Abstract
The pseudomonal phytotoxin syringomycin E, and related nonribosomal peptides, contain an L-threo-β-hydroxy-aspartyl residue at the eighth position of the lipodepsipeptide backbone as part of a conserved nonproteinogenic tripeptide motif. Informatic analysis of the P. syringae genome suggests only one putative non-heme iron hydroxylase, AspH. On heterologous expression in E.coli AspH shows robust catalytic activity with free L-Asp and L-Asp thioesters to make β-OH-Asp, but yields the erythro diastereomer rather than the threo configuration that is found in syringomycin. Further analysis of the Syr gene cluster indicated that SyrP, previously annotated as the gene regulatory protein for the five- gene Syr cluster, is actually homologous to the known non-heme mononuclear iron hydroxylase TauD. Indeed, purified SyrP acts on Asp tethered as the protein-bound S-pantetheinyl thioester on the eighth module of the SyrE megasynthetase. The hydroxylation gives the anticipated L-threo-3-OH-Asp diastereomer found in syringomycin. The knockout of syrP abolishes the production of the mature syringomycin E, while knockout of aspH has no effect on syringomycin production.
The pseudomonad genus of gram-negative bacteria includes prolific producers of nonribosomal peptide (NRP) natural products. Phytopathogenic Pseudomonas syringae strains produce a number of such plant toxins, including coronatine, syringomycins, phaseolotoxin, tabtoxin, and tagetitoxin (1-5). Syringomycins are members of a family of related cyclic lipodepsipeptides that act as plant toxins by aggregating to form pores in plant cell membranes to facilitate exit of nutrients. One hallmark of nonribosomal peptides is the content of non-proteinogenic amino acid residues. In the five known pseudomonal lipopeptides, syringomycin E, cormycin A, pseudomycin B, syringostatin A and syringotoxin B there is a conserved C-terminal tripeptide moiety in the scaffold, composed of dehydrobutyrine7, β-OH-Asp8, and 4-Cl-Thr9 (Figure 1a) (3, 6-9). This nonproteinogenic tripeptide motif, comprising residues seven-nine of syringomycin, may be consequential for the biological function of these toxins.
FIGURE 1.
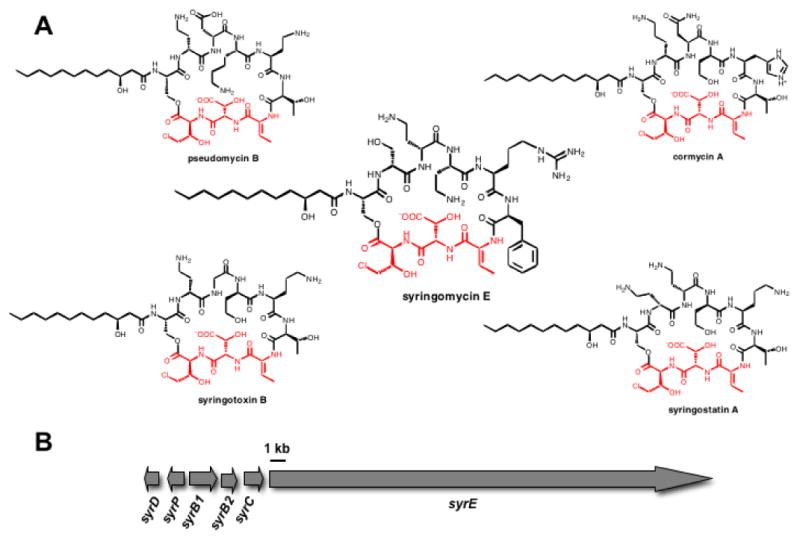
(A) Chemical structures of several pseudomonal lipopeptides containing the conserved C-terminal Z-Dhb7-L-3-OH-Asp8-L-4-Cl-Thr9 tripeptide. (B) The syringomycin E gene cluster.
