We found substantial population differentiation and recent trans-Atlantic dispersal of a high-virulence B. burgdorferi clone.
Keywords: Multilocus sequence analysis (MLSA), ospC, Lyme disease, phylogeny, recombination, clonal complexes, research
Abstract
The A and B clones of Borrelia burgdorferi sensu stricto, distinguished by outer surface protein C (ospC) gene sequences, are commonly associated with disseminated Lyme disease. To resolve phylogenetic relationships among isolates, we sequenced 68 isolates from Europe and North America at 1 chromosomal locus (16S–23S ribosomal RNA spacer) and 3 plasmid loci (ospC, dbpA, and BBD14). The ospC-A clone appeared to be highly prevalent on both continents, and isolates of this clone were uniform in DNA sequences, which suggests a recent trans-oceanic migration. The genetic homogeneity of ospC-A isolates was confirmed by sequences at 6 additional chromosomal housekeeping loci (gap, alr, glpA, xylB, ackA, and tgt). In contrast, the ospC-B group consists of genotypes distinct to each continent, indicating geographic isolation. We conclude that the ospC-A clone has dispersed rapidly and widely in the recent past. The spread of the ospC-A clone may have contributed, and likely continues to contribute, to the rise of Lyme disease incidence.
Multilocus sequence typing (MLST) is the use of DNA sequences at multiple housekeeping loci to characterize genetic variations of natural populations of a bacterial pathogen (1,2). MLST studies showed that local populations of a bacterial species typically consist of discrete clusters of multilocus sequence types called “clonal complexes,” rather than a multitude of randomly assorted genotypes (2). Remaining to be tested are how such factors as natural selection, low recombination rate, and genetic drift due to geographic structuring contribute to the formation and maintenance of these clonal complexes in natural bacterial populations (3,4). Recently, a multilocus sequence analysis approach was proposed to reconstruct phylogenetic histories of bacterial clonal complexes by using concatenated sequences of housekeeping genes when within-loci and between-loci recombinations are infrequent (5).
Lyme disease is a multisystem infection, with inflammatory complications that commonly affect the skin, joints, and central nervous system in humans (6). Its causative agent, Borrelia burgdorferi, a spirochete that parasitizes vertebrates, is transmitted by hard-bodied ticks throughout the temperate zones of the Northern Hemisphere (7). Although humans are accidental hosts of B. burgdorferi, Lyme disease is the most common vector-borne disease in the United States with >20,000 annual reported cases, 93% of which occurred in 10 northeastern, mid-Atlantic, and north-central states (8). Small mammals such as white-footed mice (Peromyscus leucopus) and eastern chipmunks (Tamias striatus) serve as the main reservoirs of B. burgdorferi (9,10). In Europe, B. burgdorferi is transmitted by Ixodes ricinus ticks (11) and is carried by a large variety of hosts, including birds and small- to medium-sized mammals (12).
B. burgdorferi sensu stricto is the primary pathogen of Lyme disease in the United States and is the only pathogenic genospecies that causes Lyme disease in both North America and Europe. More than 12 distinct outer surface protein C (ospC) major sequence types coexist in local B. burgdorferi sensu stricto populations in the northeastern United States (13–15). Sequence variability at ospC is the highest among known genomic loci and is strongly linked to variations at other genome-wide loci, with occasional recombinant genotypes caused by plasmid exchanges (16–19).
B. burgdorferi sensu stricto intraspecific clonal complexes may differ in their host specificity and degree of human pathogenecity. Different clonal complexes may prefer different host species (9). A restriction fragment length polymorphism type of intergenic spacer (IGS) sequence (corresponding to the ospC-A and -B groups) is associated with hematogenous dissemination in patients with early stage Lyme disease (20,21). Four ospC clonal complexes (A, B, I, and K groups) were found to be more likely than others to cause disseminated Lyme disease (22). Also, an association of ospC clonal types with invasive disease in humans has been found in other pathogenic genospecies such as B. afzelii and B. garinii (23,24). However, additional ospC clonal types have been isolated in patients with invasive disease (14).
Previous molecular assays found a close relationship and overlapping genotypes between the European and North American populations (25–27). These authors found greater genetic diversity among American strains than European strains and proposed a North American origin for this genospecies. Although these studies provided the first evidence for recent intercontinental migrations, they left the phylogenetic relationships among clonal complexes unresolved because of the use of either anonymous genome-wide markers (e.g., arbitrarily primed PCR), genes with a high recombination rate (e.g., ospC), or sequences at a single locus. A phylogeographic approach with multiple molecular markers provides a more robust inference on population history (28). Here we obtained a well-resolved phylogeny of B. burgdorferi sensu stricto clonal complexes by using multilocus sequence typing at housekeeping loci as well as loci under adaptive evolution. We found evidence of genetic endemism, recent migration events, and recombinant genomic types. In fact, the highly pathogenic ospC-A clone seems to have spread rapidly in recent years to infect a broad range of host species in 2 continents.
