Naturally Acquired Human Plasmodium knowlesi Infection, Singapore
Keywords: Malaria, zoonoses, parasites, dispatch
Abstract
We report a case of naturally acquired Plasmodium knowlesi in Singapore, a malaria-free country. Diagnosis was confirmed by PCR with validated species-specific primers. In industrialized countries, free-ranging primates are a potential source of P. knowlesi human infection. P. knowlesi infection is a differential diagnosis of febrile illness acquired in Singapore.
Plasmodium knowlesi is one of the simian malarias that causes human infection (1,2). All 6 published reports of naturally acquired P. knowlesi infection were in rural settings with the largest case series being reported from East Malaysia (3–8). P. knowlesi is commonly misidentified as P. malariae since the blood stages are morphologically similar on microscopy, and molecular methods of detection are necessary for accurate diagnosis (5,8).
Singapore is an urban city-state, which was declared free of human malaria by the World Health Organization in 1982 (9). However, we report a case of locally acquired P. knowlesi malaria, which indicates that this emerging zoonotic parasite should be considered as an etiologic agent of acute febrile illness acquired in Singapore, the southern-most locale reported thus far.
The Case
A previously healthy 20-year-old soldier in the Singapore Army sought treatment on April 28, 2007. He had had a fever for 4 days, along with myalgia, anorexia, nausea, and occasional vomiting. For a year leading up to his illness, he had trained in a forested area inhabited by the long-tailed macaque (Macaca fascicularis) in Lim Chu Kang, northwestern Singapore. His only travel out of Singapore was a 3-week training visit to a non–malaria-endemic foreign country in September 2006 and another visit to Bukit Batok Nature Reserve in western Singapore, an area with monkeys (M. fascicularis) 1 month before onset of symptoms. On initial examination, his temperature was 39.5°C with a pulse rate of 106 beats/min. He was lethargic with tender hepatomegaly. Laboratory investigations showed thrombocytopenia (platelet count 66 × 109/L), hyperbilirubinemia (bilirubin 33 mmol/L [reference level 7–31 mmol/L]), and mild transaminitis (alanine transaminase 64 U/L [reference level 17–63 mmol/L] and aspartate transaminase 67 U/L [reference level 15–41 mmol/L]).
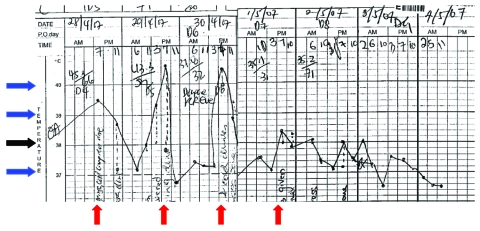
Initial diagnosis was dengue fever, which is endemic in Singapore. The patient experienced daily fever spikes from 39.5°C to 40.4°C (Figure 1). When fever persisted (40.4°C on day 6 of his illness, hospital day 3), the clinical picture was atypical for dengue fever. Blood films for malaria parasites were ordered, because introduced cases of malaria have been reported in Singapore (10). Microscopy showed Plasmodium parasitemia of 0.2% (equivalent to 7,700 parasites/mmol/L blood) with morphologic features consistent with P. malariae. Results of dengue reverse transcription–PCR (RT-PCR) on serum, 2 sets of blood cultures, and Rickettsia typhi serologic testing were negative. Results of a chest radiograph and ultrasound of the abdomen were normal.
Figure 1.
Patient’s temperature chart showing fever spikes 24 h apart at approximately 7 PM daily (red arrow). The black arrow denotes 38°C, and each blue arrow denotes a difference of 1°C from the neighboring arrow.
Oral chloroquine was started with an initial dose of 600 mg base, followed by 300 mg base 6 h later and another 2 doses over the next 2 days. He defervesced rapidly; blood smears were negative 3 days after chloroquine therapy. At 2 weeks follow-up, he was clinically well.
