Abstract
The distribution of fitness effects for beneficial mutations is of paramount importance in determining the outcome of adaptation. It is generally assumed that fitness effects of beneficial mutations follow an exponential distribution, for example, in theoretical treatments of quantitative genetics, clonal interference, experimental evolution, and the adaptation of DNA sequences. This assumption has been justified by the statistical theory of extreme values, because the fitnesses conferred by beneficial mutations should represent samples from the extreme right tail of the fitness distribution. Yet in extreme value theory, there are three different limiting forms for right tails of distributions, and the exponential describes only those of distributions in the Gumbel domain of attraction. Using beneficial mutations from two viruses, we show for the first time that the Gumbel domain can be rejected in favor of a distribution with a right-truncated tail, thus providing evidence for an upper bound on fitness effects. Our data also violate the common assumption that small-effect beneficial mutations greatly outnumber those of large effect, as they are consistent with a uniform distribution of beneficial effects.
Keywords: Fitness distribution, Extreme value theory, Adaptation, Bacteriophage, Virus
Introduction
Adaptation is a process in which beneficial mutations increase in frequency in a population. The distribution of fitness effects is central to many aspects of this process and influences, for example, the rate of adaptation (Wilke 2004) and the mean fitness increase due to the fixation of a beneficial mutation (Orr 2003). Beneficial mutations of large effect have historically been assumed to be rare relative to those of small effect, an idea propounded by Fisher (1930), and more recently it has been argued that beneficial fitness effects should in fact be approximately exponentially distributed. The exponential distribution has become a prominent assumption in theoretical studies of the genetics of adaptation, serving as the starting point for theories of quantitative genetics (Otto and Jones 2000), clonal interference (Gerrish and Lenski 1998; Rozen et al. 2002; Wilke 2004), experimental evolution (Wahl and Krakauer 2000), and the adaptation of DNA sequences (Gillespie 1983, 1984, 1991; Orr 2002, 2003; Rokyta et al. 2006). The first theoretical justification for this assumption was provided by Gillespie (1983, 1984, 1991): if beneficial mutations are rare, then the relevant portion of the full fitness distribution is the extreme right tail. If we consider the fitnesses of all possible genotypes differing from the wild type by a single nucleotide change as a large sample from an unknown fitness distribution, the vast majority of them will fall below the wild type's fitness. A fitness greater than the wild type's is a rare event and, thus, lies in the extreme right tail of the fitness distribution. Tails for many distributions have limiting forms that are only weakly dependent on the parent distribution. Furthermore, the limiting form which describes the tails of most commonly encountered distributions (e.g., normal, gamma, exponential, etc.) is, in fact, the exponential distribution. Distributions of this form belong to the Gumbel domain of attraction in extreme value theory (EVT).
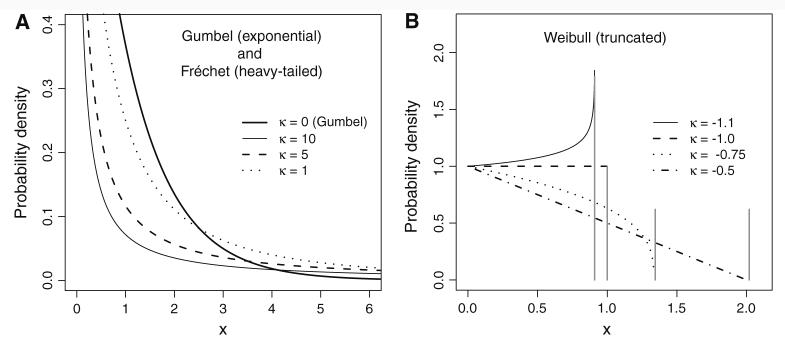
Since it is generally accepted that beneficial mutations are rare, it seems reasonable to assume that EVT can provide information regarding the distribution of beneficial fitness effects. However, according to EVT, the exponential is but one of three possible limiting tail distributions, as has been noted previously in the context of adaptation theory (Orr 2005, 2006). The others loosely correspond to distributions with heavier than exponential tails (the Fréchet domain) and distributions which are right-truncated (the Weibull domain); see Leadbetter et al. (1980) for more precise descriptions. The tails for each of these three EVT domains can all be described by the generalized Pareto distribution (GPD). Its cumulative distribution function is given by
| (1) |
with shape parameter k and scale parameter τ. The shape parameter, k, determines the domain (Pickands 1975), with k = 0 corresponding to the Gumbel domain, k > 0 corresponding to the Fréchet domain, and k < 0 corresponding the Weibull domain (illustrated in Fig. 1). This formulation describes the limiting distributions of the tail above a high threshold (set to zero here). In the context of beneficial mutations, if the threshold is set to the fitness of the wild type, the GPD would describe the distribution of beneficial fitness effects. However, the GPD shape parameter is stable with respect to changes in the threshold (Castillo and Hadi 1997), thus any high threshold is equivalent for characterizing the domain of attraction.
Fig. 1.
