Abstract
Streptococcus suis sequence type (ST) 7 has been spreading throughout China. To determine events associated with its emergence, we tested 114 isolates. In all 106 ST7 strains responsible for human outbreaks and sporadic infections, the tetracycline-resistance gene, tetM, was detected on the conjugative transposon Tn916. Horizontal transmission of tetM is suspected.
Keywords: Streptococcus suis, multilocus sequence typing (MLST), tetracycline resistance, transposon, dispatch
A large outbreak of Streptococcus suis serotype 2 infection emerged in the summer of 2005 in Sichuan Province, People’s Republic of China, and resulted in 215 cases and 38 deaths among humans (1). Sporadic infections were identified in 4 other provinces. A smaller, previously overlooked, outbreak occurred in Jiangsu Province in 1998; 25 cases and 14 deaths were reported (1,2). The causative agent of the Sichuan and Jiangsu outbreaks was identified as a clone of S. suis sequence type (ST) 7 (3). ST7 was first identified in 1996 in a patient with meningitis in Hong Kong and later caused the 1998 outbreak in Jiangsu; it spread further to cause the largest outbreak in Sichuan in 2005 (3,4). The spread of S. suis ST7 across China underscores the need to better understand the genetic and ecologic events associated with its emergence as an important pathogen in humans.
The Study
Using the MICroSTREP Plus system (Dade Behring, Deerfield, IL, USA), we tested 114 ST7 isolates from China and found that all isolates were resistant to tetracycline and susceptible to 12 of 13 antimicrobial drugs. Of these 114, 6 were isolated in 2006, 84 were from human patients and 8 from diseased pigs in the 2005 Sichuan outbreak, 7 were from sporadic human cases and 3 from diseased pigs in other provinces in 2005, and 4 were from human patients and 2 from diseased pigs in the 1998 Jiangsu outbreak (Table 1). The isolates were susceptible to penicillin, ampicillin, cefotaxime, ceftriaxone, cefepime, meropenem, levofloxacin, chloramphenicol, erythromycin, azithromycin, clindamycin, and vancomycin. In contrast, 7 of 12 S. suis serotype 2 strains from other countries and 18 of 34 serotype reference strains were resistant to tetracycline; 3 tetracycline-resistant strains were also resistant to erythromycin, azithromycin, and clindamycin.
Table 1. Source, serotype, sequence type, and tetracycline-resistant genes in Streptococcus suis strains*.
| No. strains |
Source (no.) |
Place of
isolation |
Year of
isolation (no.) |
Serotype (no.) |
ST (no.) |
tet gene (no. positive) |
Tn916 (no.) |
Virulence genes (no.) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
cps2j
|
mrp
|
sly
|
ef
|
||||||||
| 98 outbreak-associated ST7 strains in China | |||||||||||
| 84 | Human patients | Sichuan, China | 2005 | 2 | ST7 | tetM (84) | Intact (84) | + (84) | + (84) | + (84) | + (84) |
| 8 | Diseased pigs | Sichuan, China | 2005 | 2 | ST7 | tetM (8) | Intact (8) | + (8) | + (8) | + (8) | + (8) |
| 4 | Human patients | Jiangsu, China | 1998 | 4 | ST7 | tetM (4) | Intact (4) | + (4) | + (4) | + (4) | + (4) |
| 2 |
Diseased pigs |
Jiangsu, China |
1998 |
2 |
ST7 |
tetM (2) |
Intact (2) |
+ (2) |
+ (2) |
+ (2) |
+ (2) |
| 8 ST7 strains isolated from sporadic cases in China | |||||||||||