The syringomycin gene cluster contains six ORFs, syrD, P, B1, B2, C, and E. The sequence of this biosynthetic gene cluster (10) for lipo-nonapeptidolactone syringomycin (Figure 1b) suggested that residues one through eight are assembled on a multimodular megasynthetase protein SyrE. Residue nine is the 4-Cl-threonine, inserted at the end of the assembly line by the tandem action of three enzymes SyrB1, SyrB2, and SyrC (11-13). Threonine is activated by the adenylation (A) domain of SyrB1, installed as the S-pantetheinyl thioester on the adjacent T (thiolation) domain (11), and then chlorinated by the non-heme mononuclear iron-containing halogenase SyrB2 (11, 12). At that juncture SyrC acts as an acyltransferase for the 4-Cl-Thr moiety, moving it to the T9 domain of the SyrE assembly line (13). SyrD is annotated as an ABC-type export protein and SyrP as a regulatory phosphoryl transferase that is an analog to a sensor kinase (14, 15).
Having worked out the tailoring and insertion of threonine as 4-Cl-Thr9 (11, 13), we have now turned our attention to genesis of the L-threo-β-OH-Asp8 residue in syringomycin and the related pseudomonal lipodepsipeptides. Many of the proteinogenic amino acid monomers, including Asp, Asn, Glu, Pro, and Lys, undergo enzymatic β-hydroxylation, both in the posttranslational modification of ribosomal proteins (16, 17) and in the biosynthesis of nonribosomal peptides (18-22). The known hydroxylases use iron as the redox catalyst in the active site, either Fe3+ in a cytochrome P450 heme environment (19) or Fe2+ in nonheme mononuclear state at the active site (22).
Informatic analysis of the P. syringae pv. syringae B728A genome (23) turns up one ORF at the primary locus Psyr_1584, aspH, predictively annotated as a non-heme, iron-dependent aspartyl hydroxylase. To evaluate the catalytic function of AspH, we have expressed AspH in E.coli and report on its activity here. Our particular interest was 1) in the capacity of AspH to hydroxylate free Asp vs. Asp in a thioester linkage and 2) determination of the chirality of β-hydroxylation. Our unanticipated finding, that AspH generates L-erythro- rather than L-threo-3-OH-Asp, has led us in this work to re-examine the remaining ORF in the syringomycin cluster, syrP (Figure 1b). Although SyrP has been annotated as a regulatory protein for the pathway (15), we report that it is actually an aspartyl hydroxylase that generates only the threo-3-OH-Asp product, and that it is involved in syringomycin maturation.
Experimental Procedures
Materials and general methods
All radiolabeled chemicals were obtained from American Radiolabeled Chemicals Inc. (ARC). L-threo-3-OH-Asp was obtained from Tocris Pharmaceuticals, and L-erythro-3-OH-Asp was obtained from Wako Pure Chemicals, Ltd. All other chemicals used were from Sigma, unless otherwise specified. The TOP10 and BL21(DE3) competent E. coli strains were purchased from Stratagene. The PfuTurbo DNA Polymerase used in PCR was purchased from Stratagene. During protein purification, cells were lysed using an Avestin EmμLsiflex-C5 cell disruptor. SDS polyacrylamide gels were obtained from BioRad and autoradiography was performed on a Typhoon 9400 scanner (GE Healthcare). HPLC was carried out on a Beckman Coulter System Gold using a Vydac C18 column or a Phenomenex Chirex column. Radio-HPLC was performed on an identical system equipped with a β-Ram radioisotope detector (IN/US). LC-MS analysis was performed on an LCMS-QP8000a spectrometer (Shimadzu) with a Vydac C18 LC-MS column.
Cloning, overproduction, and purification of SyrE-A8T8, AspH, and SyrP
The SyrE-A8T8 construct was obtained by PCR amplification from a pET-28a plasmid containing the SyrE-A8T8C9T9TE fragment (13). The following oligonucleotide pairs were used in PCR amplification: 5′ CGG GGA TCC CAT ATG CTT GAG CAG GAT CCG 3′ and 5′ CGG ATA GAG CTC CTA ACG GCC AGA GCG ATT 3′. The aspH and syrP genes were obtained from GeneArt in synthetic form with codons optimized for expression in E. coli. All constructs were digested with NdeI and XhoI and ligated into similarly digested pET28a vectors to generate N-terminally His6-tagged constructs.
