Abstract
Activating mutations in NOTCH1 are the most prominent genetic abnormality in T-cell acute Lymphoblastic Leukemia (T-ALL) and inhibition of NOTCH1 signaling with γ-secretase inhibitors (GSIs) has been proposed as targeted therapy in this disease. However, most T-ALL cell lines with mutations in NOTCH1 fail to respond to GSI therapy. Using gene expression profiling and mutation analysis we showed that mutational loss of PTEN is a common event in T-ALL and is associated with resistance to NOTCH inhibition. Furthermore, our studies revealed that NOTCH1 induces upregulation of the PI3K-AKT pathway via HES1, which negatively controls the expression of PTEN. This regulatory circuitry is evolutionary conserved from Drosophila to humans as demonstrated by the interaction of overexpression of Delta and Akt in a model of Notch-induced transformation in the fly eye. Loss of PTEN and constitutive activation of AKT in T-ALL induce increased glucose metabolism and bypass the requirement of NOTCH1 signaling to sustain cell growth. Importantly, PTEN-null/GSI resistant T-ALL cells switch their oncogene addiction from NOTCH1 to AKT and are highly sensitive to AKT inhibitors. These results should facilitate the development of molecular therapies targeting NOTCH1 and AKT for the treatment of T-ALL.
Keywords: T-cell lymphoblastic leukemia, NOTCH1, PTEN, AKT, γ-secretase inhibitor, oncogene addiction
Targeting NOTCH1 signaling in T-ALL
T-lineage acute lymphoblastic leukemia (T-ALL) is an aggressive hematologic cancer that accounts for 10% to 15% of pediatric and 25% of adult ALL cases. 1, 2 Despite recent progress in the treatment of this disease the prognosis of T-ALL patients with primary resistant or relapsed disease is very poor, underscoring the need to develop more effective antileukemic drugs. 3-5
The identification in the last years of aberrant activation of NOTCH1 signaling in the majority of T-ALL cases and the feasibility to inhibit the activation of NOTCH receptors with small molecule inhibitors of the gamma-secretase complex have created the opportunity to develop molecular therapies targeting the NOTCH1 signaling pathway in this disease. 6, 7
NOTCH signaling plays a critical role in lineage specification decisions by which equivalent multipotential precursor cells become committed to specific cell lineages during development, and has important roles in cellular differentiation, proliferation and apoptosis. 6-9 The fundamental components of the NOTCH pathway are the DSL ligands (Delta-like 1, 3 and 4; and Jagged 1 and 2), the NOTCH receptors (NOTCH1, 2, 3 and 4), and the CSL DNA binding protein, a transcription factor which mediates the conversion of NOTCH activating signals in the cell surface into changes in gene expression in the nucleus.
Both the DSL ligands and the NOTCH receptors are type I transmembrane proteins, and activation of the NOTCH signaling pathway is triggered by the interaction of a NOTCH receptor in one cell with a DSL ligand expressed in the surface of a neighboring cell. This ligand-receptor interaction induces two consecutive proteolytic cleavages, first by an ADAM metalloprotease at the cell surface and subsequently by the γ-secretase complex, which catalyzes the final cleavage step in the transmembrane domain of the receptor. After γ-secretase cleavage the resulting activated form of NOTCH (ICN1) rapidly translocates to the nucleus where it interacts with the CSL DNA binding protein. Binding of ICN1 to CSL recruits the MAML1 coactivator to the complex and induces the transcriptional activation of NOTCH-CSL target genes. 6, 7, 10
The NOTCH signaling pathway plays a critical role in the hematopoietic system at multiple stages of T-cell development. During early hematopoiesis NOTCH signaling is required for the commitment of multipotent hematopoietic progenitors to the T-cell lineage. 2, 11-13 Immunodeficient mice reconstituted with bone marrow progenitors expressing a constitutively active form of Notch1 show ectopic T-cell development in the bone marrow and, in contrast with mice reconstituted with normal progenitor cells, fail to produce B lymphocytes. 2 Conversely, mice harboring a conditional deletion of Notch1 in hematopoietic progenitors fail to develop T-cells, and show ectopic B-cell development in the thymus. 11
In addition to this early role in T-cell lineage commitment, NOTCH signaling participates, later on, in essential processes at multiple stages of thymocyte development. 14 Thus, NOTCH1 activity is required for lineage progression through the early DN1, DN2 and DN3 stages of thymocyte development; 15 participates in the regulation of T-cell receptor loci; 16 and regulates lineage decisions between αβ vs. γδ lineages. 17
The first evidence linking aberrant NOTCH1 signaling to the pathogenesis of T-ALL came from the characterization of the t(7;9)(q34;q34.3) translocation, a rare recurrent chromosomal rearrangement present in less than 1% of human T-ALL cases. This translocation juxtaposes a truncated NOTCH1 gene next to the T-cell receptor B (TCRB) locus leading to the aberrant expression of an intracellular constitutively active form of NOTCH1.18 Following this initial finding, the causative role of activated NOTCH1 in the pathogenesis of T-ALL was further demonstrated by animal models of NOTCH1 induced leukemia. Thus, mice reconstituted with hematopoietic progenitor cells transduced with viruses driving the expression of constitutively active forms of NOTCH1 develop T-cell tumors, 19 and transgenic mice expressing dominant active forms of NOTCH1 in hematopoietic progenitor cells or in immature thymocytes developed T-cell leukemias. 19, 20
However, the role of NOTCH1 in the pathogenesis of human T-ALL was not fully realized until the identification of activating mutations in NOTCH1 in the majority of T-ALL patient samples.21 Activating mutations in NOTCH1 result in ligand independent activation of the receptor or increased ICN1 protein stability in over 50% of T-ALLs.21, 22 23 24, 25 Importantly, small molecule inhibitors of the γ-secretase complex (GSIs) effectively abrogate the function of the receptor encoded by these oncogenic NOTCH1 alleles, making NOTCH1 a promising therapeutic target for the treatment of T-ALL. Initial studies treating human T-ALL cell lines harboring activating mutations in NOTCH1 with GSIs demonstrated that inhibition of NOTCH signaling induces cell cycle arrest.21, 26, 27 However, these same studies also demonstrated that despite high prevalence of NOTCH1 mutations and the presence of high levels of ICN1 in these tumors, the majority of T-ALL cell lines failed to respond to NOTCH1 inhibition, suggesting that primary resistance to GSI treatment was readily present in the majority of these cell lines.21
Numerous mechanisms have been identified in the resistance of cancer cells to chemotherapy and molecularly targeted drugs. 28-30 General mechanisms of drug resistance typically decrease the effective intracellular concentration of the drug due to decreased drug uptake, increased drug export or drug metabolism. In the case of molecularly targeted drugs, resistance typically results from mutations that block the interaction of the drug with its specific molecular target. A common feature in all these mechanisms is that defective cellular responses are the result of persistent biochemical activity of the molecules and pathways that mediate drug response. Thus, the initial experiments aimed to identify the mechanisms of resistance to GSIs focused on the analysis of NOTCH1 activity. Surprisingly, these studies revealed that GSI treatment effectively blocked γ-secretase activity and as a result, induced decreased levels of ICN1 and transcriptional downregulation of NOTCH1 target genes in GSI-resistant T-ALL cell lines.31 This observation indicated that the lack of response to GSIs occurs despite effective NOTCH1 inhibition, and pointed to the existence of alternative pathways promoting cell growth in the absence of NOTCH1 signaling in these tumors. Thus, we postulated that understanding of the mechanisms of sensitivity and resistance to GSIs in T-ALL required the identification of the key downstream target genes and pathways responsible for NOTCH1 induced transformation.
