Abstract
Shigella sonnei UCN59, isolated during an outbreak of S. sonnei in January 2007, was resistant to azithromycin (MIC 64 mg/L). The isolate contained a plasmid-borne mph(A) gene encoding a macrolide 2′-phosphotransferase that inactivates macrolides. Emergence of the mph(A) gene in S. sonnei may limit usefulness of azithromycin for treatment of shigellosis.
Keywords: Antibiotic resistance, shigellosis, surveillance, outbreak, azithromycin, dispatch
Shigellosis remains a common gastrointestinal disease in developing and industrialized countries. It occurs mostly in children <5 years of age; Shigella sonnei is the most frequently isolated species (1). Ampicillin and trimethoprim-sulfamethoxazole alleviate the dysenteric syndrome of shigellosis and reduce the infectious period. However, current resistance patterns limit the use of these drugs (2). Although fluoroquinolones are an effective alternative for adults, they are not approved for shigellosis treatment in children <18 years of age because of their potential toxicity (2,3). Azithromycin, a macrolide, represents an attractive treatment option for several reasons. It has in vitro activity against most Shigella spp. isolates (4), can be given once a day, and attains high intracellular concentrations (5). Despite MICs from 2 to 8 mg/L for Shigella spp., sufficient concentrations of azithromycin in the colon may inhibit Shigella spp. growth (6). Azithromycin is recommended by the American Academy of Pediatrics for treatment of shigellosis in children, by the World Health Organization as a second-line treatment in adults, and, since June 2004, by the Agence Française de Sécurité Sanitaire des Produits de Santé (2,7; www.agmed.sante.gouv.fr/htm/10/filcoprs/mp040601.pdf). In 1996, 2002, 2003, and 2007, outbreaks of shigellosis caused by S. sonnei resistant to ampicillin and trimethoprim-sulfamethoxazole occurred in children in northern Paris. The outbreaks occurred in religious schools, similar to cyclic outbreaks in US Jewish schools related to secondary transmission (8,9).We report an outbreak of shigellosis in and around Paris, France, in which azithromycin failure was related to emergence of plasmid-mediated resistance to macrolides.
The Study
On January 24, 2007, S. sonnei strain UCN59 was isolated from a 4-year-old girl admitted to Robert Debré Hospital, Paris, for bloody diarrhea and fever. The strain was resistant to ampicillin, trimethoprim, sulfonamides, and cotrimoxazole but susceptible to quinolones, third-generation cephalosporins, and doxycycline according to the disk-diffusion technique. MICs of macrolides were markedly increased for S. sonnei UCN59 compared with those for a susceptible control S. sonnei UCN62 (Table). From January to April 23, 2007, a total of 50 cases of laboratory-confirmed shigellosis were identified. Isolates included, in addition to UCN59, 31 S. sonnei that had an azithromycin MIC >64 mg/L from 31 children <15 years of age, who had each been prescribed azithromycin for diarrhea. All patients lived in the Paris area and attended 8 religious schools.
Table. Macrolide susceptibility of outbreak and control Shigella isolates and Escherichia coli constructs.