The pET-28a expression vectors containing the constructs described above were transformed into BL21 (DE3) competent cells. Cultures were grown in Luria-Bertani medium supplemented with 30 μg/mL kanamycin at 37°C until the OD600 reached ∼0.3, at which time the cultures were cooled to 25°C, and grown until the OD600 reached ∼0.6. The cultures were induced with 0.1 mM IPTG and grown at 15°C overnight. Cells were harvested by centrifugation at 6,000 rpm for 30 min, flash frozen in liquid N2, and stored at -80°C until further purification.
Cells were thawed, resuspended in Buffer A (300 mM NaCl, 5 mM imidazole, 20 mM Hepps, pH 8.0), and lysed by cell disruption. Cell debris was removed from the lysate by centrifugation at 15,000 rpm for 30 min, and the supernatant was removed and bound to Ni-NTA resin by rocking at 4°C for 2 hr. The resin was added to a Bio-Rad Econo-Sphere column and washed with Buffer A. Protein was eluted with Buffer B (300 mM NaCl, 30 mM imidazole, 20 mM Hepps, pH 8.0) and Buffer C (100 mM NaCl, 200 mM imidazole, 20 mM Hepps, pH 8.0). Protein-containing fractions were identified by SDS-PAGE, combined and dialyzed overnight in 100 mM NaCl, 1 mM EDTA, 50 mM Hepps, pH 8.0 with 10% glycerol. The dialyzed protein was concentrated, flash frozen in liquid N2 and stored at -80°C.
ATP-PPi exchange assay for SyrE-A8
A typical reaction contained 10 mM amino acid, 10 mM MgCl2, 1 mM ATP, 1 mM DTT, 2 μM SyrE8,9, and 5 mM sodium [32P]-pyrophosphate in a total volume of 500 μL with 50 mM Hepes, pH 7.5. Reactions were carried out with L-Asp, L-erythro-3-OH-Asp, and L-threo-3-OH-Asp. In tandem, reactions containing no enzyme were carried out as negative controls. Reactions were initiated by the addition of enzyme and aliquots were quenched at 0, 1, 2, 5, 10, 20, 30, and 60 min by the addition of 750 μL of a solution containing 1.6% (w/v) activated charcoal, 200 mM sodium pyrophosphate, and 3.5% (v/v) perchloric acid. For each time point, the charcoal was pelleted by centrifugation and washed twice with 750 μL of a solution containing 200 mM sodium pyrophosphate with 3.5% perchloric acid (wash buffer). The charcoal pellet was then resuspended in 750 μL of wash buffer and mixed with liquid scintillation fluid. Radioactivity bound to the charcoal was measured by liquid scintillation counting.
Phosophopantetheinylation of SyrE-A8T8
In order to generate the active holo form of the T8 thiolation domain in the SyrE-A8T8 construct, a phosphopantetheinylation reaction must be carried out. In a typical reaction, 1 nmol of enzyme is incubated with MgCl2 (0.5 μmol), CoA (50 nmol), Sfp (10 nmol) in 50 mM Hepes buffer, pH 7.5, in a total reaction volume of 25 μL for 30 min at room temperature to produce the holo-enzyme.
Autoradiographic assay for autoaminoacylation of SyrE-A8T8
Holo-SyrE-A8T8 (0.6 nmol) was incubated with L-[14C]Asp (20 nmol) and ATP (100 nmol) in 50 mM Hepes buffer, pH 7.5 in a total reaction volume of 59 μL at room temperature. The reaction was initiated by the addition of ATP. At 1, 10, and 30 min, 20 μL aliquots of the reaction were quenched in 2× SDS-PAGE loading buffer. The quenched aliquots were heated to 90°C for 10 minutes and then run on a 12% SDS polyacrylamide gel. The gel was stained, destained, dried, and exposed to a phosphorimager screen for 3 days. The screen was then scanned using a Typhoon imager.