Microarray gene expression studies and ChIP-on-chip analysis have recently identified a major role of NOTCH1 in the regulation of cell growth and metabolism.32 Thus, NOTCH1 inhibition with a GSI drives a gene expression signature dominated by the downregulation of genes involved in anabolic pathways such as ribosome biosynthesis, protein translation, RNA synthesis and nucleotide metabolism in T-ALL.32-35 Notably, NOTCH1 also controls the expression of MYC, which in turn is also a broad transcriptional activator of anabolic genes and pathways involved in growth and metabolism.32 This NOTCH1-MYC transcriptional network constitutes a feed-forward-loop circuit that confers increased robustness to the transcriptional regulation of cell growth downstream of NOTCH1 signaling. In addition, this model couples cell fate and developmental signals upstream of NOTCH1 with the activation of pathways promoting cell proliferation upstream of MYC, and establishes a fundamental relationship between NOTCH1 and MYC in T-cell transformation. However, this NOTCH1-MYC feed forward loop regulatory network constitutes only a fraction of the transcriptional network that mediates the oncogenic and developmental effects of NOTCH1 signaling in T-cells. In this context, we hypothesized that aberrant activation of effector signaling pathways controlling cell growth and metabolism downstream of NOTCH1 could uncouple NOTCH1 signaling and cell proliferation rendering leukemic T-cells resistant to NOTCH inhibition with GSIs.
PTEN mutations confer resistance to GSI in T-ALL
To identify possible mechanisms of resistance to NOTCH1 inhibition in T-ALL, we analyzed global gene expression signatures associated with GSI sensitivity or resistance in a panel of leukemia cell lines using oligonucleotide microarrays. This analysis identified decreased levels of the PTEN tumor suppressor gene as the most consistent transcriptional feature associated with GSI-resistant cell lines.31 Notably, Western blot and mutation analysis showed total absence or marked decrease in PTEN protein levels and the presence of biallelic loss of function mutations in PTEN in GSI-resistant T-ALL cells, respectively. In contrast, all GSI-sensitive cell lines analyzed expressed wild type PTEN transcripts and had readily detectable levels of PTEN protein.
The PTEN tumor suppressor gene encodes a lipid phosphatase responsible for the degradation of phosphatidylinositol triphosphate (PIP3) and the inhibition of the PI3K-AKT signaling pathway, which promotes cell growth, increased glucose uptake and oxidation, cell cycle progression and cell survival through multiple direct and indirect mechanisms. 36-41 The PI3K-AKT pathway is activated upon generation of PIP3 following the engagement of growth factor receptors at the plasma membrane and the subsequent activation of class I phosphatidylinositol 3-kinases (PI3Ks). The accumulation of PIP3 at the membrane recruits the AKT kinase inducing its phosphorylation and activation by the PDK1 and the mTOR-Rictor kinases. 42, 43
Consistent with the presence of loss of function mutations in PTEN, all GSI-resistant T-ALL cell lines analyzed showed increased levels of AKT phosphorylation, indicative of constitutive PI3K-AKT activation. These results pointed to a close relationship between the PI3K-AKT signaling pathway, and GSI resistance.
Homozygous and heterozygous somatic mutations in PTEN have been described in a very broad range of human cancers. 44-46 However, PTEN mutations had only been reported sporadically in leukemias and lymphomas. 47-50 Thus, to determine whether our discovery of PTEN loss in GSI-resistant cell lines might also be relevant to primary human cancers, we examined the status of PTEN by immunohistochemistry and flow cytometry in T-cell lymphoblastic leukemia and lymphoma (T-ALL/LL) clinical samples. These analyses demonstrated complete loss of the PTEN protein in 17% of T-ALL/LL cases at diagnosis. In addition, mutation analysis demonstrated the presence of PTEN mutations in 8% of T-ALL samples. Consistent with the role of PTEN in the regulation of the PI3K-AKT signaling pathway, immunohistochemistry analysis showed increased levels of phosphoAKT in T-cell lymphoblastic tumors with loss of PTEN. Notably, the analysis of paired diagnostic and relapsed tumor samples showed patients with loss of PTEN at relapse indicating that PTEN loss can be associated with tumor progression in T-ALL. Overall these results demonstrate that PTEN loss and constitutive activation of PI3K-AKT signaling is a frequent event in T-cell lymphoblastic leukemias and lymphomas and is associated with resistance to NOTCH1 inhibition with GSIs.
These results led us to experimentally test the hypothesis that loss of PTEN and activation of AKT plays a causative role in resistance to GSIs. To address this question we first expressed a constitutively active form of AKT (MYR-AKT) in GSI-sensitive/PTEN-positive cells and showed that AKT activation is sufficient to rescue the reduction in cell growth induced by NOTCH1 inhibition with a GSI. Similarly, shRNA knock-down of PTEN in GSI-sensitive/PTEN-positive cells induced resistance to GSI treatment. These results mechanistically linked the loss of PTEN and constitutive activation of AKT with resistance to NOTCH1 inhibition in human T-ALL cell lines.