Materials and Methods
B. burgdorferi Isolates and DNA Isolation
The B. burgdorferi sensu stricto isolates were obtained from clinical and tick specimens and cultures from animals in the United States and Europe and maintained as frozen stocks at –70°C (Table 1). For in vitro propagation, a small amount of frozen culture was scraped from the surface of each sample with a sterile inoculating loop and injected into complete Barbour-Stoenner-Kelly II medium (Sigma-Aldrich Corp., St. Louis, MO, USA). Spirochetes were then cultivated at 34°C. All cultures used in this study had undergone a maximum of 2 in vitro passages after recovery from frozen stock. For isolation of genomic DNA, 10 mL of low-passage log-phase bacteria was harvested by centrifugation at 10,000 rpm for 30 min at 4°C. The bacterial pellet was washed twice with Tris-Cl buffer (10 mmol/L Tris [pH 7.5], 100 mmol/L NaCl), and resuspended in 430 μL TES (10 mmol/L Tris [pH 7.5], 100 mmol/L NaCl, 10 mmol/L EDTA). Subsequently, 10 μL of freshly prepared lysozyme (50 mg/mL), 50 μL Sarkosyl (10%), and 10 μL proteinase K (10 mg/mL) were then added, and the mixture was incubated at 50°C overnight before RNase treatment. After incubation, DNA was extracted with phenol/chloroform and chloroform, precipitated with ethanol, and finally resuspended in TE buffer (1 mmol/L Tris [pH7.5], 1 mmol/L EDTA).
Table 1. Borrelia burgdorferi isolates*.
| Isolates studied† | ospC type‡ | Biologic origin | US frequency§ | EU frequency |
|---|---|---|---|---|
| B31, CS1, CS2, CS3, 132a, 132b, IP1, IP2, IP3, Ho, HB1, Lenz, L65, PKa2, HII | A | Ixodes scapularis, human | 6 (New York) | 13 (France, Austria, Germany, Italy, Russia) |
| N40, 88a, 167bjm, SD91, NP14 | E | I. scapularis, human | 3 (New York) | 6 (Hungary) |
| 136b, 163b, 297, CS6, CS9, OEA11 | K | I. scapularis, human | 6 (New England) | 1 (Hungary) |
| 109a, 160b, 64b, CS7, MI415¶ | B1 | I. scapularis, human, Peromyscus leucopus | 5 (New York, Michigan) | 0 |
| JD1 | C | I. scapularis | 1 (Massachusetts) | 0 |
| 121a | D | Human | 1 (New York) | 0 |
| MI407 | F | P. leucopus | 1 (Michigan) | 0 |
| 72a | G | Human | 1 (New York) | 0 |
| 156a, 156b, MI403, MI411 | H | Human, Tamias striatus | 4 (New York, Michigan) | 0 |
| 86b, 97b, MI409¶ | I | Human, T. striatus | 3 (New York, Michigan) | 0 |
| 118a | J | Human | 1 (New York) | 0 |
| CS8, 80a, MI418¶ | N | I. scapularis, human, P. leucopus | 3 (New York, Michigan) | 0 |
| 94a, CS5 | U | Human, I. scapularis | 2 (New York) | 0 |
| Bol12, VS219,¶ Lx36, ZS7 | B2 | I. ricinus, human | 0 | 17 (Finland, Denmark, Switzerland, Italy, Austria, Slovakia, Germany) |
| Y1, Y10, 217–5, Bol6, Z6 | L | I. ricinus | 0 | 10 (Finland, Poland, Italy, Austria) |
| Fr-93/1, Bol15, Bol25, Bol27 | Q | I. ricinus, human | 0 | 4 (Poland, Italy) |
| Bol26,¶ Z9, PO7 | S | I. ricinus, human | 0 | 3 (Italy, Austria) |
| Bol29, Bol30 | V | Human | 0 | 15 (Italy, Switzerland, Slovenia, Germany) |
| SV1 | X | I. ricinus | 0 | 1 (Finland) |
| Ri5 | W | I. ricinus | 0 | 1 (Finland) |
*ospC, outer surface protein C; US, United States; EU, European Union. †Isolates subjected to multilocus sequence typing analysis. ‡Type names follow (13), except that B was split to B1 and B2, and 3 new types (V, X, W) were assigned to European isolates. §Number and geographic origins of an ospC type in our collection. ¶Isolates showing evidence for plasmid-chromosome recombination.
Genomic Markers, PCR Amplifications, and DNA Sequencing
PCR amplifications were attempted at 4 genomic loci for all isolates and at 6 chromosomal housekeeping loci for a genetically representative subset of isolates (Table 2). The IGS locus was chosen for its phylogenetically informative polymorphisms (16,20). The IGS locus and 6 housekeeping genes (gap, alr, glpA, xylB, ackA, tgt) were approximately evenly distributed on the main chromosome based on the B31 genome (29). The 3 plasmidborne loci were selected for their high sequence variability and for the absence of close paralogs based on a genome comparison (17,19). IGS sequences were amplified by using a nested PCR procedure (30). Because of high sequence variability, dbpA sequences were amplified by using 2 alternative forward primers. PCR amplification was performed in 50 μL containing 200 mmol/L of each dNTP, 2.0 mmol/L MgSO4, 2.5 U of Platinum Taq DNA polymerase High Fidelity (Invitrogen, Carlsbad, CA, USA), 0.5 μmol/L of each primer, and 100 ng of genomic DNA template. Following denaturation at 94°C for 1 min, samples underwent 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30s, initial extension at 68°C for 1.5 min, and a final extension step at 68°C for 10 min. PCR products were purified by GFX chromatography (Amersham Pharmacia Biotech, Inc., Piscataway, NJ, USA), resolved by agarose gel electrophoresis, and visualized by ethidium bromide staining. Purified amplicons were sequenced by using standard dideoxy terminator chemistry as outlined below with the forward and reverse PCR primers. Absence of specific PCR products, indicating potential absence of particular genetic loci or plasmids, was confirmed by follow-up amplifications of the flanking DNA segments.