| 5 | Human patients | 6 provinces, China | 2005 (2) 2006 (3) | 2 | ST7 | tetM (5) | Intact (5) | + (5) | + (5) | + (5) | + (5) |
| 3 |
Diseased pigs |
Jiangxi, China |
2005 |
2 |
ST7 |
tetM (3) |
Intact (3) |
+ 3) |
+ (3) |
+ (3) |
+ (3) |
| 7 ST1 and 1 untypeable strain isolated from sporadic cases in China | |||||||||||
| 5 | Human patients | Guizhou, Guangxi (4) | 2005 | 2 (4) 14 | ST1 | tetM (4) | Intact (4) | + (4) | + (5) | + (5) | + (5) |
| 3 |
Human patients |
3 provinces |
2006 |
2 |
ST1 (2)
UT |
tetO (2)
tetM |
Intact (1) |
+ (3) |
+ (3) |
+ (3) |
+ (3) |
| 12 serotype 2 strains from other countries | |||||||||||
| 5 | Human patients | Netherlands (2), France (3) | NA | 2 | ST1 | tetO (3) | + (5) | + (5) | + (5) | + (4) | |
| 3 | Diseased pigs | Netherlands, France, England | NA | 2 | ST1 | + (3) | + (3) | + (3) | + (2) | ||
| 4 |
Human patients (2), healthy pigs, diseased pigs |
Canada (3), England |
NA |
2 |
ST25 |
tetO (4) |
|
+ (4) |
– |
– |
– |
| 34 serotype reference strains | |||||||||||
| 1 | Human patients | Netherlands | NA | 14 | ST6 | – | – | + | – | ||
| 1 | Diseased pig | Denmark | NA | 13 | ST71 | tetM | Intact | – | – | + | – |
| 25 | Diseased pigs | Canada (11), Denmark (5), Netherlands (9) | NA | 1/2, 2–12, 15–16, 22–30, 32, 34 | ST1, ST35, ST53–55, ST68, ST69, ST72–73, ST75, ST77–78, ST80–82, ST87, ST91–92, UT (7) | tetO (11) | + (2) | + (2) | + (7) | + (1) | |
| 2 | Diseased calves | Canada, United States | NA | 20, 31 | ST70 (1), UT (1) | tetO (1) | – | – | – | – | |
| 1 | Diseased lamb | Canada | NA | 33 | UT | – | – | – | – | ||
| 4 | Healthy pigs | Canada | NA | 17–19, 21 | ST76 (2), ST79, UT | tetO (4) | – | – | + (2) | – | |
*ST, sequence type; tet, tetracycline; UT, untypeable by multilocus sequence typing; NA, not available.
Multilocus sequence typing analysis showed that of the 114 isolates from China, 106 were typed as ST7: 98 from the Sichuan and Jiangsu outbreaks, 5 from sporadic infections in other provinces in 2005 and 2006, and 3 from diseased pigs from other provinces in 2005. Of the other 8 isolates from sporadic cases in 2005 and 2006, 7 were ST1 and 1 was untypeable. Of the 12 serotype 2 strains from other countries, 8 were ST1 and 4 were ST25. Of the 34 serotype reference strains, serotype 2 strain R735 was ST1, 10 serotypes were untypeable, and 22 STs were identified as ST6 (serotypes 17 and 19), ST35, ST53-55, ST68-73, ST75-82, ST87, or ST91-2. Serotype 17 and 19 strains were identified as ST76 (Table 1) (5,6).
PCR was used to screen all isolates for tetracycline resistance genes; primers specific for tetABCDEGKLMOQS were used (7,8). Of the 114 tetracycline-resistant isolates from China, 111 (all 106 ST7 strains and 5 of 7 ST1 strains) harbored the tetM gene. The tetO gene was carried by 1 ST1 and 1 sequence-untypeable strain. All 7 tetracycline-resistant serotype 2 strains from other countries and 16 of 18 tetracycline-resistant strains in the 34 reference serotypes carried the tetO gene (Table 1) (5,6). The only other tetM-positive strain was from serotype 13, an ST71 isolated from a diseased pig in Denmark (Table 1). The PCR results were confirmed by sequencing the PCR-synthesized fragments.