Assay for aminoacylation of SyrE-A8T8 by TCA precipitation
Holo-SyrE-A8T8 (17 nmol) was incubated with L-[14C]Asp (170 nmol) and ATP (200 nmol) in 50 mM Hepes buffer, pH 7.5 in a total reaction volume of 650 μL at room temperature. The reaction was initiated by the addition of ATP. At various time points, 50 μL aliquots of the reaction were quenched in 500 μL of 10% trichloroacetic acid (TCA). To each quenched aliquot, 100 μL of bovine serμM albumin (BSA) was added, and the aliquots were incubated on ice for 5 min. To pellet the precipitated proteins, the reactions were centrifuged at 13,000 rpm for 5 min, and an additional 500 μL of 10% TCA was added to each. Centrifugation was repeated, and the supernatant was aspirated. The remaining protein pellets were redissolved in 100 μL formic acid and all samples were subjected to liquid scintillation counting.
General assay for hydroxlase activity with various substrates
In a typical 100 μL reaction, the recombinant hydroxylase (either AspH or SyrP) was incubated with substrate (250 μM), (NH4)2Fe(SO4) (0.5 mM), alpha-ketoglutarate (0.5 mM), ascorbate (0.5 mM), dithiothreitol (0.5 mM), and catalase (0.5 ng/μL) in 50 mM Hepes, pH 7.5. Reactions were incubated at room temperature for 30 min.
Hydroxylase assay with L-[14C]Asp
Recombinant hydroxylase (either AspH or SyrP) was incubated with L-[14C]Asp under the conditions described above. The reactions were quenched in 10% TCA, centrifuged at 13,000 rpm for 20 min, and the supernatant was analyzed by chiral radio-HPLC on a Phenomenex 250 × 4.6 mm 3126 Chirex column with an isocratic gradient of 95% 2 mM copper sulfate in 5% isopropanol, monitoring 14C radioactive counts. Reactions with cold L-Asp were performed in tandem for LC-MS analysis.
Hydroxylase assay with SyrE-A8T8-S-L-[14C]Asp
The SyrE-A8T8-S-L-[14C]Asp substrate was generated by phosphopantetheinylation followed by autoaminoacylation of SyrE-A8T8 with L-[14C]-Asp, as described above. The reaction mixture containing SyrE-A8T8-S-L-[14C]Asp was washed twice with 300 μL of 50 mM Hepes in a Biomax 10 kDa MWCO centrifugation filter to remove unbound L-[14C]Asp. The SyrE-A8T8-S-L-[14C]Asp substrate was then incubated with either of the recombinant hydroxylases as described, and reactions were quenched with 10% TCA, and centrifuged at 13,000 rpm for 20 min. The supernatant was aspirated, and the protein pellet was redissolved and thioester linkage hydrolyzed in 200 μL of 100 mM LiOH with incubation at 80°C for 20 min. The protein was then reprecipitated by addition of 40 μL of 50% TCA followed by centrifugation at 13,000 rpm for 20 min. The supernatant was analyzed by chiral radio-HPLC as described above.
Hydroxylase assay with L-aspartyl-S-N-acetylcysteamine
The aspartyl-S-N-acetylcysteamine (L-Asp-SNAC) substrate was synthesized as described previously (24). Recombinant hydroxylase was incubated with the L-Asp-SNAC, reactions were quenched in 10% TCA, and the thioester bond was hydrolyzed in 100 mM LiOH, as with the SyrE-A8T8-S-L-[14C]Asp substrate. Reactions were analyzed by chiral radio-HPLC as described.
Hydroxylase assay with N-acyl-nonapeptide (COOH-L-Thr-L-Asp-L-Abu-L-Phe-L-Arg-L-Dab-D-Dab-D-Ser-L-Ser-N-Ac), and N-acyl-nonapeptidyl-CoA (CoA-S-L-Thr-L-Asp-L-Abu-L-Phe-L-Arg-L-Dab-D-Dab-D-Ser-L-Ser-N-Ac)
The nonapeptide and nonapeptidyl-CoA were synthesized as described previously (13). Recombinant hydroxylase was incubated with either substrate, reactions were quenched in 10% TCA, centrifuged to remove protein, and analyzed by LC-MS and HPLC. The HPLC analysis was performed on a Vydac 250 × 4.6 mM C18 small pore column with a 0-100% acetonitrile gradient over 40 minutes beginning in 0.1 % TFA in H2O and monitoring absorbance at 220 nm.