A comparison of right tails for the three domains of attraction under the generalized Pareto distribution (GPD). The shape parameter k determines the domain, and the scale parameter τ = 1 for all examples. (a) The right tails for distributions in the Gumbel domain correspond to the GPD with k = 0 (i.e., the exponential distribution). Distributions in the Fréchet domain (k > 0) have tails that are heavier than exponential. (b) Distributions in the Weibull domain have right-truncated tails and correspond to the GPD with k < 0. Truncation points are denoted by vertical gray lines
Using a statistical method tailored to this problem described by Beisel et al. (2007), we tested the null hypothesis that the fitness distribution has an exponential tail (k = 0 under the GPD) for two sets of beneficial mutations from viruses for which the identities of the mutations were determined by sequencing. One set consisted of nine different beneficial mutations for the ssDNA bacteriophage ID11, selected for high growth rate in liquid culture at 37°C on host Escherichia coli C (Rokyta et al. 2005). The second set consisted of 16 beneficial mutations for RNA phage ϕ6, selected for ability to grow on a novel host (Ferris et al. 2007).
Materials and Methods
Likelihood Ratio Test
The likelihood ratio test (LRT) and its statistical properties have been described in detail by Beisel et al. (2007). Briefly, negative twice the difference in log likelihoods, −2logΛ, is calculated based on the GPD, comparing the null model k = 0 to the alternative k ≠ 0. Thus, the test determines whether the data are more consistent with a fitness distribution in the Gumbel domain of attraction (i.e., a distribution with an exponential tail) or either the Fréchet (k > 0) or Weibull (k < 0) domain, assuming that the data consist of values above a high threshold. Under the GPD, the parameter of interest, k, is stable with respect to changes in the threshold (Castillo and Hadi 1997). Thus, it is not necessary to use the wild type's fitness as the threshold to characterize the domain of attraction for fitness distributions. Beisel et al. (2007) argued that fitnesses should be shifted relative to the fitness of the smallest-effect mutation observed rather than the wild type, at the cost of a single degree of freedom. This reduces possible bias introduced by missing small-effect beneficial mutations in an empirical sample and ensures that the threshold is high enough for EVT to apply. Measurement error can be easily incorporated into the test, but Beisel et al. (2007) showed that doing so has no significant effect on type I error rates as long as the coefficient of variation for the data is relatively small (<∼0.2). Although the distribution of the test statistic, −2logΛ, is asymptotically , we used a parametric bootstrapping approach since our sample sizes were small. The p-values are based on 10,000 parametric bootstrap replicates. All analyses were performed using R (R Development Core Team 2006).
An Approximate Method for Estimating k
The GPD has been widely applied to problems in engineering and finance, but is not commonly encountered in biological applications (but see Orr 2006). Much of the statistical theory concerning the GPD involves asymptotic results that describe approximations to the distribution of the maximum likelihood estimator (MLE) which are valid for large sample sizes. To obtain these results, it is necessary to restrict the range of the parameter space. For example, the statistical literature on the estimation of the parameters of the GPD generally ignores the case of k < −1/2, since for −1 ≤ k < −1/2 asymptotic properties do not obtain, and for k < −1, the MLE does not exist (Castillo and Hadi 1997). However, these restrictions are artificial and are made only for mathematical convenience. In applying the test of Beisel et al. (2007) to our data, we found that k̂ = −1 (see Results), but as part of the testing procedure, it was necessary to restrict k > −1, suggesting that the true value of k could potentially be less than −1. Thus, to address parameter estimation for values in this range, we derived a simple procedure for estimating k for values near −1.
The probability density function for the GPD is given by
| (2) |
We restrict our attention to the case where k < 0 and let λ = −τ/k, which corresponds to the right truncation point. If λ is known, then the MLE for k can be calculated directly using equation (2) and is given by
| (3) |
Under these conditions, it can be shown that −ln(1 − Xi/λ) follows the exponential distribution with mean −k. This implies that E(k̂) = k;Var(k̂) = k2/n, and 2nk̂/k follows the χ2 distribution with 2n degrees of freedom.
Now consider an ordered sample of size n from the GPD, X(1), X(2),Ω, X(n), where X(n) is the largest value from the sample and X(1) is the smallest. For k ≤ −1 and moderate sample sizes, X(n) will be close to λ. Thus, we can take λ̂ = X(n) as an estimate of λ. However, if we replace λ with λ̂ = X(n) in Eq. 3 above, the sum is undefined. To circumvent this problem we simply drop the X(n) term in (3) to get the approximate estimator
| (4) |
An approximate confidence interval can be constructed by assuming that 2(n − 1)k̂/k follows a χ2 with 2(n − 1) degrees of freedom.
Performance of this new estimation procedure was assessed through simulations using R (R Development Core Team 2006). We used sample sizes of 10 and 30, and considered −3 ≤ k ≤ −1/2 with right truncation point λ = −τ/k = 10. For each combination of sample size and k, we generated 100,000 samples from the GPD and calculated k̂ and its 95% confidence interval.