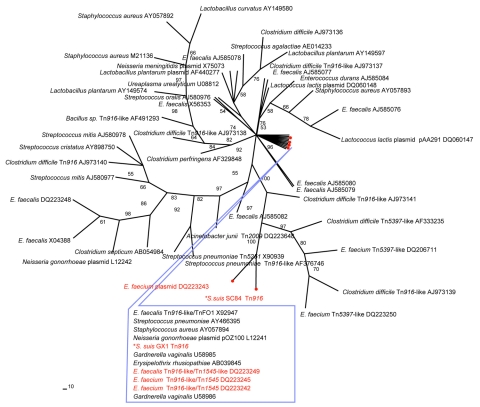
To further characterize the tetM genes, the open reading frame (ORF) was completely sequenced by using 16 selected strains: 9 isolates from humans and 1 from a pig from the Sichuan outbreak; 3 from sporadic infections in Guangxi, Jiangsu, and Guangdong; 1 from a diseased pig in Jiangxi Province in 2005; and 2 from the Jiangsu outbreak in 1998 (Table 1). Sequence alignments showed 2 groups (GenBank accession nos. EF101931, EF016118). The first group comprised 15 of the 16 isolates typed as ST7 (3). The second group had only 1 isolate, GX1, typed as ST1 (3). The sequences of tetM gene for ST7 (strain SC84) and ST1 (GX1) were 1,920 and 1,917 bp, respectively, with 90 nt variations between the 2 sequences leading to 32 aa changes. Comparison of the 53 tetM sequences with those from public databases showed that the tetM of S. suis SC84 was most related to Enterococcus faecium isolate 9830470-4 plasmid pYA470-4 (DQ223243) (7). The tetM sequence of S. suis ST1 strain GX1 was most closely related to S. pneumoniae Tn916-like/Tn2009 (AY466395) and to Gardnerella vaginali (U58986) (Figure 1) (9).
Figure 1.
Phylogenetic relationship of the tetM sequences of Streptococcus suis. An unrooted maximum-parsimony tree was based on multiple aligned partial tetM sequences of 2 S. suis (asterisk) and 53 reference sequences retrieved from GenBank. The alignment length for the analysis was 1,415 bp. If available, the designation of the tetM-carrying plasmid or transposon is indicated, followed by the GenBank accession number. Percent bootstrap support at each internal node was based on 200 replicate trees. The sequences of known pig origin are marked in red.
Because the gene tetM is reported to be associated with transposon Tn916, we designed 24 pairs of primers targeting its 24 ORFs based on published Tn916 sequences (Table 2). The complete sequence of Tn916 from SC84 was obtained by sequencing the PCR-synthesized fragments. Between Tn916 of SC84 and plasmid pYA470-4 of E. faecium, we observed 133 nt variations, 91 of which were in the tetM gene, 3 in the integrase gene, 1 in excisionase gene, and 38 in 10 additional ORFs. PCR showed that 111 isolates from China and 1 from Denmark have intact Tn916 (Table 1).
Table 2. Primers used to detect tet genes and conjugative transposon Tn916 in Streptococcus suis*.