Because P. malariae infection was not consistent with the clinical findings of the initial examination, we investigated further to determine the etiology of this case. End-point nested Plasmodium genus- and species-specific nested PCR carried out on DNA extracted from whole blood samples were positive for Plasmodium sp. but negative for the 4 species that cause human malaria (Table) (11). Similarly, the sample was negative on real-time PCR for the 4 human parasites (12). P. knowlesi species-specific PCR resulted in a 153-bp fragment indicative of P. knowlesi (5). This 153-bp PCR product was directly sequenced and verified in the BLAST database (www.ncbi.nlm.nih.gov/blast/Blast.cgi) to match only P. knowlesi small subunit ribosomal RNA (SSU rRNA).
Table. Primers used for the PCR investigation of the clinical sample from Singapore*.
| Primers | Forward | Sequence (5′ → 3’) | Reverse | Sequence (5′ → 3’) | Results |
|---|---|---|---|---|---|
| Nest 1, genus specific primers | |||||
| Genus specific (11) |
rPLU 1 |
TCA AAG AAT AAG CCA TGC AAG TGA |
rPLU 2 |
TAC CCT GTT GTT GCC TTA AAC TCC |
+ |
| Nest 2, genus- and species-specific primers | |||||
| Genus specific (11) | rPLU 3 | TTT TTA TAA GGA TAA CTA CGG AAA AGC TGT | rPLU 4 | TAC CCG TCA TAG CCA TGT TAG GCC AAT ACC | + |
| Plasmodium knowlesi specific (5) | Pmk8 | GTT AGC GAG AGC CAC AAA AAA GCG AAT | Pmkr9 | ACT CAA AGT AAC AAA ATC TTC CGT A | + |
| P. vivax specific (11) | rVIV1 | CGC TTC TAG CTT AAT CCA CAT AAC TGA TAC | rVIV2 | ACT TCC AAG CCG AAG CAA AGA AAG TCC TTA | – |
| P. falciparum specific (11) | rFAL1 | TTA AAC TGG TTT GGG AAA ACC AAA TAT ATT | rFAL2 | ACA CAA TGA ACT CAA TCA TGA CTA CCC GTC | – |
| P. malariae specific (11) | rMAL1 | ATA ACA TAG TTG TAC GTT AAG AAT AAC CGC | rMAL2 | AAA ATT CCC ATG CAT AAA AAA TTA TAC AAA | – |
| P. ovale specific (11) | rOVA1 | ATC TCT TTT GCT ATT TTT TAG TAT TGG AGA | rOVA2 | GGA AAA GGA CAC ATT AAT TGT ATC CTA GTG | – |
*PCR was carried out at the Environmental Health Institute and cycling conditions used were as described in the references shown in parentheses. +, positive; –,negative.
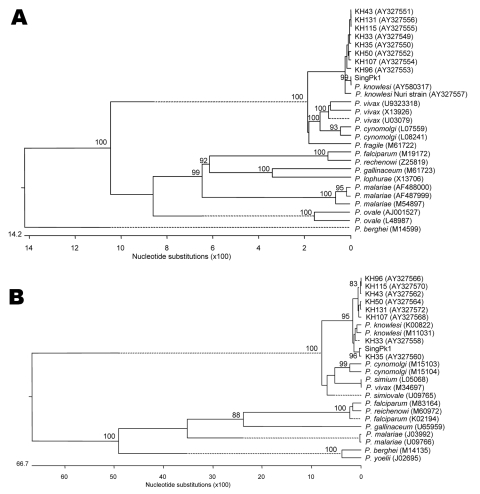
We confirmed the pathogen by using previously described approaches to compare the sequences of the 5′ and 3′ ends of the circumsporozoite protein (csp) gene (13), as well as the gene encoding of the sSSU rRNA (5) in our case sample, to other Plasmodium parasites. Sequences were obtained by direct sequencing of PCR products and aligned by using the ClustalW method (EMBL-EBI, Hixton, Cambridge, UK); we constructed phylogenetic trees by using the MegAlign software (DNASTAR Inc, Madison, WI, USA). The case sample (denoted as SingPk1) clustered with other P. knowlesi isolates and is clearly distinct from other Plasmodium species (Figure 2).
