cyst formation of its host but not transformation to the flagellate stage. Acta Protozool. 2000;39:199–207. [Google Scholar]
- 2.Greub G, Mege JL, Raoult D. Parachlamydia acanthamebae enters and multiplies within human macrophages and induces their apoptosis. Infect Immun. 2003;71:5979–85. 10.1128/IAI.71.10.5979-5985.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greub G, Raoult D. Microorganisms resistant to free-living amoebae. Clin Microbiol Rev. 2004;17:413–33. 10.1128/CMR.17.2.413-433.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greub G, Raoult D. Parachlamydiaceae: potential emerging pathogens. Emerg Infect Dis. 2002;8:625–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greub G, Berger P, Papazian L, Raoult D. Parachlamydiaceae as rare agents of pneumonia. Emerg Infect Dis. 2003;9:755–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greub G, Boyadjiev I, La Scola B, Raoult D, Martin C. Serological hint suggesting that Parachlamydiaceae are agents of pneumonia in polytraumatized intensive care patients. Ann N Y Acad Sci. 2003;990:311–9. [DOI] [PubMed] [Google Scholar]
- 7.Casson N, Medico N, Bille J, Greub G. Parachlamydia acanthamoebae enters and multiplies within pneumocytes and lung fibroblasts. Microbes Infect. 2006;8:1294–300. 10.1016/j.micinf.2005.12.011 [DOI] [PubMed] [Google Scholar]
- 8.Greub G, Raoult D. Crescent bodies of Parachlamydia acanthamoebae and its life cycle within Acanthamoeba polyphaga: an electron micrograph study. Appl Environ Microbiol. 2002;68:3076–84. 10.1128/AEM.68.6.3076-3084.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casson N, Entenza JM, Greub G. Serological cross-reactivity between different Chlamydia-like organisms. J Clin Microbiol. 2007;45:234–6. 10.1128/JCM.01867-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang R, Raoult D. Antigenic classification of Rickettsia felis by using monoclonal and polyclonal antibodies. Clin Diagn Lab Immunol. 2003;10:221–8. 10.1128/CDLI.10.2.221-228.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas V, Casson N, Greub G. Criblamydia sequanensis, a new intracellular Chlamydiales isolated from Seine river water using amoeba coculture. Environ Microbiol. 2006;8:2125–35. 10.1111/j.1462-2920.2006.01094.x [DOI] [PubMed] [Google Scholar]
- 12.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–63. 10.1093/bib/5.2.150 [DOI] [PubMed] [Google Scholar]
- 13.Everett KD, Bush RM, Andersen AA. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol. 1999;49:415–40. [DOI] [PubMed] [Google Scholar]
- 14.Jaton K, Bille J, Greub G. A novel real-time PCR to detect Chlamydia trachomatis in first-void urine or genital swabs. J Med Microbiol. 2006;55:1667–74. 10.1099/jmm.0.46675-0 [DOI] [PubMed] [Google Scholar]
- 15.Welti M, Jaton K, Altwegg M, Sahli R, Wenger A, Bille J. Development of a multiplex real-time quantitative PCR assay to detect Chlamydia pneumoniae, Legionella pneumophila and Mycoplasma pneumoniae in respiratory tract secretions. Diagn Microbiol Infect Dis. 2003;45:85–95. 10.1016/S0732-8893(02)00484-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Growth rate of Protochlamydia naegleriophila within Acanthamoeba castellanii assessed using a specific quantitative real-time PCR. Number of DNA copies present in culture are plotted according to time postinfection. Standard errors of the mean of duplicate experiments are shown. B) Indirect immunofluorescence preformed using rabbit anti-KNic antibody directly on the bronchoalveolar lavage showing the presence of few Pr. naegleriophila strain KNic or C) in clusters of this obligate intracellular bacteria. D) Immunofluorescence performed on amebal coculture showing the presence of Pr. naegleriophila. This strain was lost in subsequent passages.