| Gene | Primers (5′ → 3′) | Product size, bp | Annealing temperature, °C |
|---|---|---|---|
| tetA | GCTACATCCTGCTTGCCTTC; CATAGATCGCCGTGAAGAGG | 210 | 55 |
| tetB | TTGGTTAGGGGCAAGTTTTG; GTAATGGGCCAATAACACCG | 659 | 55 |
| tetC | CTTGAGAGCCTTCAACCCAG; ATGGTCGTCATCTACCTGCC | 418 | 55 |
| tetD | AAACCATTACGGCATTCTGC; GACCGGATACACCATCCATC | 787 | 55 |
| tetE | AAACCACATCCTCCATACGC; AAATAGGCCACAACCGTCAG | 278 | 55 |
| tetG | GCTCGGTGGTATCTCTGCTC; AGCAACAGAATCGGGAACAC | 468 | 55 |
| tetK | TCGATAGGAACAGCAGTA; CAGCAGATCCTACTCCTT | 169 | 55 |
| tetL | TCGTTAGCGTGCTGTCATTC; GTATCCCACCAATGTAGCCG | 267 | 55 |
| tetM | GTGGACAAAGGTACAACGAG; CGGTAAAGTTCGTCACACAC | 406 | 55 |
| tetO | AACTTAGGCATTCTGGCTCAC; TCCCACTGTTCCATATCGTCA | 515 | 55 |
| tetQ | TTATACTTCCTCCGGCATCG; ATCGGTTCGAGAATGTCCAC | 904 | 55 |
| tetS | CATAGACAAGCCGTTGACC; ATGTTTTTGGAACGCCAGAG | 667 | 55 |
| ORF 24 | ATGAGGTGTCTATTTTTTTA; TTATTGGCTGAATGAATGTT | 120 | 52 |
| ORF 23 | AATTTGTGATTCCCAACATG; CGTCAGCATGTAAAAGGTAA | 315 | 52 |
| ORF 22 | ATGATGAGATTAGCAAATGG; CTATTTGTCTTGTGTCGGTT | 387 | 52 |
| ORF 21 | TTTCATTTTACGATAGCGTC; GTCGCCTGCGTGGACTGTCT | 1,308 | 55 |
| ORF 20 | ATGCTGTTTGATTATGTAAG; TTATTTTTTTGTTGTTATCA | 990 | 52 |
| ORF 19 | ATGAATTTTGGACAAAACCT; TTAAGCACCAATAATGCGAT | 222 | 52 |
| ORF 18 | TTTAGGCAAATACAATGAGG; GATTGGTTGAGATAAACGTT | 443 | 56 |
| ORF 17 | ATGGGATTTTTGAAATCGTC; TTAATTGGATATGCCATAAA | 507 | 52 |
| ORF 16 | ATGGCATATCCAATTAAATA; TTACACCTCTTTTCGCACAG | 2,448 | 52 |
| ORF 15 | ATGTGAAACCATCAATAGTA; TCATCTGAAAATAAAATGGC | 2,265 | 52 |
| ORF 14 | ATGAAGTTGAAAACTTTAGT; TCATTGTTTGATTCGTCCTG | 1,002 | 52 |
| ORF 13 | AGAAAAACAGATACCAAAGG; CGTTCTTTTCAAGTACCAAA | 860 | 54 |
| ORF 12-tetM | ATGCTTTGTATGCCTATGGT; CTAAGTTATTTTATTGAACA | 2022 | 54 |
| ORF 5–10 | ATTATAAACTACAAGTGGAT; TTCGTTTAATCTGAATACGA | 2233 | 52 |
| Xis-Tn | ATGAAGCAGACTGACATTCC; TTCGTTTAATCTGAATACGA | 204 | 52 |
| Int-Tn | GACTGGAGAGAGCCAACGAA; CATCATGCCGTTGTAATCAC | 925 | 54 |
*tet, tetracycline; ORF, open reading frame.
The most recognized virulence genes of S. suis, including mrp, sly, and ef, were detected by PCR in all 114 Chinese isolates tested in this study. Of the 12 serotype 2 strains from other countries, 6 of 8 ST1 strains were positive for all 3 virulence genes. However, none of the 4 ST25 strains tested positive (Table 1). Of the 34 serotype reference strains, serotype 2 strain R735 was positive for mrp and sly. The sly gene was detected in 10 reference strains that were typed serotype 1/2 as untypeable, serotype 2 as ST1, serotype 4 as ST54, 5 as ST53, 7 as untypeable, 13 as ST71, 14 as ST6, 16 as ST73, 17 as ST76, and 18 as ST79 (Table 1).