Two alternative models emerged from these results. First, it is possible that NOTCH1 and PI3K-AKT signaling are parallel pathways controlling cell growth in leukemic lymphoblasts and that constitutive activation of AKT in PTEN-null cells provides alternative trophic signals that sustain cell growth upon inhibition of NOTCH1 signaling with GSIs. Alternatively, it is possible that NOTCH1 and PI3K-AKT signaling are closely interlinked in the control of cell growth and proliferation during normal T-cell development and that constitutive activation of NOTCH1 signaling promotes cell growth in part through PI3K-AKT. According to the latter model, mutational loss of PTEN would bypass the requirement of NOTCH1 to sustain cell growth.
NOTCH1 regulatesPTENexpression and the activity of the PI3K-AKT pathway via HES1 and MYC
Although the induction of resistance to NOTCH1 inhibition by constitutive activation of the AKT pathway does not strictly require a functional connection between NOTCH and the PI3K-AKT pathways, the effects of GSI treatment in T-ALL cells closely resembles the growth defect induced by nutrient deprivation, cytokine withdrawal and inhibition of the PI3K pathway. In addition, NOTCH1 activation has recently been shown to be essential to sustain AKT activation and glucose metabolism during thymocyte development.51 These observations, together with the close association between the presence of PTEN mutations and GSI resistance led us to hypothesize that PTEN might be functionally linked to NOTCH1 signaling in T-ALL.
To test this possibility we analyzed the effects of NOTCH1 inhibition with a GSI in the PI3K-AKT pathway in PTEN-positive/GSI-sensitive T-ALL cells. These studies showed that blocking the activity of oncogenic NOTCH1 was followed by transcriptional upregulation of PTEN, increased PTEN protein levels and decreased activity of the PI3K-AKT pathway as shown by decreased phosphorylation of AKT and AKT targets. Furthermore, analysis of NOTCH1 inhibition in primary T-cell progenitors by removal of the NOTCH ligand Delta-like 1 results in downregulation of NOTCH1 target and induces transcriptional upregulation of Pten. These results demonstrate that NOTCH1 signaling regulates the PI3K-AKT pathway in human leukemic cells expressing mutant NOTCH1 receptors and also in primary mouse thymocytes expressing wild type NOTCH1. In addition, the effects of NOTCH signaling on PTEN expression are evidenced by pharmacologic inhibition of NOTCH signaling with a GSI and also upon withdrawal of NOTCH signals by removal of the stimulus provided by NOTCH ligands.
How does NOTCH signaling regulate the expression of PTEN? Given the well-established role of NOTCH1 as a transcriptional activator, we proposed that downregulation of PTEN transcripts by NOTCH1 signaling could be mediated by a transcriptional repressor downstream of NOTCH1.
Among the numerous signaling molecules and transcriptional factors controlled by NOTCH1 we selected HES1, as a possible mediator of the effects of NOTCH1 on PTEN expression. HES1 is a transcriptional repressor directly controlled by NOTCH1 52 and has been shown to mediate important NOTCH1 functions in T-cell development 53. In addition, analysis of PTEN regulatory sequences across different species demonstrated the presence of a conserved MYC-MAX canonical sequence in the PTEN proximal promoter, suggesting that NOTCH1 could regulate PTEN expression through MYC. ChIP-on-chip analysis of HES1 and MYC showed that both factors bind to the PTEN promoter in T-ALL cells. Notably, HES1 induced a marked reduction in the activity of the PTEN promoter in reporter assays, while MYC expression induced a moderate increase in the PTEN promoter activity, which was effectively abrogated by HES1 expression. Thus transcriptional regulation of PTEN downstream of NOTCH1 seems to be controlled by a dual input consisting of negative signals from HES1 and positive signals by MYC. The balance between these two factors and the dominant effect of HES1 over MYC in the activity of the PTEN promoter is consistent with our observation of a moderate and slow upregulation of PTEN upon NOTCH1 inhibition and illustrates the essential role of combinatorial transcriptional regulatory systems to tune the kinetics and intensity of potentially oncogenic pathways controlling cell growth. Thus, the combined action of HES1 and MYC downstream of NOTCH1 in T-cell progenitors increases the activity of the PI3K-AKT signaling pathway in response to extracellular stimuli and promotes cell growth without inducing full oncogenic activation of AKT. In contrast, mutational loss of PTEN eliminates this transcriptional regulatory node, uncouples the PI3K-AKT pathway from extracellular signals, induces oncogenic levels of AKT, and bypasses the requirement of NOTCH1 signaling to maintain cell growth in T-ALL.
NOTCH1 and the PI3K-AKT pathways regulate glucose metabolism
The PI3K-AKT pathway controls numerous cellular functions associated with cell growth and regulates glucose metabolism at multiple levels 36, 54. First, AKT activation promotes glucose uptake by the GLUT family of glucose transporters 55-58 and activates glycolysis by hexokinase and phosphofructokinase 59, 60. Additionally, AKT inhibition of glycogen synthase kinase1 promotes glycogen synthesis 61 and blocks gluconeogenesis 58. Most importantly, constitutive AKT signaling results in constitutive activation of aerobic glycolysis in tumor cells, a phenomenon known as the Warburg effect. 62, 63 The potential relevance of glucose metabolism in the sensitivity and resistance to NOTCH1 inhibition in T-ALL is highlighted by the prominent reduction in cell size of PTEN-positive/GSI-sensitive T-ALL cells treated with GSI and by evidence of a major role of NOTCH signaling in promoting glucose metabolism in developing thymocytes.51
Analysis of glucose metabolism in T-ALL cells showed that PTEN-null/GSI-resistant cells have higher levels of glucose uptake and glucose oxidation compared with PTEN-positive/GSI-sensitive cell lines. Furthermore, GSI treatment of PTEN-positive/GSI sensitive cells induced further reduction in glucose uptake and glucose oxidation, which were unaltered upon inhibition of NOTCH1 signaling in PTEN-null/GSI-resistant cells. Finally, treatment with methyl pyruvate, a membrane permeable metabolite that bypasses glycolysis and directly feeds the Krebs cycle, rescues the defects in cell size and cell cycle progression induced by GSI treatment in PTEN-positive/GSI-sensitive T-ALL cells (P. Real and A. Ferrando unpublished results). Overall, these results show a fundamental role for glucose metabolism in the maintenance of cell growth in T-ALL and mechanistically link the antitumor effects of GSIs with the inhibition of glucose uptake and oxidation in leukemic lymphoblasts.