| Strain | MIC, mg/L |
|||
|---|---|---|---|---|
| Erythromycin | Clarithromycin | Azithromycin | Telithromycin | |
| Shigella sonnei UCN 62 | 64 | 32 | 2 | 8 |
| S. sonnei UCN 59 | 1,024 | 1,024 | 64 | 64 |
| Escherichia coli K12 AG100A | 2 | 2 | 2 | 1 |
| E. coli K12 AG100A/pUV21 | 512 | 512 | 8 | 32 |
| E. coli K12 AG100A/pUC18Ωmph(A) | >1,024 | 512 | 128 | 512 |
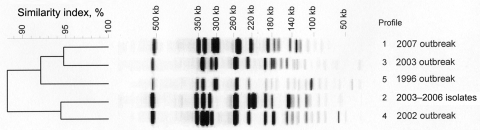
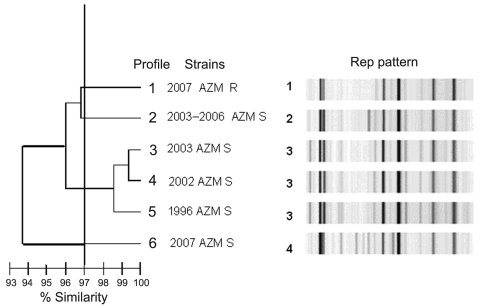
Typing by pulsed-field gel electrophoresis (PFGE) and repetitive sequence–based PCR (rep-PCR) by using the automated DiversiLab system (bioMérieux, La-Balme-les-Grottes, France) (10) was performed on the 32 azithromycin-resistant and on 11 azithromycin-susceptible (MIC <16 mg/L) sporadic or outbreak isolates obtained during 1996–2007 in the Paris area. Five different PFGE patterns were obtained by using the enzyme BlnI. All the 2007 outbreak isolates, including the 32 azithromycin-resistant isolates and 2 azithromycin-susceptible isolates, were clustered into a single profile, profile 1 (Figure 1). The presence of azithromycin-susceptible isolates with profile 1 was detected among the 1996 and 2002–2006 outbreak isolates (data not shown), showing the persistence of this clonal type over 10 years in this area of Paris. Other isolates displayed PFGE types 2 to 5. Low diversity of PFGE profiles was consistent with isolation of strains in the same area and for most of them from the same community. Four different patterns with <97% similarity were distinguished by rep-PCR (Figure 2). Again, all the 2007 azithromycin-resistant isolates were clustered; however, they could be distinguished from the 2007 azithromycin-susceptible isolates. In contrast to PFGE findings, rep-PCR showed that isolates representative of the 1996, 2002, and 2003 outbreaks were genetically related.
Figure 1.
Pulsed-field gel electrophoresis–generated dendrogram for 43 Shigella sonnei isolates obtained from sporadic or outbreak cases during 1996–2007 in the Paris area. Profile 1) representative isolates from the 2007 outbreak, including 32 isolates with azithromycin MIC >256 mg/L by Etest and 2 isolates with azithromycin MIC <16 mg/L. Profile 2) 6 representative isolates from sporadic cases (2003–2006) with azithromycin MIC <16 mg/L. Profile 3) representative isolate Shi 03-3580 from 2003 outbreak with azithromycin MIC <16 mg/L by Etest. Profile 4) representative isolate Shi 02-9633 from 2002 outbreak with azithromycin MIC <16 mg/L by Etest. Profile 5) representative isolate Shi 96 1420 from 1996 outbreak with azithromycin MIC <16 mg/L by Etest.
Figure 2.
Repetitive sequence–based, PCR–generated dendrogram for 43 Shigella sonnei isolates obtained from sporadic or outbreak cases during 1996–2007 in the Paris area. Isolates with >97% similarity were considered to be closely genetically related. Profile 1) representative of the 32 isolates of the 2007outbreak with azithromycin (MIC >256 mg/L by Etest) . Profile 2) 1 of 6 isolates from sporadic cases (2003–2006) with azithromycin MIC <16 mg/L. Profile 3) representative isolate Shi 03-3580 from 2003 outbreak with azithromycin MIC <16 mg/L by Etest. Profile 4) representative isolate Shi 02-9633 from 2002 outbreak with azithromycin MIC <16 mg/L by Etest. Profile 5) representative isolate Shi 96 1420 from 1996 outbreak with azithromycin MIC <16 mg/L by Etest. Profile 6) isolate from 2007 with azithromycin MIC <16 mg/L by Etest; another AZM S 2007 isolate had an identical profile. AZM, azithromycin; R, resistant; S, sensitive; Rep, repetitive sequence-based PCR.
The mph(A) gene, which encodes a macrolide 2′-phosphotransferase that inactivates macrolide antimicrobial drugs, was amplified from S. sonnei UCN59 DNA by PCR (11). PCR was negative for the erm(A), erm(TR), erm(B), erm(C), and erm(X) methylase genes; the ere(A), ere(B) genes encoding esterases; the mph(B) gene encoding a phosphotransferase; and the efflux genes mef(A) and msr(A). Sequence of the genes that encode ribosomal structures composing the target of macrolides, rrl, rplD, and rplV genes in S. sonnei UCN59, did not display any mutation in the critical bases of resistance to macrolides.