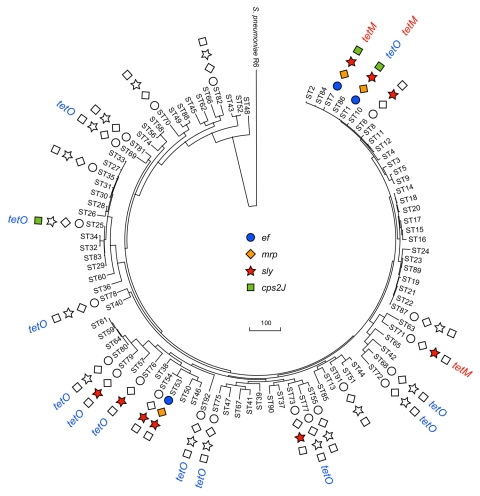
To determine the significance of horizontal gene transfer of Tn916 with the tetM and virulence genes, we constructed a rooted phylogenetic tree by using the maximum-parsimony method. The sequence of S. pneumoniae R6 was chosen as the outgroup that is closely related to S. suis (10). The data suggest S. suis ST7 evolved originally from ST1 and ST48. The horizontal transfer of tested virulence genes and tetM occurred in various stages of the evolution of S. suis and played a major role in the emergence of ST1 and ST7 (Figure 2).
Figure 2.
Horizontal transfer events of virulence genes and conjugative transposon Tn916 with tetM in Streptococcus suis sequence types. The rooted maximum-parsimony tree was based on the concatenated sequences of 7 housekeeping genes used for multilocus sequence typing analysis of S. suis by using S. pneumoniae R6 as the outgroup. Virulence genes acquired by strains of various sequence types were from this study and other published papers. The colored labels indicate positive detection; uncolored labels indicate negative results for the virulence gene; no label indicates that the strain was not available for testing. The scale bar indicates a branch length corresponding to 100 character-state changes.
Conclusions
We report that S. suis ST7 was responsible for 2 large outbreaks and sporadic infections in several provinces of China and has recently acquired the tetracycline resistance gene, tetM, associated with the conjugative transposon Tn916 (3,7). Horizontal transfer of Tn916 with the tetM gene occurred in at least 3 STs located at various stages in the constructed phylogenetic tree and played a central role in the evolution of the epidemic S. suis ST7 clone. All 3 virulence genes tested in this study were shown to be transferred horizontally (11–13).
Our data support the contention that Tn916 with tetM acts as an important selective factor that provides considerable advantages for the clone emergence and spread of S. suis ST7 (3). The widespread use of tetracycline in swine feed could provide the selective pressure for clone amplification and spreading, thus contributing to the outbreak of S. suis ST7 through Tn916 (14). The countrywide spread of S. suis ST7-tetM represents a model of selective pressure leading to the emergence of a bacterium as a virulent pathogen in humans. The case of S. suis ST7 is a sign that pathogens present in food animals can result in substantial public health problems if no action is taken to prevent the indiscriminate use of antimicrobial drugs in animal feed (15).
Acknowledgments
This work was supported by grants (2005CB522904 and 2003BA712A02 to J.X.) from the Ministry of Science and Technology, People’s Republic of China.
Biography
Dr Ye is a microbiologist at the State Key Laboratory for Infectious Disease Prevention and Control, Beijing. Her main research interest is emerging infectious diseases. She is a group leader for the study of S. suis infection in China.