Construction of the aspH gene disruption cassette
The aspH gene was PCR-amplified from genomic DNA from Pseudomonas syringae pv. syringae B301D (obtained as a gift from Dennis C. Gross, Texas A&M University, College Station, Texas) using the following primers: 5′-CCGGATCCATGACCCTTTCATTTGTTGCCAAGGC-3′ and 3′-CCAAGCTTTTAACCAAACAACCAGTAGCCCAG-5′ containing the BamHI and HindIII sites, respectively. Amplification was carried out using Finnzyme Phusion DNA polymerase (New England Biolabs), following the manufacturer's instructions. The resulting fragment was digested with BamHI/HindIII and ligated into the similarly digested pEX18Gm gene replacement vector (25) to produce the pEX18GmA1 plasmid. A 559 bp central fragment was excised from the aspH gene within pEX18GmA1 using NruI and NsiI, and the vector fragment was purified and its 5′ and 3′ termini were blunted using the NEB Quick Blunting Kit.
The cat gene was PCR-amplified from pKD3 (26) using the following primers: 5′CCGAATTCATGGAGAAAAAAATCACTGGATATACCAC3′ and 3′CCGAATTCTCATCGCAGTACTGTTGTATTCATTAAGC5′ both encoding for EcoRI sites. The resulting fragment was EcoRI digested and ligated into the similarly digested pPS854 vector between the FRT-encoding sites in the multiple-cloning site of the vector to generate the pPS854-FCF vector. The cat gene flanked by the two FRT sites was excised from pPS854-FCF with SacI, and the resulting FRT-cat-FRT fragment was gel purified and its 5′ and 3′ termini were blunted as described above. The FRT-cat-FRT fragment was then ligated via blunt ends into the pEX18GmA1 linear fragment described above to generate the pEX18GmAKO gene replacement vector.
Construction of the syrP gene disruption cassette
The syrP gene was PCR-amplified from genomic DNA from Pseudomonas syringae pv. syringae B301D using the following primers: 5′-CCGGATCCATGAGTCTTCAAGCTAACACTGCGCC-3′ and 3′-CCAAGCTTTCAGGCCGGTTGCCAAACGTCGCC-5′ containing the BamHI and HindIII sites, respectively. Amplification was carried out as described above. The resulting fragment was digested with BamHI/HindIII and ligated into the similarly digested pET28a vector to produce the pET28aP vector. A central 300 bp segment of the syrP gene in pET28aP was excised with BstBI and the linear vector fragment was gel-purified and its 5′ and 3′ termini were blunted as described. Blunt-end ligation was carried out between this fragment and the FRT-cat-FRT, generated as described in the previous section to generate the pET28aPFCFP vector. Using the BamHI and HindIII sites in this plasmid, the 1.6 kb fragment containing syrP interrupted by the FRT-cat-FRT cassette (ie. syrP-FRT-cat-FRT-syrP) was excised and ligated into the pEX18Gm gene transfer vector to produce the pEX18GmPKO gene replacement vector.