Conservation of the NOTCH-AKT interplay in Drosophila
The growth and morphogenesis of the Drosophila eye depends on the activation of the Notch receptor by its ligands Delta and Serrate/Jagged in a conserved dorsal-ventral organizer, a specialized region of the eye imaginal disc that controls the growth and organization of the fly eye during development 64-66. Ubiquitous overexpression of Delta in the developing fly eye induces a mild overgrowth which has been exploited to identify genes that interact the Delta-Notch pathway and induce tumors using forward genetic screens 64, 67. In this screen, gain-of-expression mutations induced at random in the fly genome using the Gene Search (GS) system were combined with a gain of expression of Delta to isolate genetic alterations that synergize with Notch hyperactivation and induce an overt tumor phenotype 67.
A remarkable finding in this screen was the identification of a gain-of-expression mutation within the Akt locus that induced a tumor invasion phenotype when combined with Delta overexpression. Flies from this Akt mutant line show no spontaneous tumor phenotype. In contrast, when crossed with the Delta expressing line, activation of Akt1 induces massive tumor overgrowths with a 100% penetrance and distal metastasis in 8% of the insects. Similarly, overexpression of the PI3K homolog (Dp110) with Delta also induced tumors showing that activation of Notch and PI3K-Akt have synergistic effects in tumor development in this model. A requirement of Akt for Notch-induced growth in non-tumoral conditions was also demonstrated by experiments in which Notch induced overgrowth was completely abrogated by inhibiting the PI3K/AKT pathway through overexpression of Pten. In addition, we showed using a null Akt allele (analyzed in mosaic patches of mutant cells), that eye overgrowth induced by expression of Delta is almost totally prevented in cells null for Akt. Finally, we showed also that, misexpression of the Notch modulator fringe induces a small eye phenotype, which can be rescued by overexpression of Akt. Thus Akt activation is necessary for NOTCH induced growth and also sufficient to restore cell growth upon inhibition of Notch signaling, an outcome that perfectly recapitulates the results obtained in the analysis of leukemic cells and shows that the functional interaction between Notch and AKT is evolutionary conserved.
Oncogene addiction
The model that emerges from these results places NOTCH1 at the center of a transcriptional regulatory network controlling cell growth in T-ALL. Thus, NOTCH1 directly controls the expression of a broad number of genes involved in cell anabolic pathways, an effect that is further enhanced by direct transcriptional upregulation of the MYC oncogene and indirect transcriptional downregulation of the PTEN tumor suppressor gene via HES1. Given the redundancy of mechanisms promoting cell growth downstream of NOTCH1, it will not be surprising that the identification of additional edges and nodes in this network shows new effector genes and pathways involved in T-cell transformation. Nevertheless, the basic observation that PTEN loss is coupled with fundamental metabolic changes induced by constitutive activation of AKT suggests that GSI resistance may require significant rewiring of the metabolic machinery of these cells. Thus, we hypothesized that GSI resistance could occur at the expense of making PTEN-null T-ALL cells addicted to constitutive AKT signaling. In agreement with this model, retroviral expression of PTEN-induced decreased cell size and cell cycle arrest in PTEN-null/GSI-resistant cells. In addition, treatment of T-ALL cells with a phosphatidylinositol analog inhibitor of AKT demonstrated that GSI-resistant/PTEN-null T-ALL cells are significantly more sensitive to AKT inhibition than GSI-sensitive/PTEN-positive cells. These results suggest that, indeed, aberrant activation of the PI3K-AKT pathway makes PTEN-null T-ALL cells dependent on high levels of AKT signaling in a clear example of secondary oncogene addiction.
Concluding remarks and future directions
The introduction of highly intensive therapy has led to significant progress in the treatment of childhood leukemias over the last decades. However, 20 % of pediatric and 50% of adult T-ALL cases still relapse and ultimately die because of refractory disease. Furthermore, the intensified therapies used in the treatment of T-ALL are frequently associated with life-threatening and debilitating toxicities adding significant acute and chronic morbidity to this disease. In this context, the identification of activating mutations in NOTCH1 present in over 50% of T-ALL patients at diagnosis 21 has brought enormous interest for the development of molecularly tailored therapies in T-ALL and prompted the initiation of a clinical trial to test the effectiveness of blocking NOTCH1 signaling with a small molecule GSI in this disease. Furthermore, a growing body of literature supports a role for aberrant NOTCH signaling in the pathogenesis of breast cancer, medulloblastoma, lung cancer, colon carcinomas, multiple myeloma and pancreatic cancer; 68-83 and GSIs targeting NOTCH signaling are in clinical trials for breast cancer and medulloblastoma. Thus, the successful development of anti NOTCH therapies and overcoming the resistance to NOTCH inhibitors in T-ALL may ultimately have a broad impact in the treatment of numerous human cancers.
Our studies have offered new insights on the role of NOTCH1 signaling in the regulation of the PTEN/AKT pathway and the mechanisms of resistance to GSIs in T-ALL. The identification of PTEN mutations as a frequent event in T-ALL highlights the importance of this tumor suppressor gene in the pathogenesis of T-ALL. Furthermore, the association of mutational loss of PTEN with resistance to GSIs should be tested prospectively in clinical trials testing the efficacy of GSIs in the treatment of T-ALL. Finally, the increased sensitivity of T-ALL lines resistant to GSI to inhibitors of the AKT pathway should be further studied using adequate animal models to analyze the underlying mechanisms and to test potentially new targeted therapies for the treatment of T-ALL.