Data Sets
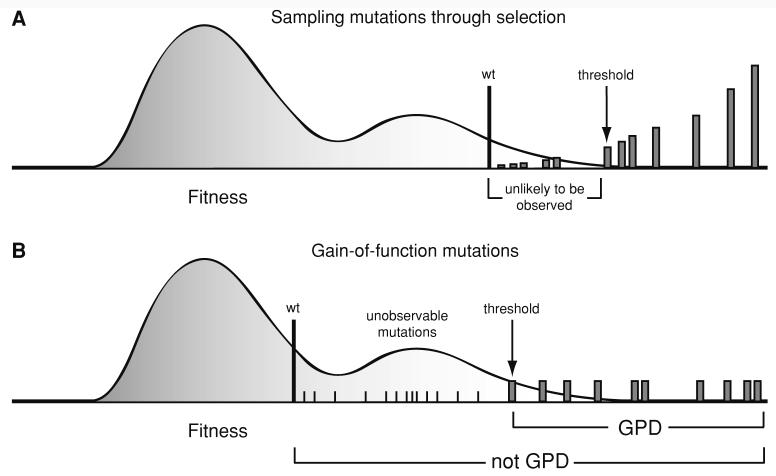
The details of the experimental protocols for isolating, identifying, and measuring the fitnesses of the nine beneficial mutations for the DNA phage ID11 are described by Rokyta et al. (2005). The raw fitness data and identities of the mutations are listed in Table 1. Briefly, 20 replicate lineages were selected for increased growth rate in liquid culture on host E. coli C at 37°C. Populations were repeatedly bottlenecked to ∼104 phages to minimize the effects of clonal interference. For each lineage, a single beneficial mutation was allowed to fix and was identified through full genome sequencing, yielding a total of nine unique beneficial mutations. The fitness of each unique mutation was measured in 10 replicates as the log2 increase in the phage population per 15 min (approximately one generation). We ignored the frequencies at which the various beneficial mutations fixed, and considered the unique fixed beneficial mutations to be a biased sample from the distribution of new beneficial mutations. This approach was discussed at length by Beisel et al. (2007) and is addressed further in the Discussion and in Fig. 2a. Although selection will bias the sample toward mutations of larger effect, shifting the fitnesses to be relative to the fitness of the smallest-effect mutation observed should largely eliminate this issue.
Table 1.
The nine beneficial mutations for the phage ID11 selected for rapid growth at 37°C on Escherichia coli C (Rokyta et al. 2005)
| Genome positiona |
Nucleotide substitution |
Amino acid positionb |
Amino acid substitution |
Fitnessc | SEd |
|---|---|---|---|---|---|
| Ancestor | 3.65 | 0.15 | |||
| 3864 | A → G | F421 | D → G | 4.06 | 0.16 |
| 3567 | A → G | F322 | N → S | 4.19 | 0.12 |
| 2609 | G → T | F3 | V → F | 4.39 | 0.13 |
| 3857 | A → G | F419 | T → A | 4.76 | 0.14 |
| 3543 | C → T | F314 | A → V | 4.78 | 0.15 |
| 2520 | C → T | J15 | A → V | 4.82 | 0.15 |
| 3850 | G → A/T | F416 | M → I | 4.86 | 0.15 |
| 3665 | C → T | F355 | P → S | 5.01 | 0.14 |
| 2534 | G → T | J20 | V → L | 5.08 | 0.13 |
Position corresponds to the published sequence of ID11 (GenBank accession number AY751298)
The letter indicates the gene name and the number gives the amino acid position
Fitness is measured as the log2 increase in population size per 15 min
Based on 10 replicates
Fig. 2.
Two methods of sampling beneficial mutations. The shaded curve represents a hypothetical fitness distribution, and the vertical gray bars represent a sample of beneficial mutations. The locations of the vertical bars on the horizontal axis represent their fitnesses, and their heights represent their hypothetical relative probabilities of being observed in a single experiment under the sampling strategy. Note that only the fitnesses, and not their probabilities of being observed, are considered in the test. (a) Under selection, the wild type fitness lies in the tail but sampling is biased toward large-effect mutations. Repeated sampling will yield multiple observations of the same mutations, though those of very small effect may not be observed in a reasonable number of replicates. Shifting alleviates the bias due to missing very small-effect mutations. (b) In gain-of-function experiments, the wild type fitness will not lie in the tail and may in fact be zero, and some beneficial mutations may not have effects large enough to be observed. Mutations with effects above some threshold will have equal probabilities of being sampled, and as these are the fittest mutations, they will lie in the tail. The fitness of the smallest-effect mutation from those sampled provides a threshold in the tail of the distribution
The protocols for isolating and characterizing the beneficial mutations for RNA phage ϕ6 have been described in detail elsewhere (Ferris et al. 2007). Briefly, ϕ6 clones were plated on the nonpermissive host Pseudomonas syringae pv. glycinea strain R4a such that only phages with host range mutations were able to form plaques. For each of 40 replicates, a single randomly selected plaque was chosen, and its P3 gene was sequenced, as its gene product has been previously associated with host range expansion (Duffy et al. 2005). Nineteen unique genotypes were identified, though we excluded three from our analyses. Two genotypes were excluded because they were found to have two different mutations in P3, and the other was excluded since its mutation gave rise to the same amino acid substitution as another. Thus, this data set consisted of 16 unique host range mutations (Table 2). Fitness was measured in six replicates as log10 of the number of progeny per initial plaque after 24 h of growth on plates on the novel host. As these are gain-of-function mutations, the wild type has a fitness of zero under the new conditions. In addition, there is a potential bias in that only mutations with large enough beneficial effects to allow the formation of a visible plaque on a plate will be sampled. Shifting the fitnesses relative to the fitness of the smallest-effect mutation observed alleviates both of these issues. The use of this type of data for testing the domain of attraction for fitness distributions was discussed in detail by Beisel et al. (2007) and is addressed further in the Discussion and in Fig. 2b.