Introduction of disrupted aspH and syrP genes into P. syringae pv. syringae
The pEX18GmAKO and pEX18GmPKO gene replacement vectors were transformed into the E. coli S17.7λ (27) mating strain by electroporation (28). Conjugation was performed using the filter membrane mating technique (29), using the donor strain described above and a rifampicin-resistant variant of P. syringae pv. syringae B301D as the recipient strain. Briefly, the donor strain carrying the desired delivery plasmid was grown overnight at 37 °C in 2 mL of LB mediμM containing 10 μg/mL gentamycin and 25 μg/mL chloramphenicol. The recipient strains were grown overnight in 2 mL of LB medium without selection. The donor cells were harvested and washed twice with 2 mL of 1% NaCl to remove the excess antibiotics. Ten μL of the donor cells and 100 μL of the recipient cells were added to 5 mL of 1% NaCl, vortexed, then filtered through a 25 mm type HA 0.45 μm Millipore membrane (Bedford, MA). The filter was placed on LB solid medium and incubated at 30 °C for 12 hours. The filter was then immersed in 5 mL of 1% NaCl and vortexed to resuspend the mating culture. Primary integrants were selected by plating 10-500 μL aliquots of the mating resuspension on LB solid medium containing 25 μg/mL rifampicin and 25 μg/mL chloramphenicol. Successful intergration events were confirmed by testing for ampicillin sensitivity. The second homologous crossover was selected for by dual resistance to sucrose and chloramphenicol resulting from the loss of the sacB counterselectable marker. Total genomic DNA was isolated from the sucrose and chloramphenicol resistant colonies and PCR was used to confirm the presence of the FRT-cat-FRT cassette in the aspH or syrP gene within the P. syringae chromosome (i.e. aspH∷cat and syrP∷cat).
Removal of the cat selection marker from the resulting P. syringae knockout clones was performed by transformation of the latter with the Flp recombinase-encoding plasmid pFLP2 (25). Transformants were selected for by ampicillin resistance and were grown in liquid culture. The Flp-catalyzed excision of the cat selection marker was confirmed by chloramphenicol sensitivity. Total genomic DNA was isolated from sensitive colonies and PCR was used to confirm loss of the cat element and the presence of the truncated syrP and aspH genes.
Syringomycin production, extraction, and analysis
The syrP∷cat, ΔsyrP, and wild type strains of P. syringae pv. syringae B301D were grown in 200 mL of PDB media in still culture at 25°C for 6 days, and the ΔaspH strain was grown for 8 days due to delayed growth. Cultures were quenched with 1:1 volumes of ice-cold acetone, acidified to pH 2.5, and stored at 4°C overnight. Cell debris was removed by centrifugation and acetone was removed by rotary evaporation under vacuum. The water residue was then extracted thrice in water-saturated n-butanol and the butanol phase was evaporated by rotary evaporation under vacuum. The remaining water residue was lyophilized and redissolved in 5 mL ddH2O pH 2.0. The extracts were analyzed both by LC-MS and by C18 HPLC. The HPLC analysis was performed on a Vydac 250 × 4.6 mM C18 small pore column with a 0-100% acetonitrile gradient over 50 minutes beginning in 0.1 % TFA in H2O and monitoring absorbance at 220 nm.
Results
Overproduction and purification of AspH and SyrE-A8T8
A search of the annotated genome of P. syringae pv. syringae B728a led to the identification of a single aspartyl/asparaginyl beta-hydroxylase at the primary locus Psyr_1584 (GenBank ID AAY36632.1), which we hereafter term aspH. Comparison of the protein sequence of AspH to other known bacterial iron and alpha-ketoglutarate-dependent aspartyl/asparaginyl hydroxylases indicates that it contains the conserved His2-Asp facial triad responsible for iron-binding as well as the two arginines required for binding alpha-ketoglutarate. Indeed, AspH shows significant homology to known aspartyl beta-hydroxylases from Alcanivorax and Ralstonia, as well as to LpxO, an iron and alpha-KG dependent dioxygenase from S. typimurium, that hydroxylates lipid A (30). To circumvent difficulties in recombinant expression, aspH was obtained as a synthetic gene with codons optimized for expression in E. coli. The N-terminally His6-tagged 35.7 kDa protein AspH was expressed in E. coli at 15°C following induction with 0.1 mM IPTG and purified using nickel-affinity chromatography to give a yield of 0.5 mg/ L of culture (Figure 2a).
FIGURE 2.