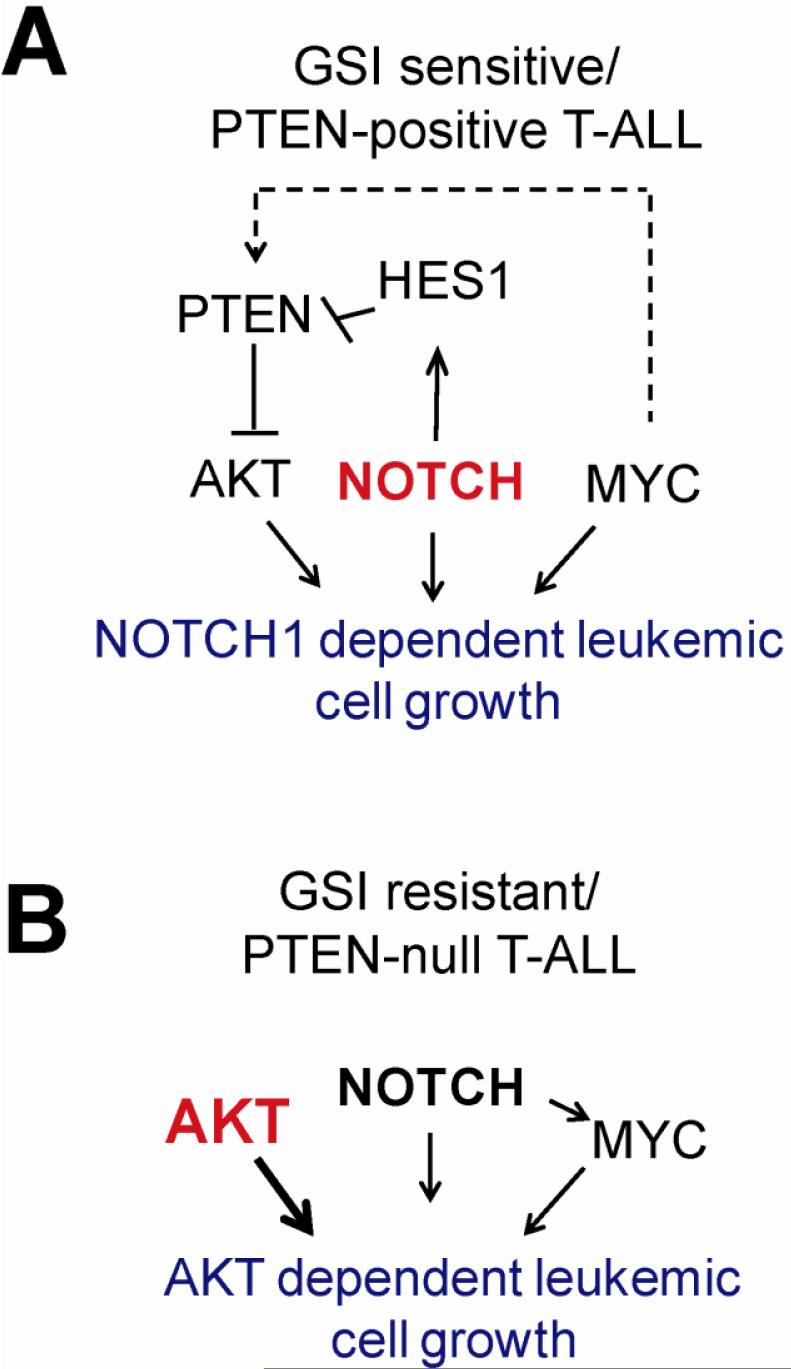
Figure 1. PTEN loss disrupts the circuitry controlling oncogenic cell growth in T-ALL downstream of NOTCH1 in T-ALL.
A. NOTCH1 controls leukemic cell growth through multiple overlapping mechanisms including activation of target genes, upregulation of MYC and downregulation of PTEN via HES1. B. Mutational loss of PTEN induces constitutive activation of AKT and uncouples NOTCH1 signaling from the PI3K-AKT pathway. Thus, PTEN-null T-ALL cells become insensitive to inhibition of NOTCH1 signaling with GSIs.
Acknowledgments
Acknowledgements
Supported by the Alex's Lemonade Stand Foundation (TP), the Spanish Ministerio de Educacion y Ciencia and Asociación Española Contra el Cancer (MD), the Cancer Research Institute, the WOLF Foundation, the Swim Across America Foundation, the Leukemia and Lymphoma Society (grants 1287-08 and 6237-08) and NIH grant CA120196, (AF). Adolfo Ferrando is a Leukemia and Lymphoma Society Scholar
Glossary
Abbreviations
- T-ALL
T-cell acute lymphoblastic leukemia
- GSI
Gamma Secretase Inhibitor
- DSL
DELTA/Serrate/Lag1 ligands
- ICN1
Intracellular NOTCH1
References
- 1.Ferrando AA, Look AT. Clinical implications of recurring chromosomal and associated molecular abnormalities in acute lymphoblastic leukemia. Semin Hematol. 2000;37:381–95. doi: 10.1016/s0037-1963(00)90018-0. [DOI] [PubMed] [Google Scholar]
- 2.Pui CH, Sandlund JT, Pei D, Campana D, Rivera GK, Ribeiro RC, Rubnitz JE, Razzouk BI, Howard SC, Hudson MM, Cheng C, Kun LE, Raimondi SC, Behm FG, Downing JR, Relling MV, Evans WE. Improved outcome for children with acute lymphoblastic leukemia: results of Total Therapy Study XIIIB at St Jude Children's Research Hospital. Blood. 2004;104:2690–6. doi: 10.1182/blood-2004-04-1616. [DOI] [PubMed] [Google Scholar]
- 3.Biggs JC, Horowitz MM, Gale RP, Ash RC, Atkinson K, Helbig W, Jacobsen N, Phillips GL, Rimm AA, Ringden O, et al. Bone marrow transplants may cure patients with acute leukemia never achieving remission with chemotherapy. Blood. 1992;80:1090–3. [PubMed] [Google Scholar]
- 4.Barrett AJ, Horowitz MM, Pollock BH, Zhang MJ, Bortin MM, Buchanan GR, Camitta BM, Ochs J, Graham-Pole J, Rowlings PA, et al. Bone marrow transplants from HLA-identical siblings as compared with chemotherapy for children with acute lymphoblastic leukemia in a second remission. N Engl J Med. 1994;331:1253–8. doi: 10.1056/NEJM199411103311902. [DOI] [PubMed] [Google Scholar]
- 5.Schroeder H, Gustafsson G, Saarinen-Pihkala UM, Glomstein A, Jonmundsson G, Nysom K, Ringden O, Mellander L. Allogeneic bone marrow transplantation in second remission of childhood acute lymphoblastic leukemia: a population-based case control study from the Nordic countries. Bone Marrow Transplant. 1999;23:555–60. doi: 10.1038/sj.bmt.1701617. [DOI] [PubMed] [Google Scholar]
- 6.Aster JC, Pear WS, Blacklow SC. Notch Signaling in Leukemia. Annu Rev Pathol. 2007 doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2006;6:347–59. doi: 10.1038/nrc1880. [DOI] [PubMed] [Google Scholar]
- 8.Greenwald I. LIN-12/Notch signaling: lessons from worms and flies. Genes Dev. 1998;12:1751–62. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- 9.Maillard I, Pear WS. Notch and cancer: best to avoid the ups and downs. Cancer Cell. 2003;3:203–5. doi: 10.1016/s1535-6108(03)00052-7. [DOI] [PubMed] [Google Scholar]
- 10.Wu L, Sun T, Kobayashi K, Gao P, Griffin JD. Identification of a family of mastermind-like transcriptional coactivators for mammalian notch receptors. Mol Cell Biol. 2002;22:7688–700. doi: 10.1128/MCB.22.21.7688-7700.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–58. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 12.Jaleco AC, Neves H, Hooijberg E, Gameiro P, Clode N, Haury M, Henrique D, Parreira L. Differential effects of Notch ligands Delta-1 and Jagged-1 in human lymphoid differentiation. J Exp Med. 2001;194:991–1002. doi: 10.1084/jem.194.7.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pear WS, Radtke F. Notch signaling in lymphopoiesis. Semin Immunol. 2003;15:69–79. doi: 10.1016/s1044-5323(03)00003-4. [DOI] [PubMed] [Google Scholar]
- 14.Pear WS, Aster JC. T cell acute lymphoblastic leukemia/lymphoma: a human cancer commonly associated with aberrant NOTCH1 signaling. Curr Opin Hematol. 2004;11:426–33. doi: 10.1097/01.moh.0000143965.90813.70. [DOI] [PubMed] [Google Scholar]
- 15.Schmitt TM, Ciofani M, Petrie HT, Zuniga-Pflucker JC. Maintenance of T cell specification and differentiation requires recurrent notch receptor-ligand interactions. J Exp Med. 2004;200:469–79. doi: 10.1084/jem.20040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfer A, Wilson A, Nemir M, MacDonald HR, Radtke F. Inactivation of Notch1 impairs VDJbeta rearrangement and allows pre-TCR-independent survival of early alpha beta Lineage Thymocytes. Immunity. 2002;16:869–79. doi: 10.1016/s1074-7613(02)00330-8. [DOI] [PubMed] [Google Scholar]