Table 2. Genomic markers and PCR primers.
| Locus* | Primer sequence (5′ → 3′)† | Location‡ |
|---|---|---|
| BB0057 (gap) | F-ATGAAATTGGCTATTAATGG, R-TTGAGCAAGATCAACCACTC | Main chromosome (52.5 K) |
| BB0160 (alr) | F-ATGTATAATAATAAAACAATGG, R-ATTTTCTCTTTTCGTATTTTCC | Main chromosome (160 K) |
| BB0243 (glpA) | F-ATGGAGGAATATTTAAATTTC, R-GTTCATTTTTCCACTCTTC | Main chromosome (249 K) |
| IGS (rrs-rrlA) | 1st round§: F-GGTATGTTTAGTGAGGG, R-GGTTAGAGCGCAGGTCTG; 2nd round: F-CGTACTGGAAAGTGCGGCTG, R-GATGTTCAACTCATCCTGGTCCC | Main chromosome (444 K) |
| BB0545 (xylB) | F-ATGAATGCTCTTAGTATTG, R-CCCGTTAACAAATAGAC | Main chromosome (555 K) |
| BB0622 (ackA) | F-TTGTCAAATACAAAAGG, R-AATGTCTTCAAGAATGG | Main chromosome (649 K) |
| BB0809 (tgt) | F-ATGTTTAGTGTAATCAAGAATG, R-ATCGAAATTTTCCTCTTCATAC | Main chromosome (855 K) |
| BBA24 (dpbA) | F1-TAATGTTATGATTAAATG, F2-ATGAATAAATATCAAAAAAC, R-GAAATTCCAAATAACATC | lp54 |
| BBB19 (ospC) | F-CCGTTAGTCCAATGGCTCCAG, R-ATGCAAATTAAAGTTAATATC | cp26 |
| BBD14 | F-ATGATAATAAAAATAAAAAATAATG, R-ATTTTGATTAATTTTAATTTTGCTG | lp17 |
Automated DNA sequencing of both strands of each fragment was performed by the Stony Brook University Core DNA Sequencing Facility (Stony Brook, NY, USA) by using the dye-terminator method with the same oligonucleotide primers used for PCR amplification or, where required, appropriate internal primers. Sequences were inspected and assembled with the aid of the Sequencher program (Gene Codes, Inc., Ann Arbor, MI, USA). DNA sequences were analyzed by using the BLASTN program through GenBank at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov). Nucleotide and protein sequence alignments were performed with MacVector version 6.5 (MacVector, Inc., Cary, NC, USA). New sequences were deposited to GenBank under accession nos. EF537321–EF537573.
Phylogenetic Inference and Tests of Population Differentiation
The IGS sequences were used to resolve intraspecific phylogenetic relationships among B. burgdorferi isolates (16,20). Two highly divergent tick isolates from Finland (SV1 and Ri5) were used as outgroups for rooting the phylogenetic tree. IGS sequences were aligned by using ClustalW (31). A Bayesian majority-rule consensus tree was estimated by using MrBayes (version 2.1) (32) as described previously (19). Sequences at the 3 plasmid-borne protein–coding loci were translated into protein sequences and aligned in a pairwise fashion with ClustalW (31). Nucleotide alignments were obtained according to the protein alignments. Neighbor-joining trees based on pairwise nucleotide sequence distances were inferred by using PHYLIP (33) and plotted by using the APE package of the R statistical package (34). Genetic differentiation among geographic populations was tested by using the analysis of molecular variance (AMOVA) method implemented in the software package Arlequin 3.1 (35). The 6 housekeeping genes were used to infer the overall within- and between-genospecies phylogeny. Sequences of strains B31 and PBi (B. garinii) were downloaded from GenBank (29,36). Sequences of N40, JD1, DN127 (B. bissettii), and PKo (B. afzelii) were from draft genomes (S. Casjens, pers. comm.). The 6 alignments were concatenated and tested for the presence of gene conversion by using GENECONV with the “within-group fragments only” option (37). Two approaches, a Bayesian method with codon site-specific evolutionary rates (using MrBayes) and the other maximum likelihood method with 100 bootstrapped alignments (using DNAML in PHYLIP) (33), were used for phylogenetic reconstruction based on concatenated sequences. Branch supports were measured by the posterior probabilities in the Bayesian method and the bootstrap values in the maximum likelihood method.
Results and Discussion
AMOVA Tests of Geographic Differentiation
We sequenced 68 isolates (including 30 from northeastern United States, 6 from the midwestern United States, and 32 from Europe) at a single chromosomal locus (IGS) and 3 plasmid loci (ospC, dbpA, and BBD14). Using AMOVA, we evaluated the genetic differentiation among geographic samples and found significant genetic differentiation between the North American and European populations at IGS, ospC, and dbpA, but not BBD14 (Table 3). Among these loci, IGS is the most informative in reflecting the effect of genetic drift caused by geographic isolation because sequence variations at IGS are likely to be selectively neutral. In addition, IGS is on the main chromosome and less likely to be subject to gene conversion. Genetic variations at 3 plasmid loci are more likely to be influenced by natural selection such as adaptation to local vector and host species. Also, plasmid genes are more likely to be transferred so that footprints of geographic isolation might be obscured by gene flow between populations. Natural selection can both enhance and reduce geographic differentiation. With adaptation to local habitats, natural selection acts to enhance the geographic divergence, especially at target loci. On the other hand, diversifying selection within populations inflates within-population diversity, which results in lack of differentiation within populations relative to the within-population polymorphism.