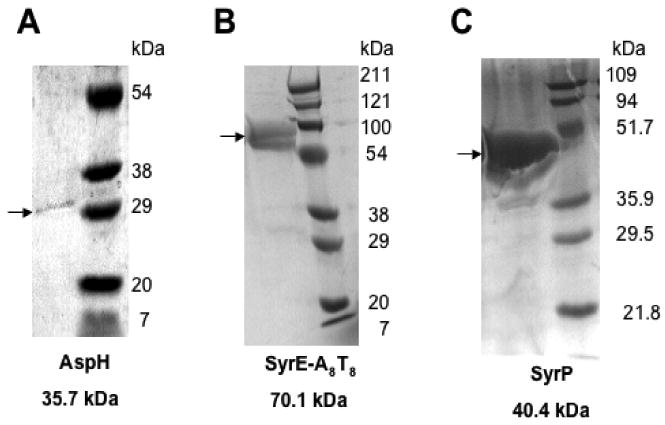
SDS-PAGE analysis of purified (A) AspH, 35.7 kDa, (B) SyrE-A8T8, 70.1 kDa, (C) SyrP, 40.4 kDa.
In addition, a construct comprising part of the eighth module of the SyrE megasynthetase, namely, SyrE- A8T8, was cloned into the pET-28a vector and expressed in E. coli as described above. With this 70.1 kDa construct, yields of 8-10 mg protein/ L of culture were obtained (Figure 2b).
ATP-PPi exchange assays to evaluate SyrE-A8 substrate specificity
In order to obtain a first approximation of the timing of aspartyl hydroxylation in syringomycin biosynthesis, the ATP-PPi exchange assay was performed to examine the substrate preference of the eighth adenylation domain of SyrE (SyrE-A8). The ability of the adenylation domain of SyrE-A8T8 to activate L-Asp, L-erythro-3-OH-Asp, and L-threo-3-OH-Asp was assayed. It was found that A8 preferentially activates unmodified L-Asp (kobs = 6.3 min-1) in comparison to the erythro (kobs = 0.1 min-1) or threo (kobs = 0.2 min-1) 3-OH-Asp substrates (Figure 3). These results indicate that hydroxylation of Asp8 in syringomycin biosynthesis occurs either on the assembly line, or after cyclization and release of the cyclic lipopeptidolactone, but is unlikely to occur on free aspartic acid.
FIGURE 3.
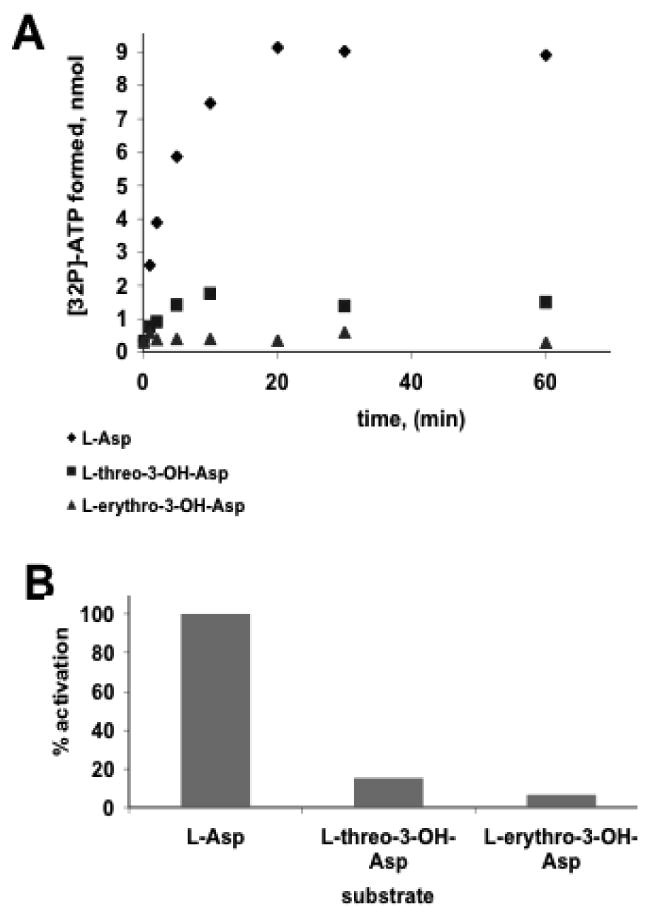
Results of the ATP-PPi exchange assay to determine the substrate specificity of SyrE-A8 (A) Determination of kobs for three substrates: black diamonds, L-Asp, kobs = 6.3 min-1; gray squares, L-threo-3-OH-Asp, kobs = 0.2 min-1; white circles, L-erythro-3-OH-Asp, kobs = 0.1 min-1. (B) Comparison of % activation by SyrE-A8 of L-Asp, L-threo-3-OH-Asp, L-erythro-3-OH-Asp.