Table 2.
The 16 beneficial mutations for the phage ϕ6 selected for the ability to grow on novel host Pseudomonas syringae pv. glycinea (Ferris et al. 2007)
| Mutant ID |
Gene position |
Nucleotide substitution |
Amino acid position |
Amino acid substitution |
Fitnessa | SEb |
|---|---|---|---|---|---|---|
| G19 | Not in P3 | Unknown | Unknown | Unknown | 5.53 | 0.11 |
| g18 | 1603 | G → A | 535 | D → N | 6.23 | 0.07 |
| g27 | 1016 | C → T | 339 | P → H | 6.41 | 0.14 |
| g25 | 23 | A → G | 8 | E → G | 6.48 | 0.05 |
| g24 | 22 | G → A | 8 | E → K | 6.60 | 0.10 |
| g9 | 1661 | A → C | 554 | D → A | 6.73 | 0.11 |
| g2 | 1661 | A → T | 554 | D → V | 6.78 | 0.07 |
| g6 | 1661 | A → G | 554 | D → G | 6.83 | 0.05 |
| g14 | 1663 | C → T | 555 | L → F | 6.86 | 0.05 |
| g22 | 13 | G → A | 5 | G → S | 6.94 | 0.09 |
| g36 | 1660 | G → A | 554 | D → N | 6.97 | 0.06 |
| g15 | 1598 | A → C | 533 | D → A | 7.02 | 0.10 |
| g8 | 434 | A → G | 145 | D → G | 7.03 | 0.10 |
| g5 | 437 | A → G | 146 | N → S | 7.19 | 0.11 |
| g20 | 1546 | A → G | 516 | T → A | 7.26 | 0.06 |
| g29 | 534 | G → C | 178 | E → D | 7.56 | 0.10 |
Fitness was measured as the log10 of the number of phages per plaque per 24 h on plates
Based on six replicates
Results
Performance of the New Estimator for k
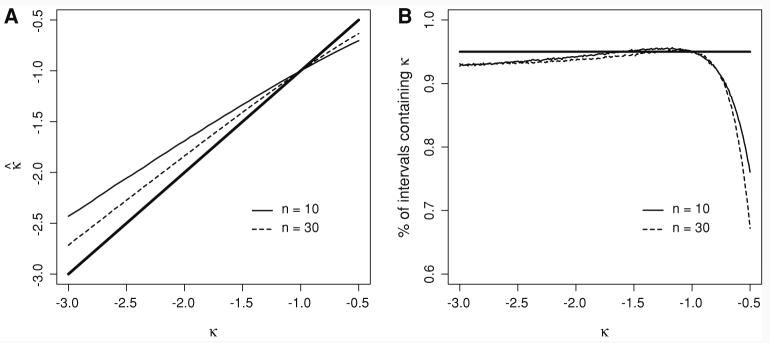
We explored the behavior of our new estimator of the shape parameter of the generalized Pareto distribution (GPD) using simulations with sample sizes of 10 and 30 (Fig. 3). We considered −3 ≤ k ≤ −1/2 with right truncation point λ = −τ/k set to 10. The behavior at each combination of k and sample size was approximated by 100,000 simulated data sets. Estimates of the shape parameter, k̂, are slightly biased toward −1 for k ≠ 1,with bias increasing with the distance from −1 and decreasing with increasing sample size (Fig. 3a). The approximate 95% confidence intervals performed as expected for −2 < k < −0.8 (Fig. 3b). For −3 < k < −2, the confidence intervals captured the truth ∼93% of the time, and for −0.8 < k < −0.5, their performance was poor. These results were similar across both sample sizes considered.
Fig. 3.
Performance of the approximate estimator for k. Each point represents the average of 100,000 simulated data sets. (a) Bias in k̂. The bold solid line gives the expectation for an unbiased estimate. For k ≠ −1, k̂ is biased toward −1. (b) Performance of 95% confidence intervals. The intervals were symmetric, with 2.5% upper and 2.5% lower tail probabilities. The bold solid line denotes a 95% probability of containing the true value of k
Analysis of Two Viral Data Sets
For both data sets, we analyzed measures of log fitness, or Malthusian fitness, as this is a more appropriate measure when reproduction does not occur at discrete times (i.e., log fitness is the appropriate parameter for continuous growth models). To account for the possible empirical absence of small-effect beneficial mutations due to sampling strategies, we tested the distribution of fitness effects shifted relative to the fitness of the smallest-effect mutation observed rather than the wild type, as described by Beisel et al. (2007). Thus, rather than using the wild type's fitness as the threshold, we used the fitness of the smallest-effect mutation. The coefficients of variation for both data sets were ∼0.07, thus we ignored measurement error in our analyses as suggested by Beisel et al. (2007). Including measurement error had no effect on our results (not shown).