Figure 2.
Phylogenetic trees comparing our case sample (denoted as SingPk1) with other Plasmodium species, based on SSU rRNA (A) and csp (B) sequences. Species and sequences used were selected to match those previously reported (5). Figures on the branches are bootstrap percentages based on 1,000 replicates, and only those above 80% are shown. GenBank accession numbers are in parentheses.
Conclusions
We describe an unequivocal case of P. knowlesi infection supported by clinical findings and laboratory diagnostics classic for this pathogen. Similar to our patient, the classic scenario that raises the suspicion of P. knowlesi infection is a blood smear consistent with P. malariae but with parasitemia exceeding 5,000 per mmol/L blood, daily fever spikes, and pronounced symptoms, features atypical for P. malariae infection (1,5). The daily fever spike is due to the P. knowlesi 24-hour asexual life cycle, the shortest of all primate malarias (8). P. malariae has a 72-hour asexual life cycle and manifests as chronic, asymptomatic infection with low level parasitemia (5).
As in this case, P. knowlesi is commonly mistaken for P. malariae by microscopy due to similarity of the blood stages (5). P. knowlesi can be misidentified as P. falciparum if only ring forms are identified (5). The P. knowlesi–specific primers used by both independent laboratories have previously been shown not to detect any of the 4 Plasmodium species that cause human infection or the 3 agents that cause simian malaria: P. cynomolgi, P. fieldi, and P. fragile (5). PCR detection using P. knowlesi–specific primers, followed by sequencing and phylogenetic analyses of the csp and SSU rRNA genes confirmed P. knowlesi infection in our patient. This report extends the range of natural P. knowlesi human infection from East Malaysia, peninsular Malaysia, Thailand, and Myanmar to Singapore, an industrialized country that had been declared malaria-free by WHO (3–8).
Our patient likely acquired the infection in the forested area in Lim Chu Kang where he had been training for the entire year before his illness. Experimental P. knowlesi studies show a prepatent period of 9–12 days in humans (14). P. knowlesi has no liver hypnozoite stage and does not cause relapse (1). The patient’s pevious overseas travel 7 months before and his visit to Bukit Batok Nature Reserve a month before onset of illness are beyond the incubation period.
P. knowlesi’s natural hosts are the macaques, M. fascicularis and Macaca nemestrina (1). Notably, the first studies on P. knowlesi were on a parasite isolated from a macaque imported into India from Singapore (2). M. fascicularis and Presbytis femoralis are the 2 native monkeys in Singapore, with M. fascicularis being the only species in Lim Chu Kang and Bukit Batok Nature Reserve (15). Mosquitoes of the Anopheles leucophyrus group have been identified as vectors of P. knowlesi and are present in surrounding countries in southeast Asia (1,8). Studies are ongoing to determine potential mosquito vectors and whether macaques are hosts of P. knowlesi in Singapore.
Our patient’s condition was diagnosed within 6 days of illness, and the infection responded rapidly to oral chloroquine. Although most patients’ infections respond well to antimalarial agents, 4 fatal cases of P. knowlesi infection were reported recently in patients ages 39 to 69 years, whose conditions were all diagnosed within 7 days of symptom onset (8). Common clinical features included fever, abdominal pain, thrombocytopenia (platelet count <30 × 109/ μL), renal impairment, and jaundice. All of the patients received a misdiagnosis of P. malariae infection.
P. knowlesi infection should be considered as an etiologic agent of malaria acquired in Singapore, particularly in cases with daily fever spikes and blood smears suggestive of P. malariae. Epidemiologic studies into the parasite’s reservoir and mosquito vector will be important in the prevention of this emerging zoonotic disease.
Acknowledgments
We thank Professor Balbir Singh for his useful comments and advice on the revision of our manuscript; David Lye for comments on the manuscript; Sharon Chan for sharing her data on monkeys in Singapore; Kuperan Ponnudurai for the photomicrograph; and Victor Koh, Janet Chew, and Shirley Seah for their assistance with molecular and sequence analyses.