Table 3. Analysis of molecular variance results*†.
| Locus | Molecular variance, % |
Nucleotide diversity, π |
Fixation index (FST)‡ | |||
|---|---|---|---|---|---|---|
| Between continents | Within continents | North America | Europe | |||
| IGS | 19.5 | 80.5 | 0.0253 | 0.0243 | 0.1952§ | |
| ospC | 3.13 | 96.87 | 0.2066 | 0.1900 | 0.0313¶ | |
| dbpA | 26.5 | 73.5 | 0.1480 | 0.0999 | 0.2650§ | |
| BBD14 | 2.54 | 97.46 | 0.0834 | 0.1333 | 0.0254 (NS) | |
*IGS, intergenic spacer; ospC, outer surface protein C; NS, not significant (p>0.05). †Results were obtained by using Arlequin 3.1 (35). Samples were 66 IGS sequences divided into 2 continental populations: North America (36 sequences from New York, Connecticut, Massachusetts, and Michigan) and Europe (30 sequences from Italy, Austria, France, Germany, Switzerland, Poland, Hungary, Slovenia, and Finland). Two outgroup sequences (SV1 and Ri5) were excluded from the European sample. Genetic distances between haplotypes were based on the Kimura 2-parameter model. ‡Levels of significance were obtained by 1,000 permutations. §p<0.001. ¶0.01<p<0.05.
The low level of geographic differentiation at ospC showed the divergence-reducing effect of natural selection. Genetic variability of ospC is as high within populations as between populations and is caused by diversifying natural selection (9,13). In such a case, summary statistics such as AMOVA fixation index (FST) are misleading because sequence cluster analysis showed that most ospC alleles have geographically restricted distributions (Figure 1, panel B). The insignificant AMOVA result at BBD14 might be due to a similar effect of high within-population polymorphisms as a result of diversifying selection. In contrast, dbpA showed the divergence-enhancing effect of natural selection. The dbpA locus showed the highest level of geographic differentiation, owing to a shared allelic type among B2, L, S, Q, and V clonal groups in Europe (Table 3; Figure 1, panel C). An adaptive sweep likely has homogenized these divergent European lineages at dbpA.
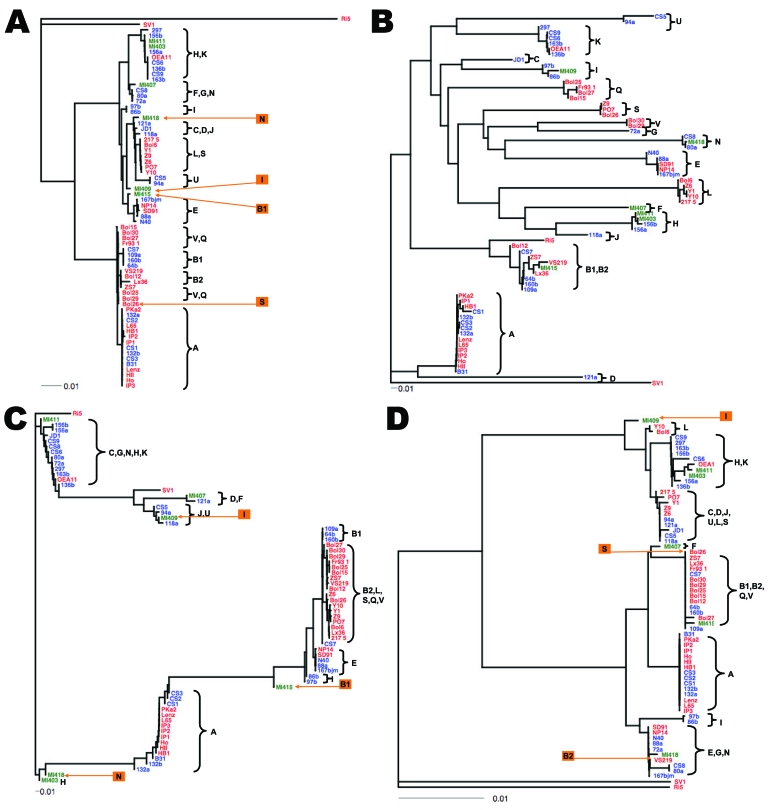
Figure 1.
Gene trees showing nucleotide sequence clusters of 68 Borrelia burgdorferi isolates at 1 chromosomal locus (panel A: rrs-rrlA spacer, or intergenic spacer [IGS]) and 3 plasmid loci (panels B, C, and D: ospC on cp26, dbpA on lp54, and BBD14 on lp17, respectively). Trees were inferred based on nucleotide sequence alignments and were rooted by using the Ri5, SV1, or both, sequences as outgroups. The DNADIST and neighbor-joining programs of the PHYLIP package (33) were used for distance calculation and the APE software package (34) was used for tree plotting. Isolates were grouped as clonal groups (A through U), which are named by their typical ospC alleles. Five isolates (Bol26, VS219, MI409, MI415, and MI418) showing atypical allelic associations with ospC alleles, likely caused by recombination, were labeled in orange. Red, European isolates; blue, northeastern US isolates; green, midwestern US isolates. Scale bars indicate number of nucleotide substitutions per site.