The genes conferring resistance to ampicillin and erythromycin were transferred en bloc by conjugation from S. sonnei UCN59 to a macrolide-susceptible mutant Escherichia coli AG100A at a frequency of ≈10–3 per donor cell-forming unit after the mating period. A single plasmid was extracted from a transconjugant E. coli AG100A/pUV21. After restriction analysis, its size was estimated at ≈90 kb. PCR experiments showed that this plasmid belonged to incompatibility group I (12). MICs of macrolides for E. coli AG100A/pUV21 confirmed that this plasmid conferred cross-resistance to macrolides (Table).
EcoRI-restricted fragments of plasmid pUV21 were transferred to a nylon membrane and hybridized to an mph(A) probe. The mph(A) gene was borne by an ≈20-kb EcoRI fragment, confirming that resistance to azithromycin was plasmid mediated.
After plasmid digestion with PstI enzyme, a DNA fragment that conferred resistance to erythromycin was cloned in plasmid pUC18 and introduced by transformation into E. coli AG100A to generate E. coli K12 AG100A/pUC18Ωmph(A). Sequence of the inserted DNA was determined. The fragment contained 4 open reading frames (ORFs) in the same orientation: mph(A), mrx that putatively encodes a membrane protein, mphR(A) that regulates the expression of mph(A), and an ORF of unknown function. This series of ORFs was flanked by a copy of IS26 at the 5′ end and a copy of IS6100 at the 3′ end. BLAST analysis (www.ncbi.nlm.nih.gov/blast/Blast.cgi) showed that the nucleotide sequence was nearly identical to that of fragments of plasmid pU302L from Salmonella enterica serotype Typhimurium (C.Y. Chen et al., unpub. data, GenBank accession no. NC_006816), of Shigella flexneri transposon TnSF1 (J.H. Chen and J.Y. Chen, unpub. data, GenBank accession no. AF188331), and of plasmids pRSB101 and pSRB107 (13,14).
Conclusions
Few data are available on azithromycin resistance in Shigella spp. A recent report from Bangladesh mentioned that 16% of Shigella isolates were resistant to azithromycin and that 62% had intermediate resistance according to the Clinical Laboratory Standards Institute breakpoints recommended for streptococci (>1 mg/L, resistant; <0.25 mg/L, susceptible) (15). However, the MIC90 (MIC at which 90% are susceptible) of 8 mg/L displayed by the microorganisms was within the normal range of MICs for this microorganism; no isolate had an azithromycin MIC >24 mg/L, which suggests that none had acquired resistance to azithromycin. Surveillance for resistance to azithromycin in Shigella spp. requires specific breakpoints for this species (3).
The mph(A) gene has been detected in the sequence of transposon TnSF1 isolated from S. flexneri (J.H. Chen and J.Y. Chen JY, unpub. data, GenBank accession no. AF188331). The mph(A) gene was first reported in an E. coli isolate from Japan (10). Since then, the gene has been found in Aeromonas hydrophila, Pseudomonas spp., Stenotrophomonas spp., and a variety of enterobacteria (listed at http://faculty.washington.edu/marilynr/ermweb4.pdf).
Azithromycin was used to treat shigellosis in France only after the release of the French recommendations in 2004. Subsequent rapid emergence of azithromycin-resistant isolates may be a limitation for the use of macrolides in shigellosis. Because use of azithromycin is proposed for treatment of shigellosis, susceptibility of the isolates to azithromycin should be routinely tested.
Biography
Dr Boumghar-Bourtchai is pursuing a PhD degree at the University of Caen; she completed this work as part of her PhD program. Her main research interest is emergence of unusual mechanisms of resistance to macrolides in various bacteria, such as S. sonnei, Turicella otitidis, and other gram-positive organisms.