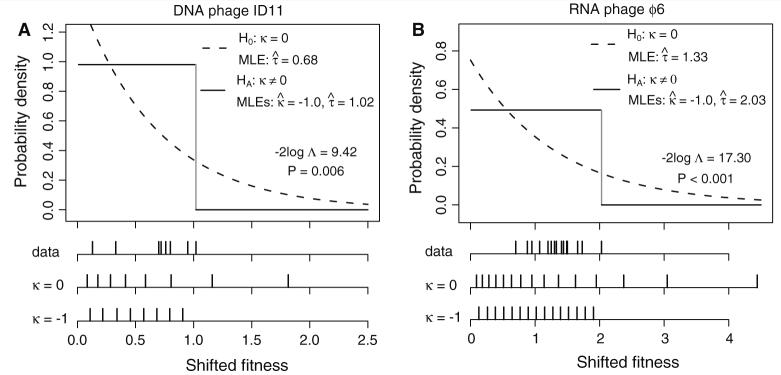
Despite the small samples sizes, the Gumbel domain (k = 0) was rejected using the LRT in favor of the Weibull domain (k < 0) for both data sets (Fig. 4, Table 3). This result indicates that a fitness distribution with a right-truncated tail gives a better fit to both data sets than a distribution with an exponential tail. Furthermore, the Gumbel domain is still rejected for the DNA phage ID11 data set if the threshold is set to the fitness of the second or third smallest-effect mutation and for the RNA phage ϕ6 data set with the threshold set to the fitness of the second smallest-effect mutation (Table 3), providing stronger evidence that missed small-effect mutations are not responsible for this result.
Fig. 4.
The distributions of shifted fitness effects for two viruses. The null hypothesis (H0: k = 0) is that the fitness distribution belongs to the Gumbel domain of attraction. The alternative hypothesis (HA: k ≠ 0) is that the distribution is in either the Weibull (k < 0) or Fréchet (k > 0) domain. The dashed and solid lines depict the fitted densities under the null and alternative models respectively. p-values are based on 10,000 parametric bootstrap replicates. The shifted empirical fitnesses, plotted as vertical lines, are compared to their expected values under the fitted null (vertical lines with k = 0) and alternative (vertical lines with k = −1) models. (a) The nine beneficial mutations for the DNA phage ID11. (b) The 16 host range mutations for RNA phage ϕ6
Table 3.
A summary of the statistical results for two viral data sets
| Phage | Obs. | Shifta | H0: k = 0 (τ̂) | HA: k ≠ 0 (k̂, τ̂) | Likelihood ratio testb |
Approximate methodb |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| −2logΛ | df | p | k̂ | df | 95% CI | |||||
| ID11 | 9 | 1 | 0.68 | (−1.0, 1.02) | 9.42 | 8 | 0.006 | −1.06 | 7 | (−2.64, −0.57) |
| 2 | 0.63 | (−1.0, 0.89) | 9.04 | 7 | 0.006 | −1.08 | 6 | (−2.93, −0.55) | ||
| 3 | 0.50 | (−1.0, 0.69) | 8.01 | 6 | 0.009 | −1.00 | 5 | (−3.08, −0.49) | ||
| ϕ6 | 16 | 1 | 1.33 | (−1.0, 2.03) | 17.30 | 15 | <0.001 | −1.00 | 14 | (−1.83, −0.63) |
| 2 | 0.68 | (−1.0, 1.33) | 8.98 | 14 | 0.021 | −0.65 | 13 | (−1.22, −0.40) | ||
This designates the threshold used for the analysis. 1 indicates the fitness of the lowest fitness mutant; 2, the second lowest; etc. Shifting further than shown results in a failure to reject at the 5% significance level
The likelihood ratio test and approximate method are described under Materials and Methods
In applying the LRT, it is necessary to restrict k ≥ −1, since for k < −1, the likelihood can become infinite, and thus the maximum likelihood estimates do not exist. This restriction makes the test conservative, yet the fact that k̂ ≈ −1 suggests that the best estimate of k is ≤ 1. We developed a novel estimation method appropriate for k near −1, which also provides confidence intervals (Materials and Methods). Applying this method with the threshold set as the fitness of the smallest-effect mutation, we estimated k̂ = −1.06 (ID11; 95% CI, −2.64 < k < −0.57) and k̂ = −1.00 (ϕ6; 95% CI, −1.83 < k < −0.63), in close agreement with the results from the LRT (Table 3). These confidence intervals encompass a wide range of tail behaviors (see Fig. 1), yet all are characterized by a right-truncated distribution and differ substantially from the exponential distribution.
Discussion
We have shown, using collections of beneficial mutations from a DNA virus and an RNA virus, that at least some fitness distributions do not belong to the Gumbel domain of attraction from extreme value theory (EVT). The wide-spread assumption of exponentially distributed beneficial fitness effects in theories of adaptation is strongly rejected for our data. Both data sets suggest that fitness distributions can instead belong to the Weibull domain of attraction, which implies that the distribution of beneficial fitness effects is right-truncated. In fact, the fitted distributions (GPD with k = −1) correspond to a uniform distribution, which has been considered an unrealistic distribution for beneficial fitness effects (Wilke 2004) and is at variance with the common observation that small-effect beneficial mutations greatly outnumber those of large effect (Imhof and Schlötterer 2001; Kassen and Bataillon 2006; Perfeito et al. 2007; Sanjuán et al. 2004).