In summary, on the basis of the neutral genetic variations at IGS, we conclude that the European and North American populations of B. burgdorferi sensu stricto have diverged significantly because of genetic drift. Plasmid genes evolved independently and showed various effects of adaptive divergence and diversifying selection. At all 4 loci, genetic variations within the 2 continents contributed to most (>70%) of the total sequence diversity, which suggests recent common ancestry, migration, or both, between the European and North American populations.
Endemic and Shared ospC Alleles
Gene trees showed more detailed pictures of geographic variations at each locus (Figure 1). Among the 17 major sequence groups of ospC, 2 minor sequence variants of major-group allele B were geographically distinct and thereby named B1 in North America and B2 in Europe. Three ospC alleles (A, E, and K) were observed in both continents, 5 (B2, S, L, Q, and V) exclusively in Europe (not including the outgroup Ri5 and SV1 alleles), and 10 (B1, C, D, F, G, H, I, J, N, and U) exclusively in North America (Table 1). Although the sample sizes of the North America isolates were small, the same set of ospC alleles has repeatedly been identified in surveys of natural populations (14–16,38). These isolates are therefore a reasonably complete representation of ospC diversity in North America. How well our European samples represent the overall ospC diversity in Europe is less certain because the European isolates were from an archived collection rather than from systematic surveys of natural populations. For instance, ospC alleles J, P, and R have been identified in Europe (26). Nonetheless, ospC-A appeared to be the only allele that is highly prevalent on both continents (Table 1). An earlier study showed that ospC-A and ospC-B alleles existed in both continents, whereas other ospC alleles were geographically distinct (K, J, F in North America and P, Q, R, S in Europe) (24). Our results further suggested that the ospC-B clonal group had 2 geographically distinct subtypes (Figure 1, panel B).
Recombinant Genotypes
Previous MLST studies showed complete linkage between ospC and other loci on plasmids or the main chromosome in the North American populations (15,16). This finding is consistent with our study, in which allelic types at IGS, dbpA, and BBD14 of the 68 isolates were almost entirely predictable from their ospC types. Because of the nearly complete linkage between ospC and a locus, individual clonal complexes could conveniently be named after their ospC alleles. However, 5 isolates showed alleles at non-ospC loci inconsistent with allelic types typically associated with their ospC alleles, including MI409, MI415, and MI418 from the midwestern United States and Bol26 and VS219 from Europe (Figure 1). Because these genotypes were new combinations of allelic types found elsewhere, they are more likely to be recombinant genotypes caused by plasmid exchanges, rather than locally evolved new genotypes (17). Notably, these probable recombinants were from samples from either the midwestern United States or Europe, and none were from the intensively surveyed northeastern United States. A higher number of clones in the northeastern United States than elsewhere could be understood because B. burgdorferi populations in that region are evolutionarily young and show an epidemic population structure (15,19). On the basis of the presence of allele types at 4 loci, we determined preliminarily that Bol26 is a group Q or V clone with a transferred ospC-S allele because Bol26 clustered with group Q and V isolates at IGS, dbpA, and BBD14 (Figure 1). By the same reasoning, VS219 is a group B2 clone with a transferred BBD14 allele. We are currently investigating the donor and recipient genomic types of these recombinant isolates by sequencing 6 additional loci.
Recent Trans-Oceanic Dispersals
Three clonal complexes (A, E, K) are distributed in both continents (Table 1). For the A clonal group, 6 isolates from the United States and 11 isolates from Europe were sequenced at 4 loci. The 4-locus sequences of the isolates between the 2 continents were identical (Figure 1). Thus, the A clonal complex likely was dispersed across the Atlantic Ocean rather recently. To verify the genetic homogeneity of group A isolates from the 2 continents, we randomly selected 4 group A isolates (B31 and 132b from the United States; IP1 and PKa2 from Europe) for further sequencing at an additional 6 chromosomal loci. No fixed sequence differences between 2 continental samples were found, which lends further support for the recent trans-oceanic migration of the A clone (Figure 2). Similarly, the 4-loci sequences of E and K isolates between the 2 continental samples were identical, indicating recent migration of these clonal groups as well (Figure 1). However, the E and K groups seemed less prevalent in Europe than the A group (Table 1). Because individual ticks and hosts are commonly infected with multiple B. burgdorferi clones, any migration, whether by natural or human-facilitated mechanisms, is likely to involve a mixture of clonal groups, rather than a single clone. Upon their arrival, however, clonal groups may differ in their ability to colonize a new niche consisting of novel vector and host species. By this reasoning, the A clone is the most ecologically successful strain, able to thrive in a new niche with little genetic change. This conclusion is supported by surveys that showed a broad range of host species for this clonal group (9,10).