- 17.Washburn T, Schweighoffer E, Gridley T, Chang D, Fowlkes BJ, Cado D, Robey E. Notch activity influences the alphabeta versus gammadelta T cell lineage decision. Cell. 1997;88:833–43. doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]
- 18.Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–61. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 19.Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J, Baltimore D. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–91. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deftos ML, Huang E, Ojala EW, Forbush KA, Bevan MJ. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 2000;13:73–84. doi: 10.1016/s1074-7613(00)00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 22.Zhu YM, Zhao WL, Fu JF, Shi JY, Pan Q, Hu J, Gao XD, Chen B, Li JM, Xiong SM, Gu LJ, Tang JY, Liang H, Jiang H, Xue YQ, Shen ZX, Chen Z, Chen SJ. NOTCH1 mutations in T-cell acute lymphoblastic leukemia: prognostic significance and implication in multifactorial leukemogenesis. Clin Cancer Res. 2006;12:3043–9. doi: 10.1158/1078-0432.CCR-05-2832. [DOI] [PubMed] [Google Scholar]
- 23.Mansour MR, Duke V, Foroni L, Patel B, Allen CG, Ancliff PJ, Gale RE, Linch DC. Notch-1 mutations are secondary events in some patients with T-cell acute lymphoblastic leukemia. Clin Cancer Res. 2007;13:6964–9. doi: 10.1158/1078-0432.CCR-07-1474. [DOI] [PubMed] [Google Scholar]
- 24.Mansour MR, Linch DC, Foroni L, Goldstone AH, Gale RE. High incidence of Notch-1 mutations in adult patients with T-cell acute lymphoblastic leukemia. Leukemia. 2006;20:537–9. doi: 10.1038/sj.leu.2404101. [DOI] [PubMed] [Google Scholar]
- 25.Breit S, Stanulla M, Flohr T, Schrappe M, Ludwig WD, Tolle G, Happich M, Muckenthaler MU, Kulozik AE. Activating NOTCH1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lymphoblastic leukemia. Blood. 2006;108:1151–7. doi: 10.1182/blood-2005-12-4956. [DOI] [PubMed] [Google Scholar]
- 26.Palomero T, Barnes KC, Real PJ, Bender JL, Sulis ML, Murty VV, Colovai AI, Balbin M, Ferrando AA. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006;20:1279–87. doi: 10.1038/sj.leu.2404258. [DOI] [PubMed] [Google Scholar]
- 27.Lewis HD, Leveridge M, Strack PR, Haldon CD, O'Neil J, Kim H, Madin A, Hannam JC, Look AT, Kohl N, Draetta G, Harrison T, Kerby JA, Shearman MS, Beher D. Apoptosis in T cell acute lymphoblastic leukemia cells after cell cycle arrest induced by pharmacological inhibition of notch signaling. Chem Biol. 2007;14:209–19. doi: 10.1016/j.chembiol.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 28.Apperley JF. Part I: mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007;8:1018–29. doi: 10.1016/S1470-2045(07)70342-X. [DOI] [PubMed] [Google Scholar]
- 29.Ferrer-Soler L, Vazquez-Martin A, Brunet J, Menendez JA, De Llorens R, Colomer R. An update of the mechanisms of resistance to EGFR-tyrosine kinase inhibitors in breast cancer: Gefitinib (Iressa) -induced changes in the expression and nucleo-cytoplasmic trafficking of HER-ligands (Review) Int J Mol Med. 2007;20:3–10. [PubMed] [Google Scholar]
- 30.Mellor HR, Callaghan R. Resistance to Chemotherapy in Cancer: A Complex and Integrated Cellular Response. Pharmacology. 2008;81:275–300. doi: 10.1159/000115967. [DOI] [PubMed] [Google Scholar]
- 31.Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, Caparros E, Buteau J, Brown K, Perkins SL, Bhagat G, Agarwal AM, Basso G, Castillo M, Nagase S, Cordon-Cardo C, Parsons R, Zuniga-Pflucker JC, Dominguez M, Ferrando AA. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007 doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, Barnes KC, O'Neil J, Neuberg D, Weng AP, Aster JC, Sigaux F, Soulier J, Look AT, Young RA, Califano A, Ferrando AA. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103:18261–6. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klinakis A, Szabolcs M, Politi K, Kiaris H, Artavanis-Tsakonas S, Efstratiadis A. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc Natl Acad Sci U S A. 2006;103:9262–7. doi: 10.1073/pnas.0603371103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, Li Y, Wolfe MS, Shachaf C, Felsher D, Blacklow SC, Pear WS, Aster JC. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma VM, Calvo JA, Draheim KM, Cunningham LA, Hermance N, Beverly L, Krishnamoorthy V, Bhasin M, Capobianco AJ, Kelliher MA. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol. 2006;26:8022–31. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katso R, Okkenhaug K, Ahmadi K, White S, Timms J, Waterfield MD. Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2001;17:615–75. doi: 10.1146/annurev.cellbio.17.1.615. [DOI] [PubMed] [Google Scholar]