Sampling Procedures
The methods used for collecting the ID11 and ϕ6 data sets may at first seem to be at odds with the theory being tested. The beneficial mutations for ID11 were sampled only after they had fixed in an evolving population and, thus, represent a nonrandom sample of new beneficial mutations. For the ϕ6 data set, the wild type was unable to grow under the selective conditions, and thus its fitness cannot be assumed to be in the tail of the fitness distribution. However, the statistical methodology we apply was developed specifically for these types of data. Beisel et al. (2007) provide extensive discussions of the appropriateness of beneficial mutations collected through selection experiments (ID11) and gain-of-function experiments (ϕ6). Both cases rely on the fact that the GPD shape parameter is not altered by a change in threshold, and since the domain of attraction is specified by the shape parameter, we are able to change the threshold to alleviate sampling bias or to be certain that the threshold is far enough into the tail for EVT to apply. We briefly reiterate the arguments of Beisel et al. (2007) in the context of our two data sets.
Selection is a biased method for sampling from the distribution of new beneficial mutations. The experiments of Rokyta et al. (2005) involved 20 samples from the distribution of new beneficial mutations, biased by requiring these mutations to survive drift and fix in an evolving population to be observed. Many of the observed mutations were sampled multiple times, and this frequency data was ignored in our analysis. Only the fitnesses of unique mutations were included. Using selection as a sampling strategy implies that we are likely to thoroughly sample only large-effect mutations, and those of very small effect are likely to be missed. Figure 2 illustrates a hypothetical example. The locations of the vertical gray bars on the horizontal axis are the fitnesses of the genotypes possessing beneficial mutations. We assume a small number of beneficial mutations. Thus, under selection, which could include the effects of clonal interference, the distribution of fixed effects is a discrete probability mass function on the fitnesses of the mutants (represented in Fig. 2a by the heights of the gray bars). Higher-fitness mutants are more likely to be observed in any given replicate, but repeated experiments will give thorough sampling, especially of the largest-effect mutations. For our analysis, we shift further into the right tail to compensate for this bias and demonstrate that the threshold can be set to the smallest-, second smallest-, and third smallest-effect beneficial mutation while still rejecting the Gumbel domain (Table 3). Thus, we only need to assume that we have an adequate sample of mutations that have larger effects than the third smallest-effect mutation. Seventeen of the 20 sampled mutations described by Rokyta et al. (2005) fall into this range, which includes 6 unique mutations. Furthermore, as is clear in Fig. 4a, the discrepancy between the data and the exponential distribution actually involves the large-effect mutations, i.e., those most likely to have been observed. Under the exponential distribution, we would have expected to see well-spaced mutations of very large effect, which were not observed, and adding a handful of small-effect mutations would not have affected our result.
The use of gain-of-function mutations for testing the domain of attraction for fitness distributions involves a conceptual departure from the modeling framework to which the Gumbel assumption is generally applied. For example, the results of Orr (2002) rely on the assumption that the wild type fitness is in the tail of the distribution. However, this assumption need not hold to test the domain of attraction of a fitness distribution. If we consider the fitness of wild type ϕ6 and all of its single-mutation neighbors in sequence space as a sample from the fitness distribution, it is clear that wild type fitness (zero) does not lie in the tail and thus would not serve as an adequate threshold (Fig. 2b). However, we can be confident that the fitness of the smallest-effect mutation is in the tail, since only a small number of fitnesses were larger. Thus, shifting relative to the fitness of the smallest-effect mutation assures us that we are dealing with the tail of the fitness distribution, though this particular fitness distribution may not be amenable to the predictions of population genetic models such as those of Orr (2002). Our analysis focuses solely on the Gumbel assumption for fitness distributions and testing this assumption only requires that the threshold be in the tail.
The Domain of Attraction for Fitness Distributions
Although we have demonstrated that beneficial fitness effects in two viral systems are characterized by a right-truncated distribution, it remains to be determined whether the Weibull domain of attraction will apply generally, or whether distributions will vary among different organisms in different environments. Prior empirical attempts to characterize distributions of beneficial fitness effects for microbes did not distinguish between alternative EVT domains of attraction. A study of beneficial mutations in vesicular stomatitis virus by Sanjuán et al. (2004) rejected the exponential distribution in favor of the gamma distribution, whereas a study of beneficial mutations in Pseudomonas fluorescens by Kassen and Bataillon (2006) failed to reject the exponential in favor of the gamma. However, the gamma itself belongs to the Gumbel domain of attraction. Since the assumption of an exponential distribution for beneficial fitness effects is justified by EVT, it is more natural to turn to EVT to provide the appropriate alternative hypotheses. Various limitations of these data sets (e.g., sample size for the Sanjuán et al. data set and the unknown number of unique mutations in the Kassen and Bataillon data set) prevented us from reanalyzing them in the context of EVT. Theoretical support in favor of the Gumbel domain was provided by Orr (2006) through an analysis of the distribution of fitness effects under Fisher's (1930) geometrical model of adaptation. While the generality of our results remains uncertain, we have clearly demonstrated that not all fitness distributions fall within the Gumbel domain of attraction.