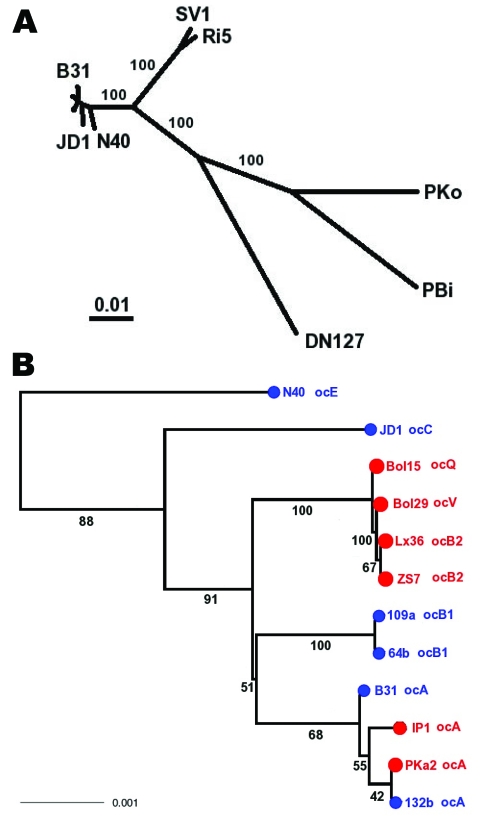
Figure 2.
Species phylogeny based on concatenated sequences at housekeeping loci. Seventeen isolates include 1 Borrelia garinii strain (PBi), 1 B. afzelii strain (PKo), 1 B. bissettii strain (DN127), 2 strains of an unnamed genomic species (SV1 and Ri5), and 12 B. burgdorferi sensu stricto isolates. These strains were selected for reconstructing interspecies phylogeny (hence species samples), as well as for resolving the clade containing clonal groups A and B (A, B1, and B2 are represented by 2 isolates). Sequences at 6 chromosomal housekeeping loci (gap, alr, glpA, xylB, ackA, and tgt) were obtained for each strain, with B31 and PBi sequences from published genomes (29,36), N40, JD1, PKo, and DN127 sequences from draft genomes (S. Casjens et al. pers. comm.). Sequences of the remaining strains were obtained by direct sequencing. The total length of concatenated alignment is 7,509 nt. A) Consensus of maximum likelihood trees obtained by using DNAML of the PHYLIP package (33). Branch support values are based on 100 bootstrapped replicates of the original alignment. B) Enlarged view of B. burgdorferi sensu stricto subtree. Tips were colored by geographic origin of the isolate (blue, North America; red, Europe) and were labeled with ospC major-group allele type. Scale bars indicate number of nucleotide substitutions per site.
We could not determine conclusively the direction, timing, or number of the trans-oceanic dispersals. Assuming that the chromosomal gene tree in Figure 2, panel B, is an accurate representation of the phylogeny of these clonal groups, a parsimonious scenario is that an early migrant from Europe was the ancestor of the North American clade consisting of the A and B1 groups, and a more recent migration has introduced the A group to Europe. However, none of the basal branches of this gene tree was well supported (Figure 2). Multilocus sequencing of more loci, especially rapidly evolving plasmid loci, of group A isolates will help find more conclusive answers to these questions. To estimate the time of the A clone migration, we noted that no fixed differences in nucleotides occurred within a total of 11,167 aligned bases at 7 chromosomal and 3 plasmid loci. If one assumes a neutral evolutionary rate on the order of 1 substitution per site per million years, the Poisson zero-term probabilities that no fixed difference has occurred within 11,167 bases in the past 50, 100, and 200 years are 0.33, 0.10, and 0.011, respectively. Therefore, the trans-oceanic migration of clone A likely occurred more recently than 200 years ago. More realistic estimates would depend on studies of the neutral mutation rate and generation time of B. burgdorferi in the wild.
Phylogenetic Heterogeneity of Group B Isolates
The ospC-B clonal group is another highly virulent strain identified by association studies (20–22,24). Initially, group B seemed to be another clone that is distributed in both continents with a few sequence differences at IGS and ospC (Figure 1). Sequencing at additional 6 housekeeping loci, however, showed deep phylogenetic heterogeneity of the B group, while the A group remained homogeneous (Figure 2). The 2 B clonal complexes (B1 in North America and B2 in Europe) do not form a monophyletic clade (Figure 2). Rather, B2 clusters with other European clones (V and Q). Also, clones B1 and A, the 2 closest North American relatives, do not form a well-supported clade (only 51% bootstrap support). Clearly, unlike the A clone, the bicontinental distribution of the B clone is not due to recent migration. Sharing of similar ospC B alleles between the 2 continents may be due to stabilizing selection or lateral transfer. Because few synonymous changes have occurred between the B1 and B2 alleles, lateral transfer is a more likely cause.
The B2/Q/V showed as a European clade with nearly uniform chromosomal sequences, although it had highly divergent ospC alleles (Figure 2). This evidence, based on chromosome-wide genes, strengthens the conclusions of an earlier study that adaptive, large sequence variations at ospC are associated with incipient genome divergence (19).
Finally, the overall genospecies phylogeny based on MLST showed that the 2 European isolates (Ri5 and SV1) that we used as outgroups may be a new genospecies (Figure 2). This phylogeny is robust because tests of recombination using GENECONV showed no statistically significant gene conversion within the 6 chromosomal housekeeping loci (37). The hypothetical genospecies represented by Ri5 and SV1 is more closely related to B. burgdorferi sensu stricto than B. bissettii (represented here by DN127) is to B. burgdorferi sensu stricto. Thus, the MLST phylogeny suggests a possibility that Europe, rather than North America, may be the origin of B. burgdorferi sensu stricto, despite a higher contemporary genetic heterogeneity in North America than in Europe.