- 37.Sulis ML, Parsons R. PTEN: from pathology to biology. Trends Cell Biol. 2003;13:478–83. doi: 10.1016/s0962-8924(03)00175-2. [DOI] [PubMed] [Google Scholar]
- 38.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6:184–92. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 39.Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5:921–9. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 40.Samuels Y, Ericson K. Oncogenic PI3K and its role in cancer. Curr Opin Oncol. 2006;18:77–82. doi: 10.1097/01.cco.0000198021.99347.b9. [DOI] [PubMed] [Google Scholar]
- 41.Hay N. The Akt-mTOR tango and its relevance to cancer. Cancer Cell. 2005;8:179–83. doi: 10.1016/j.ccr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 42.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 43.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 44.Wang SI, Parsons R, Ittmann M. Homozygous deletion of the PTEN tumor suppressor gene in a subset of prostate adenocarcinomas. Clin Cancer Res. 1998;4:811–5. [PubMed] [Google Scholar]
- 45.Celebi JT, Shendrik I, Silvers DN, Peacocke M. Identification of PTEN mutations in metastatic melanoma specimens. J Med Genet. 2000;37:653–7. doi: 10.1136/jmg.37.9.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bussaglia E, del Rio E, Matias-Guiu X, Prat J. PTEN mutations in endometrial carcinomas: a molecular and clinicopathologic analysis of 38 cases. Hum Pathol. 2000;31:312–7. doi: 10.1016/s0046-8177(00)80244-0. [DOI] [PubMed] [Google Scholar]
- 47.Chang H, Qi XY, Claudio J, Zhuang L, Patterson B, Stewart AK. Analysis of PTEN deletions and mutations in multiple myeloma. Leuk Res. 2006;30:262–5. doi: 10.1016/j.leukres.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 48.Aggerholm A, Gronbaek K, Guldberg P, Hokland P. Mutational analysis of the tumour suppressor gene MMAC1/PTEN in malignant myeloid disorders. Eur J Haematol. 2000;65:109–13. doi: 10.1034/j.1600-0609.2000.90181.x. [DOI] [PubMed] [Google Scholar]
- 49.Sakai A, Thieblemont C, Wellmann A, Jaffe ES, Raffeld M. PTEN gene alterations in lymphoid neoplasms. Blood. 1998;92:3410–5. [PubMed] [Google Scholar]
- 50.Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A, O'Neil J, Gutierrez A, Ivanova E, Perna I, Lin E, Mani V, Jiang S, McNamara K, Zaghlul S, Edkins S, Stevens C, Brennan C, Martin ES, Wiedemeyer R, Kabbarah O, Nogueira C, Histen G, Aster J, Mansour M, Duke V, Foroni L, Fielding AK, Goldstone AH, Rowe JM, Wang YA, Look AT, Stratton MR, Chin L, Futreal PA, Depinho RA. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007 doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ciofani M, Zuniga-Pflucker JC. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat Immunol. 2005;6:881–8. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 52.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–8. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 53.Tomita K, Hattori M, Nakamura E, Nakanishi S, Minato N, Kageyama R. The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev. 1999;13:1203–10. doi: 10.1101/gad.13.9.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plas DR, Thompson CB. Akt-dependent transformation: there is more to growth than just surviving. Oncogene. 2005;24:7435–42. doi: 10.1038/sj.onc.1209097. [DOI] [PubMed] [Google Scholar]
- 55.Sano H, Kane S, Sano E, Miinea CP, Asara JM, Lane WS, Garner CW, Lienhard GE. Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J Biol Chem. 2003;278:14599–602. doi: 10.1074/jbc.C300063200. [DOI] [PubMed] [Google Scholar]
- 56.Katome T, Obata T, Matsushima R, Masuyama N, Cantley LC, Gotoh Y, Kishi K, Shiota H, Ebina Y. Use of RNA interference-mediated gene silencing and adenoviral overexpression to elucidate the roles of AKT/protein kinase B isoforms in insulin actions. J Biol Chem. 2003;278:28312–23. doi: 10.1074/jbc.M302094200. [DOI] [PubMed] [Google Scholar]
- 57.Edinger AL, Thompson CB. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol Biol Cell. 2002;13:2276–88. doi: 10.1091/mbc.01-12-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barthel A, Schmoll D. Novel concepts in insulin regulation of hepatic gluconeogenesis. Am J Physiol Endocrinol Metab. 2003;285:E685–92. doi: 10.1152/ajpendo.00253.2003. [DOI] [PubMed] [Google Scholar]
- 59.Birnbaum MJ. On the InterAktion between hexokinase and the mitochondrion. Dev Cell. 2004;7:781–2. doi: 10.1016/j.devcel.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 60.Fernandez de Mattos S, de los Pinos EE, Joaquin M, Tauler A. Activation of phosphatidylinositol 3-kinase is required for transcriptional activity of F-type 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase: assessment of the role of protein kinase B and p70 S6 kinase. Biochem J. 2000;349:59–65. doi: 10.1042/0264-6021:3490059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–9. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 62.Warburg O. On the origin of cancer cells. Science. 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 63.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–9. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 64.Dominguez M, de Celis JF. A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature. 1998;396:276–8. doi: 10.1038/24402. [DOI] [PubMed] [Google Scholar]