Distributions in the Gumbel and Weibull domains of attraction differ qualitatively in the key characteristic used to predict the rate and pattern of adaptive evolution—the fitness spacings between beneficial mutations, which allow direct calculation of fitness effects and selection coefficients (Orr 2005). Under the exponential distribution, the fitness spacings between adjacently ranked beneficial mutations are independent exponential random variables. If the mean of the parent distribution is μ, then the expected difference between the largest and the second largest observations is μ, the expected difference between the second and the third largest is μ/2, the expected difference between the third and the fourth largest is μ/3, etc. Thus, the fitness effects of beneficial mutations tend to accumulate near low values, matching Fisherian intuition. This pattern holds several consequences for adaptive evolution; for example, it mitigates the effects of clonal interference in the adaptation of asexual organisms (Kim and Orr 2005), and it causes natural selection to behave, on average, halfway between perfect adaptation, where the best available mutation is always fixed, and random adaptation, where all available beneficial mutations have equal probabilities of being fixed (Orr 2002). This pattern of spacings does not hold under the GPD with k ≈ −1 (see Fig. 4), and for k < −1 the pattern begins to reverse such that fitness effects tend to accumulate near the right truncation point. As a consequence, our finding that at least some fitness distributions are not in the Gumbel domain will have a substantial impact on our understanding of the rate and pattern of adaptive evolution.
Small-Effect vs. Large-Effect Mutations
Our results appear to violate the long-held intuition that mutations of large benefit should be less common than those of small benefit. The logic behind this intuition dates back to Fisher (1930) and has been supported by recent empirical and theoretical work (Imhof and Schlötterer 2001; Kassen and Bataillon 2006; Perfeito et al. 2007; Sanjuán et al. 2004). However, both of our data sets are consistent with a uniform distribution of beneficial fitness effects, implying that small-effect and large-effect beneficial mutations are equally common for these two systems. How do we reconcile these conflicting results? First, it is important to differentiate between the distribution of beneficial fitness effects fixed over the course of an adaptive walk and the beneficial effects of potential single-step mutations. The former deals with mutations in a variety of genetic backgrounds, and each fixed mutation could conceivably give rise to an entirely new distribution of single-step mutations. Much of the work purporting to support an excess of small-effect mutations (e.g., Imhof and Schlötterer 2001; Perfeito et al. 2007) has examined the distribution over multiple steps in adaptation. Except under the assumption of strict additivity these two distributions will not be the same, and it is clear that biological systems are not strictly additive. For a single ancestral genotype, there are many contexts in which fitness might be improved greatly through the alteration of a single biochemical property (e.g., increased protein stability or drug resistance), and any of several mutations may confer roughly the same large effect. However, once a large step is taken to achieve this phenotypic change, the remaining first-step mutations may have much smaller effects or even be neutral or deleterious when combined with the fixed beneficial mutation. Furthermore, such large-effect mutations may require compensatory changes to overcome pleiotropic effects. Thus, there is little reason to expect the distribution of beneficial fitness effects from a single ancestral genotype to resemble the distribution of effects over multiple steps in an adaptive walk.
A uniform distribution of beneficial effects, and similarly a right-truncated distribution, is certainly not unreasonable at the biochemical level. For example, even if the phenotypic effects of mutations showed an excess of small-effects, the translation of phenotype into fitness could produce a more uniform distribution. A pattern of diminishing returns (i.e., a concave mapping of phenotype into fitness) such as is commonly seen for biochemical reactions and metabolic flux (Hartl et al. 1985), could conceivably yield both an apparent right truncation point and a uniform or even reversed-tailed distribution of fitness effects, regardless of the underlying distribution of phenotypic effects. Likewise, mutations can have a large effect on a phenotype such as host attachment, but the extent to which that phenotype can increase fitness may be limited (Pepin et al. 2006). This too might yield a uniform distribution of fitness effects. Thus, a more complete understanding of the distribution of fitness effects and the relative abundance of large-effect beneficial mutations may require a better understanding of the biochemical nature of adaptation.
Acknowledgments
The authors would like to thank J. J. Bull for comments and discussions. This work was supported by grants from the National Institutes of Health to P.J. and H.A.W. (R01GM076040) and to C.L.B. (R01GM067940). C.J.B. was supported in part by NIHP20 RR16448 and a grant from the National Science Foundation (DEB-0515738) to P.J. D.R.R. was supported in part by NIH P20 RR16454. Analytical resources were provided by NIH P20 RR16448 and NIH P20 RR16454.