Conclusions
To summarize, the present study used 7 chromosomal loci (IGS and 6 housekeeping genes) to reconstruct the intra- and interspecific phylogeographic histories of B. burgdorferi sensu stricto. Although the standard MLST scheme based on housekeeping genes enables estimates of recombination and mutation rates as well as intraspecific phylogenies (2,5), our approach of including plasmidborne loci under positive selection helped identify the selective causes of bacterial lineage divergence. Our results showed significant endemic lineage diversification among regional populations, discovered recombinant genotypes, and strongly indicated migrations between North American and European populations in modern times. The highly pathogenic clonal complex A has a prominent presence in both continents, which suggests its success in finding ecologic niches that enable it to infect a broad range of host and vector species. The same genetic basis of the ecologic invasiveness of the ospC-A clone may be underlying its high virulence to humans. The emergence of Lyme disease in North America since the 1970s has been attributed to an increasing overlap of human and B. burgdorferi habitats (39). On the basis of our evidence of migration events, we propose that the trans-oceanic dispersal and colonization of ecologically highly successful clonal complexes (e.g., the A group) may also have played a substantial role.
Acknowledgment
We acknowledge the Borrelia sequencing team of Sherwood R. Casjens, John J. Dunn, Benjamin J. Luft, Claire M. Fraser, Weigang Qiu, and Steven E. Schutzer, working under grants from the Lyme Disease Association and National Institutes of Health (AI37256 and AI49003), for access to unpublished sequence information.
Other supports from this work include grants GM083722-01 (toW.-G.Q.) and RR03037 (to Hunter College) from the National Institutes of Health.
Biography
Dr Qiu is an assistant professor in the Department of Biological Sciences at Hunter College and the Biology Department in the Graduate Center of the City University of New York. His research interests include the evolution and population biology of infectious diseases, comparative genomics, and bioinformatics tool development.
Footnotes
Suggested citation for this article: Qiu W-G, Bruno JF, McCaig WD, Xu Y, Livey I, Schriefer ME, et al. Wide distribution of a high-virulence Borrelia burgdorferi clone in Europe and North America. Emerg Infect Dis [serial on the Internet]. 2008 Jul [date cited]. Available from http://www.cdc.gov/EID/content/14/7/1097.htm
References
- 1.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95:3140–5. 10.1073/pnas.95.6.3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feil EJ. Small change: keeping pace with microevolution. Nat Rev Microbiol. 2004;2:483–95. 10.1038/nrmicro904 [DOI] [PubMed] [Google Scholar]
- 3.Cohan FM. Concepts of bacterial biodiversity for the age of genomics. In: Fraser CM, Read TD, Nelson KE, editors. Microbial genomes. Totwa (NJ): Humana Press, Inc.; 2004. p. 175–94. [Google Scholar]
- 4.Fraser C, Hanage WP, Spratt BG. Neutral microepidemic evolution of bacterial pathogens. Proc Natl Acad Sci U S A. 2005;102:1968–73. 10.1073/pnas.0406993102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, et al. Opinion: re-evaluating prokaryotic species. Nat Rev Microbiol. 2005;3:733–9. 10.1038/nrmicro1236 [DOI] [PubMed] [Google Scholar]
- 6.Steere AC, Coburn J, Glickstein L. The emergence of Lyme disease. J Clin Invest. 2004;113:1093–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piesman J, Gern L. Lyme borreliosis in Europe and North America. Parasitology. 2004;129(Suppl):S191–220. 10.1017/S0031182003004694 [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. Lyme disease—United States, 2003–2005. MMWR Morb Mortal Wkly Rep. 2007;56:573–6. [PubMed] [Google Scholar]
- 9.Brisson D, Dykhuizen DE. ospC diversity in Borrelia burgdorferi: different hosts are different niches. Genetics. 2004;168:713–22. 10.1534/genetics.104.028738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanincova K, Kurtenbach K, Diuk-Wasser M, Brei B, Fish D. Epidemic spread of Lyme borreliosis, northeastern United States. Emerg Infect Dis. 2006;12:604–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rauter C, Hartung T. Prevalence of Borrelia burgdorferi sensu lato genospecies in Ixodes ricinus ticks in Europe: a metaanalysis. Appl Environ Microbiol. 2005;71:7203–16. 10.1128/AEM.71.11.7203-7216.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gern L, Estrada-Pena A, Frandsen F, Gray JS, Jaenson TG, Jongejan F, et al. European reservoir hosts of Borrelia burgdorferi sensu lato. Zentralbl Bakteriol. 1998;287:196–204. [DOI] [PubMed] [Google Scholar]