- 65.Cho KO, Choi KW. Fringe is essential for mirror symmetry and morphogenesis in the Drosophila eye. Nature. 1998;396:272–6. doi: 10.1038/24394. [DOI] [PubMed] [Google Scholar]
- 66.Papayannopoulos V, Tomlinson A, Panin VM, Rauskolb C, Irvine KD. Dorsal-ventral signaling in the Drosophila eye. Science. 1998;281:2031–4. doi: 10.1126/science.281.5385.2031. [DOI] [PubMed] [Google Scholar]
- 67.Ferres-Marco D, Gutierrez-Garcia I, Vallejo DM, Bolivar J, Gutierrez-Avino FJ, Dominguez M. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature. 2006;439:430–6. doi: 10.1038/nature04376. [DOI] [PubMed] [Google Scholar]
- 68.Ayyanan A, Civenni G, Ciarloni L, Morel C, Mueller N, Lefort K, Mandinova A, Raffoul W, Fiche M, Dotto GP, Brisken C. Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism. Proc Natl Acad Sci U S A. 2006;103:3799–804. doi: 10.1073/pnas.0600065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dakubo GD, Mazerolle CJ, Wallace VA. Expression of Notch and Wnt pathway components and activation of Notch signaling in medulloblastomas from heterozygous patched mice. J Neurooncol. 2006;79:221–7. doi: 10.1007/s11060-006-9132-2. [DOI] [PubMed] [Google Scholar]
- 70.Dang TP, Gazdar AF, Virmani AK, Sepetavec T, Hande KR, Minna JD, Roberts JR, Carbone DP. Chromosome 19 translocation, overexpression of Notch3, and human lung cancer. J Natl Cancer Inst. 2000;92:1355–7. doi: 10.1093/jnci/92.16.1355. [DOI] [PubMed] [Google Scholar]
- 71.Efstratiadis A, Szabolcs M, Klinakis A. Notch, Myc and breast cancer. Cell Cycle. 2007;6:418–29. doi: 10.4161/cc.6.4.3838. [DOI] [PubMed] [Google Scholar]
- 72.Fan X, Matsui W, Khaki L, Stearns D, Chun J, Li YM, Eberhart CG. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006;66:7445–52. doi: 10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- 73.Hopfer O, Zwahlen D, Fey MF, Aebi S. The Notch pathway in ovarian carcinomas and adenomas. Br J Cancer. 2005;93:709–18. doi: 10.1038/sj.bjc.6602719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu C, Dievart A, Lupien M, Calvo E, Tremblay G, Jolicoeur P. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am J Pathol. 2006;168:973–90. doi: 10.2353/ajpath.2006.050416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jundt F, Probsting KS, Anagnostopoulos I, Muehlinghaus G, Chatterjee M, Mathas S, Bargou RC, Manz R, Stein H, Dorken B. Jagged1-induced Notch signaling drives proliferation of multiple myeloma cells. Blood. 2004;103:3511–5. doi: 10.1182/blood-2003-07-2254. [DOI] [PubMed] [Google Scholar]
- 76.Kimura K, Satoh K, Kanno A, Hamada S, Hirota M, Endoh M, Masamune A, Shimosegawa T. Activation of Notch signaling in tumorigenesis of experimental pancreatic cancer induced by dimethylbenzanthracene in mice. Cancer Sci. 2007;98:155–62. doi: 10.1111/j.1349-7006.2006.00369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nefedova Y, Cheng P, Alsina M, Dalton WS, Gabrilovich DI. Involvement of Notch-1 signaling in bone marrow stroma-mediated de novo drug resistance of myeloma and other malignant lymphoid cell lines. Blood. 2004;103:3503–10. doi: 10.1182/blood-2003-07-2340. [DOI] [PubMed] [Google Scholar]
- 78.Park JT, Li M, Nakayama K, Mao TL, Davidson B, Zhang Z, Kurman RJ, Eberhart CG, Shih Ie M, Wang TL. Notch3 gene amplification in ovarian cancer. Cancer Res. 2006;66:6312–8. doi: 10.1158/0008-5472.CAN-05-3610. [DOI] [PubMed] [Google Scholar]
- 79.Shi W, Harris AL. Notch signaling in breast cancer and tumor angiogenesis: cross-talk and therapeutic potentials. J Mammary Gland Biol Neoplasia. 2006;11:41–52. doi: 10.1007/s10911-006-9011-7. [DOI] [PubMed] [Google Scholar]
- 80.Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006;66:1517–25. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- 81.van Es JH, Clevers H. Notch and Wnt inhibitors as potential new drugs for intestinal neoplastic disease. Trends Mol Med. 2005;11:496–502. doi: 10.1016/j.molmed.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 82.Wang Z, Banerjee S, Li Y, Rahman KM, Zhang Y, Sarkar FH. Down-regulation of notch-1 inhibits invasion by inactivation of nuclear factor-kappaB, vascular endothelial growth factor, and matrix metalloproteinase-9 in pancreatic cancer cells. Cancer Res. 2006;66:2778–84. doi: 10.1158/0008-5472.CAN-05-4281. [DOI] [PubMed] [Google Scholar]
- 83.Wu F, Stutzman A, Mo YY. Notch signaling and its role in breast cancer. Front Biosci. 2007;12:4370–83. doi: 10.2741/2394. [DOI] [PubMed] [Google Scholar]