Footnotes
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Beisel CJ, Rokyta DR, Wichman HA, Joyce P. Testing the extreme value domain of attraction for distributions of beneficial fitness effects. Genetics. 2007;176:2441–2449. doi: 10.1534/genetics.106.068585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo E, Hadi AS. Fitting the generalized Pareto distribution to data. J Am Stat Assoc. 1997;92:1609–1620. [Google Scholar]
- Duffy S, Turner PE, Burch CL. Pleiotropic costs of niche expansion in the RNA bacteriophage ϕ6. Genetics. 2005;172:751–757. doi: 10.1534/genetics.105.051136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris MT, Joyce P, Burch CL. High frequency of mutations that expand the host range of an RNA virus. Genetics. 2007;176:1013–1022. doi: 10.1534/genetics.106.064634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The genetical theory of natural selection. Oxford University Press; Oxford, UK: 1930. [Google Scholar]
- Gerrish PJ, Lenski RE. The fate of competing beneficial mutations in an asexual population. Genetica. 1998;102(103):127–144. [PubMed] [Google Scholar]
- Gillespie JH. A simple stochastic gene substitution model. Theor Popul Biol. 1983;23:202–215. doi: 10.1016/0040-5809(83)90014-x. [DOI] [PubMed] [Google Scholar]
- Gillespie JH. Molecular evolution over the mutational landscape. Evolution. 1984;38:1116–1129. doi: 10.1111/j.1558-5646.1984.tb00380.x. [DOI] [PubMed] [Google Scholar]
- Gillespie JH. The causes of molecular evolution. Oxford University Press; New York: 1991. [Google Scholar]
- Hartl DL, Dykhuizen DE, Dean AM. Limits of adaptation: the evolution of selective neutrality. Genetics. 1985;111:655–674. doi: 10.1093/genetics/111.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhof M, Schlötterer C. Fitness effects of advantageous mutations in evolving Escherichia coli populations. Proc Natl Acad Sci USA. 2001;98:1113–1117. doi: 10.1073/pnas.98.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassen R, Bataillon T. Distribution of fitness effects among beneficial mutations before selection in experimental populations of bacteria. Nature Genet. 2006;38:484–488. doi: 10.1038/ng1751. [DOI] [PubMed] [Google Scholar]
- Kim Y, Orr HA. Adaptation in sexuals vs asexuals: clonal interference and the Fisher-Muller model. Genetics. 2005;171:1377–1386. doi: 10.1534/genetics.105.045252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadbetter MR, Lindgren G, Rootzén H. Extremes and related properties of random sequences and processes. Springer-Verlag; New York: 1980. [Google Scholar]
- Orr HA. The population genetics of adaptation: the adaptation of DNA sequences. Evolution. 2002;56:1317–1330. doi: 10.1111/j.0014-3820.2002.tb01446.x. [DOI] [PubMed] [Google Scholar]
- Orr HA. The distribution of fitness effects among beneficial mutations. Genetics. 2003;163:1519–1526. doi: 10.1093/genetics/163.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. The genetic theory of adaptation: a brief history. Nat Rev Gen. 2005;6:119–127. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- Orr HA. The distribution of beneficial fitness effects among beneficial mutations in Fisher's geometric model of adaptation. J Theor Biol. 2006;238:279–285. doi: 10.1016/j.jtbi.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Otto SP, Jones CD. Detecting the undetected: estimating the total number of loci underlying a quantitive trait. Genetics. 2000;156:2093–2107. doi: 10.1093/genetics/156.4.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepin KM, Samuel MA, Wichman HA. Variable pleiotropic effects from mutations at the same locus hamper prediction of fitness from a fitness component. Genetics. 2006;172:2047–2056. doi: 10.1534/genetics.105.049817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfeito L, Fernandes L, Mota C, Gordo I. Adaptive mutations in bacteria: high rate and small effects. Science. 2007;317:813–815. doi: 10.1126/science.1142284. [DOI] [PubMed] [Google Scholar]
- Pickands J., III Statistical inference using extreme order statistics. Ann Statist. 1975;3:119–131. [Google Scholar]
- R Development Core Team . R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2006. Available at: http://www.R-project.org) [Google Scholar]
- Rokyta DR, Joyce P, Caudle SB, Wichman HA. An empirical test of the mutational landscape model of adaptation using a single-stranded DNA virus. Nature Genet. 2005;37:441–444. doi: 10.1038/ng1535. [DOI] [PubMed] [Google Scholar]
- Rokyta DR, Beisel CJ, Joyce P. Properties of adaptive walks on uncorrelated landscapes under strong selection and weak mutation. J Theor Biol. 2006;243:114–120. doi: 10.1016/j.jtbi.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Rozen DE, de Visser JAGM, Gerrish PJ. Fitness effects of fixed beneficial mutations in microbial populations. Curr Biol. 2002;12:1040–1045. doi: 10.1016/s0960-9822(02)00896-5. [DOI] [PubMed] [Google Scholar]
- Sanjuán R, Moya A, Elena SF. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc Natl Acad Sci USA. 2004;101:8396–8401. doi: 10.1073/pnas.0400146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl LM, Krakauer DC. Models of experimental evolution: the role of genetic chance and selective necessity. Genetics. 2000;156:1437–1448. doi: 10.1093/genetics/156.3.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke CO. The speed of adaptation in large asexual populations. Genetics. 2004;167:2045–2053. doi: 10.1534/genetics.104.027136. [DOI] [PMC free article] [PubMed] [Google Scholar]