- 13.Wang IN, Dykhuizen DE, Qiu W, Dunn JJ, Bosler EM, Luft BJ. Genetic diversity of ospC in a local population of Borrelia burgdorferi sensu stricto. Genetics. 1999;151:15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alghaferi MY, Anderson JM, Park J, Auwaerter PG, Aucott JN, Norris DE, et al. Borrelia burgdorferi ospC heterogeneity among human and murine isolates from a defined region of northern Maryland and southern Pennsylvania: lack of correlation with invasive and noninvasive genotypes. J Clin Microbiol. 2005;43:1879–84. 10.1128/JCM.43.4.1879-1884.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu WG, Dykhuizen DE, Acosta MS, Luft BJ. Geographic uniformity of the Lyme disease spirochete (Borrelia burgdorferi) and its shared history with tick vector (Ixodes scapularis) in the northeastern United States. Genetics. 2002;160:833–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bunikis J, Garpmo U, Tsao J, Berglund J, Fish D, Barbour AG. Sequence typing reveals extensive strain diversity of the Lyme borreliosis agents Borrelia burgdorferi in North America and Borrelia afzelii in Europe. Microbiology. 2004;150:1741–55. 10.1099/mic.0.26944-0 [DOI] [PubMed] [Google Scholar]
- 17.Qiu WG, Schutzer SE, Bruno JF, Attie O, Xu Y, Dunn JJ, et al. Genetic exchange and plasmid transfers in Borrelia burgdorferi sensu stricto revealed by three-way genome comparisons and multilocus sequence typing. Proc Natl Acad Sci U S A. 2004;101:14150–5. 10.1073/pnas.0402745101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stevenson B, Miller JC. Intra- and interbacterial genetic exchange of Lyme disease spirochete erp genes generates sequence identity amidst diversity. J Mol Evol. 2003;57:309–24. 10.1007/s00239-003-2482-x [DOI] [PubMed] [Google Scholar]
- 19.Attie O, Bruno JF, Xu Y, Qiu D, Luft BJ, Qiu WG. Co-evolution of the outer surface protein C gene (ospC) and intraspecific lineages of Borrelia burgdorferi sensu stricto in the northeastern United States. Infect Genet Evol. 2007;7:1–12. 10.1016/j.meegid.2006.02.008 [DOI] [PubMed] [Google Scholar]
- 20.Wormser GP, Liveris D, Nowakowski J, Nadelman RB, Cavaliere LF, McKenna D, et al. Association of specific subtypes of Borrelia burgdorferi with hematogenous dissemination in early Lyme disease. J Infect Dis. 1999;180:720–5. 10.1086/314922 [DOI] [PubMed] [Google Scholar]
- 21.Jones KL, Glickstein LJ, Damle N, Sikand VK, McHugh G, Steere AC. Borrelia burgdorferi genetic markers and disseminated disease in patients with early Lyme disease. J Clin Microbiol. 2006;44:4407–13. 10.1128/JCM.01077-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seinost G, Dykhuizen DE, Dattwyler RJ, Golde WT, Dunn JJ, Wang IN, et al. Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect Immun. 1999;67:3518–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baranton G, Seinost G, Theodore G, Postic D, Dykhuizen D. Distinct levels of genetic diversity of Borrelia burgdorferi are associated with different aspects of pathogenicity. Res Microbiol. 2001;152:149–56. 10.1016/S0923-2508(01)01186-X [DOI] [PubMed] [Google Scholar]
- 24.Lagal V, Postic D, Ruzic-Sabljic E, Baranton G. Genetic diversity among Borrelia strains determined by single-strand conformation polymorphism analysis of the ospC gene and its association with invasiveness. J Clin Microbiol. 2003;41:5059–65. 10.1128/JCM.41.11.5059-5065.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foretz M, Postic D, Baranton G. Phylogenetic analysis of Borrelia burgdorferi sensu stricto by arbitrarily primed PCR and pulsed-field gel electrophoresis. Int J Syst Bacteriol. 1997;47:11–8. [DOI] [PubMed] [Google Scholar]
- 26.Marti Ras N, Postic D, Baranton G. Borrelia burgdorferi sensu stricto, a bacterial species “Made in the U.S.A.”? Int J Syst Bacteriol. 1997;47:1112–7. [DOI] [PubMed] [Google Scholar]
- 27.Postic D, Ras NM, Lane RS, Humair P, Wittenbrink MM, Baranton G. Common ancestry of Borrelia burgdorferi sensu lato strains from North America and Europe. J Clin Microbiol. 1999;37:3010–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avise JC. Phylogeography: the history and formation of species. Cambridge (MA): Harvard University Press; 2000. [Google Scholar]
- 29.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–6. 10.1038/37551 [DOI] [PubMed] [Google Scholar]
- 30.Liveris D, Varde S, Iyer R, Koenig S, Bittker S, Cooper D, et al. Genetic diversity of Borrelia burgdorferi in Lyme disease patients as determined by culture versus direct PCR with clinical specimens. J Clin Microbiol. 1999;37:565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–5. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- 33.Felsenstein J. PHYLIP—phylogeny inference package. Cladistics. 1989;5:164–6. [Google Scholar]
- 34.Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–90. 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- 35.Excoffier L, Laval G, Schneider S. Arlequin ver 3.0: an integratred software package for population data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 36.Glockner G, Lehmann R, Romualdi A, Pradella S, Schulte-Spechtel U, Schilhabel M, et al. Comparative analysis of the Borrelia garinii genome. Nucleic Acids Res. 2004;32:6038–46. 10.1093/nar/gkh953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawyer S. Statistical tests for detecting gene conversion. Mol Biol Evol. 1989;6:526–38. [DOI] [PubMed] [Google Scholar]
- 38.Wang G, van Dam AP, Dankert J. Evidence for frequent OspC gene transfer between Borrelia valaisiana sp. nov. and other Lyme disease spirochetes. FEMS Microbiol Lett. 1999;177:289–96. 10.1111/j.1574-6968.1999.tb13745.x [DOI] [PubMed] [Google Scholar]
- 39.Barbour AG, Fish D. The biological and social phenomenon of Lyme disease. Science. 1993;260:1610–6. 10.1126/science.8503006 [DOI] [PubMed] [Google Scholar]