Footnotes
Suggested citation for this article: Boumghar-Bourtchai L, Mariani-Kurkdjian P, Bingen E, Filliol I, Dhalluin A, Ait Ifrane S, et al. Macrolide-resistant Shigella sonnei. Emerg Infect Dis [serial on the Internet]. 2008 Aug [date cited]. Available from http://www.cdc.gov/EID/content/14/8/1297.htm
References
- 1.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, et al. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Outbreaks of multidrug-resistant Shigella sonnei gastroenteritis associated with day care centers—Kansas, Kentucky, and Missouri, 2005. MMWR Morb Mortal Wkly Rep. 2006;55:1068–71. [PubMed] [Google Scholar]
- 3.Jain SK, Gupta A, Glanz B, Dick J, Siberry GK. Antimicrobial-resistant Shigella sonnei: limited antimicrobial treatment options for children and challenges of interpreting in vitro azithromycin susceptibility. Pediatr Infect Dis J. 2005;24:494–7. 10.1097/01.inf.0000164707.13624.a7 [DOI] [PubMed] [Google Scholar]
- 4.Gordillo ME, Singh KV, Murray B. In vitro activity of azithromycin against bacterial enteric pathogens. Antimicrob Agents Chemother. 1993;37:1203–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gladue RP, Bright GM, Isaacson RE, Newborg MF. In vitro and in vivo uptake of azithromycin (CP-62,993) by phagocytic cells: possible mechanism of delivery and release at sites of infection. Antimicrob Agents Chemother. 1989;33:277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan WA, Seas C, Dhar U, Salam MA, Bennish ML. Treatment of shigellosis: V. Comparison of azithromycin and ciprofloxacin. A double-blind, randomized, controlled trial. Ann Intern Med. 1997;126:697–703. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization. Guidelines for the control of shigellosis, including epidemics due to Shigella dysenteriae type 1 [cited 2008 Apr 1]. Available from http://whqlibdoc.who.int/publications/2005/9241592330.pdf
- 8.Garrett V, Bornschlegel K, Lange D, Reddy V, Kornstein L, Komblum J, et al. A recurring outbreak of Shigella sonnei among traditionally observant Jewish children in New York City: the risks of daycare and household transmission. Epidemiol Infect. 2006;134:1231–6. 10.1017/S0950268806006182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sobel J, Cameron DN, Ismail J, Strockbine N, Williams M, Diaz PS, et al. A prolonged outbreak of Shigella sonnei infections in traditionally observant Jewish communities in North America caused by a molecularly distinct bacterial subtype. J Infect Dis. 1998;177:1405–9. [DOI] [PubMed] [Google Scholar]
- 10.Healy M, Huong J, Bittner T, Lising M, Frye S. Microbial DNA typing by automated repetitive-sequence-based PCR. J Clin Microbiol. 2005;43:199–207. 10.1128/JCM.43.1.199-207.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noguchi N, Emura A, Matsuyama H, O'Hara K, Sasatsu M, Kono M. Nucleotide sequence and characterization of erythromycin resistance determinant that encodes macrolide 2′-phosphotransferase I in Escherichia coli. Antimicrob Agents Chemother. 1995;39:2359–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–28. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- 13.Szczepanowski R, Krahn I, Linke B, Goesmann A, Pühler A, Schlüter A. Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology. 2004;150:3613–30. 10.1099/mic.0.27317-0 [DOI] [PubMed] [Google Scholar]
- 14.Szczepanowski R, Braun S, Riedel V, Schneiker S, Krahn I, Pühler A, et al. The 120 592 bp IncF plasmid pRSB107 isolated from a sewage-treatment plant encodes nine different antibiotic-resistance determinants, two iron-acquisition systems and other putative virulence-associated functions. Microbiology. 2005;151:1095–111. 10.1099/mic.0.27773-0 [DOI] [PubMed] [Google Scholar]
- 15.Rahman M, Shoma S, Rashid H, El Arifeen S, Baqui AH, Siddique AK, et al. Increasing spectrum in antimicrobial resistance of Shigella isolates in Bangladesh: resistance to azithromycin and ceftriaxone and decreased susceptibility to ciprofloxacin. J Health Popul Nutr. 2007;25:158–67. [PMC free article] [PubMed] [Google Scholar]