Biography
Dr Ng is currently a registrar at the Communicable Disease Centre in Singapore. His research interests include HIV medicine and tropical disease.
Footnotes
Suggested citation for this article: Ng OT, Ooi EE, Lee CC, Jarrod LP, Ng LC, Wong PS, et al. Naturally acquired human Plasmodium knowlesi infection, Singapore. Emerg Infect Dis [serial on the Internet]. 2008 May [date cited]. Available from http://www.cdc.gov/EID/content/14/5/814.htm
References
- 1.Coatney GR, Collins WE, Warren M, Contacos PG. The primate malarias [CD-ROM; original book published 1971]. Version 1.0. Atlanta: Centers for Disease Control and Prevention; 2003. [Google Scholar]
- 2.Knowles R, Das Gupta BM. A study of monkey-malaria and its experimental transmission to man. Ind Med Gaz. 1932;67:301–20. [PMC free article] [PubMed] [Google Scholar]
- 3.Chin W, Contacos PG, Coatney GR, Kimball HR. A naturally acquired quotidian-type malaria in man transferable to monkeys. Science. 1965;149:865. 10.1126/science.149.3686.865 [DOI] [PubMed] [Google Scholar]
- 4.Fong YL, Cadigan FC, Coatney GR. A presumptive case of naturally occurring Plasmodium knowlesi in man in Malaysia. Trans R Soc Trop Med Hyg. 1971;65:839–40. 10.1016/0035-9203(71)90103-9 [DOI] [PubMed] [Google Scholar]
- 5.Singh B, Kim Sung L, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–24. 10.1016/S0140-6736(04)15836-4 [DOI] [PubMed] [Google Scholar]
- 6.Jongwutiwes S, Putaporntip C, Iwasaki T, Sata T, Kanbara H. Naturally acquired Plasmodium knowlesi malaria in human, Thailand. Emerg Infect Dis. 2004;10:2211–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu HM, Li J, Zheng H. Human natural infection of Plasmodium knowlesi. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2006;24:70–1. [PubMed] [Google Scholar]
- 8.Cox-Singh J, Davis TME, Lee KS, Shamsul SSG, Matusop A, Ratnam S, et al. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin Infect Dis. 2008;46:165–71. 10.1086/524888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goh KT. Eradication of malaria from Singapore. Singapore Med J. 1983;24:255–68. [PubMed] [Google Scholar]
- 10.Chiam PT, Oh HM, Ooi EE. Localised outbreak of Falciparum malaria in Singapore. Singapore Med J. 2003;44:357–8. [PubMed] [Google Scholar]
- 11.Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60:687–92. [DOI] [PubMed] [Google Scholar]
- 12.Rougemont M, Van Saanen M, Sahli R, Hinrikson HP, Bille J, Haton K. Detection of four Plasmodium species in blood from humans by 18S rRNA gene subunit-based and species specific real-time PCR assays. J Clin Microbiol. 2004;42:5636–43. 10.1128/JCM.42.12.5636-5643.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCutchan TF, Kissinger JC, Touray MG, Rogers MJ, Li J, Sullivan M, et al. Comparison of circumsporozoite proteins from avian and mammalian malarias: biological and phylogenetic implications. Proc Natl Acad Sci U S A. 1996;93:11889–94. 10.1073/pnas.93.21.11889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chin W, Contacos PG, Collins WE, Jether MH, Alpert E. Experimental mosquito-transmission of Plasmodium knowlesi to man and monkey. Am J Trop Med Hyg. 1968;17:355–8. [DOI] [PubMed] [Google Scholar]
- 15.Yang CMMK. Yong K, Lim KKP. Wild mammals of Singapore. In: Chou LM, PKL Ng. Essays in zoology, papers commemorating the 40th anniversary of the Department of Zoology. Singapore: National University of Singapore; 1990. p. 1–24. [Google Scholar]