Footnotes
Suggested citation for this article: Ye C, Bai X, Zhang J, Jing H, Zheng H, Du H, et al. Spread of Streptococcus suis sequence type 7, China. Emerg Infect Dis [serial on the Internet]. 2008 May [date cited]. Available from http://www.cdc.gov/EID/content/14/5/787.htm
References
- 1.Yu H, Jing H, Chen Z, Zheng H, Zhu X, Wang H, et al. Human Streptococcus suis outbreak, Sichuan, China. Emerg Infect Dis. 2006;12:914–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu F, Yang H, Hu X, Wang H, Wang G, Song Y, et al. Homogeneity study on the Streptococcus suis isolated from human and swine [in Chinese]. Zhonghua Liu Xing Bing Xue Za Zhi. 2000;21:427–9. [PubMed] [Google Scholar]
- 3.Ye C, Zhu X, Jing H, Du H, Segura M, Zheng H, et al. Streptococcus suis sequence type 7 outbreak, Sichuan, China. Emerg Infect Dis. 2006;12:1203–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King SJ, Leigh JA, Heath PJ, Luque I, Tarradas C, Dowson CG, et al. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J Clin Microbiol. 2002;40:3671–80. 10.1128/JCM.40.10.3671-3680.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gottschalk M, Higgins R, Jacques M, Mittal KR, Henrichsen J. Description of 14 new capsular types of Streptococcus suis. J Clin Microbiol. 1989;27:2633–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottschalk M, Higgins R, Jacques M, Beaudoin M, Henrichsen J. Characterization of six new capsular types (23 through 28) of Streptococcus suis. J Clin Microbiol. 1991;29:2590–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agerso Y, Pedersen AG, Aarestrup FM. Identification of Tn5397-like and Tn916-like transposons and diversity of the tetracycline resistance gene tet(M) in enterococci from humans, pigs and poultry. J Antimicrob Chemother. 2006;57:832–9. 10.1093/jac/dkl069 [DOI] [PubMed] [Google Scholar]
- 8.Ng LK, Martin I, Alfa M, Mulvey M. Multiplex PCR for the detection of tetracycline resistant genes 20. Mol Cell Probes. 2001;15:209–15. 10.1006/mcpr.2001.0363 [DOI] [PubMed] [Google Scholar]
- 9.Morse SA, Johnson SR, Biddle JW, Roberts MC. High-level tetracycline resistance in Neisseria gonorrhoeae is result of acquisition of streptococcal tetM determinant 4. Antimicrob Agents Chemother. 1986;30:664–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoskins J, Alborn WE Jr, Arnold J, Blaszczak LC, Burgett S, DeHoff BS, et al. Genome of the bacterium Streptococcus pneumoniae strain R6. J Bacteriol. 2001;183:5709–17. 10.1128/JB.183.19.5709-5717.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okwumabua O, O’Connor M, Shull E. A polymerase chain reaction (PCR) assay specific for Streptococcus suis based on the gene encoding the glutamate dehydrogenase. FEMS Microbiol Lett. 2003;218:79–84. 10.1111/j.1574-6968.2003.tb11501.x [DOI] [PubMed] [Google Scholar]
- 12.Silva LM, Baums CG, Rehm T, Wisselink HJ, Goethe R, Valentin-Weigand P. Virulence-associated gene profiling of Streptococcus suis isolates by PCR. Vet Microbiol. 2006;115:117–27. 10.1016/j.vetmic.2005.12.013 [DOI] [PubMed] [Google Scholar]
- 13.Wisselink HJ, Joosten JJ, Smith HE. Multiplex PCR assays for simultaneous detection of six major serotypes and two virulence-associated phenotypes of Streptococcus suis in tonsillar specimens from pigs. J Clin Microbiol. 2002;40:2922–9. 10.1128/JCM.40.8.2922-2929.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klare I, Konstabel C, Badstubner D, Werner G, Witte W. Occurrence and spread of antibiotic resistances in Enterococcus faecium 6. Int J Food Microbiol. 2003;88:269–90. 10.1016/S0168-1605(03)00190-9 [DOI] [PubMed] [Google Scholar]
- 15.Yang H, Chen S, White DG, Zhao S, McDermott P, Walker R, et al. Characterization of multiple-antimicrobial-resistant Escherichia coli isolates from diseased chickens and swine in China. J Clin Microbiol. 2004;42:3483–9. 10.1128/JCM.42.8.3483-3489.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]