Phosphopantetheinylation and auto-aminoacylation of SyrE-A8T8
While the ATP-PPi exchange assay demonstrated the activity and substrate specificity of the adenylation domain of the SyrE-A8T8 construct, further work was necessary to prove the competence of the thiolation domain for covalent thioesterification with aspartic acid. Reaction of SyrE- A8T8 with Coenzyme A and the phosphopantetheinyl transferase Sfp yielded the holo-form of the thiolation domain. Further incubation with L-[14C]-Asp and ATP led to activation of the substrate and subsequent aminoacylation of the thiolation domain. Auto-aminoacylation was monitored over time by TCA precipitation followed by liquid scintillation counting as well as by autoradiography (Figure 4).
FIGURE 4.
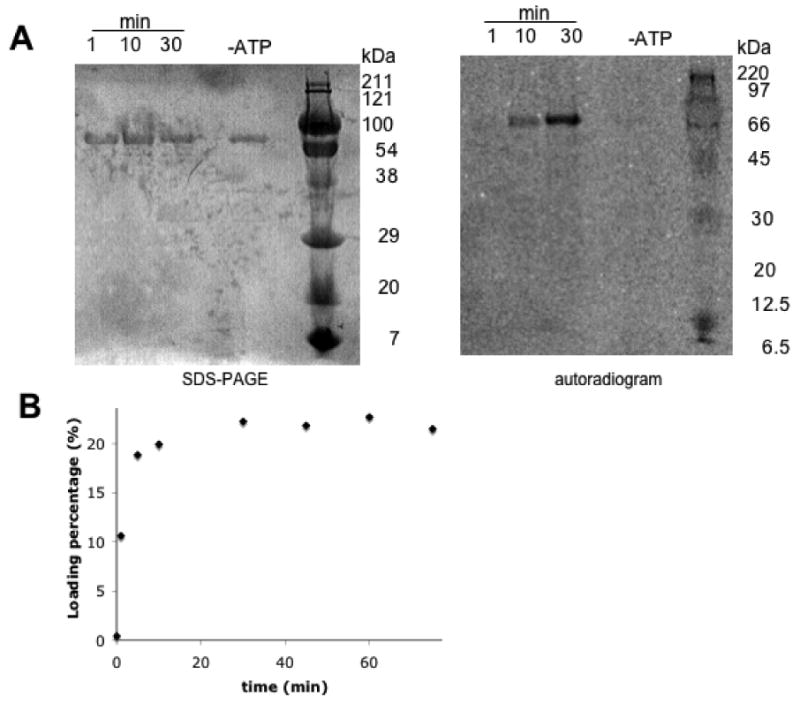
Auto-aminoacylation of SyrE-A8T8 with L-[14C]Asp. (A) 12% SDS-PAGE and autoradiogram showing an ATP-dependence of auto-aminoacylation. (B) Graphical representation of time-dependence of auto-aminoacylation, as measured by a TCA precipitation assay.
Characterization of AspH dioxygenase activity with free amino acids
In order to determine the timing of hydroxylation carried out by AspH, a number of substrates were used to test AspH activity. A chiral radio-HPLC assay was developed in order to monitor product formation and determine product chirality. In a typical assay, 1 μM AspH was incubated with 250 μM substrate, 0.5 mM (NH4)2Fe(SO4), 0.5 mM alpha-KG, 0.5 mM ascorbate, 0.5 mM DTT, and 0.2 ng/μL catalase in 50 mM Hepes buffer for 30 min. In all cases, control reactions were carried out without enzyme, without (NH4)2Fe(SO4), and without alpha-KG. Upon incubating AspH with L-[14C]Asp under the conditions described above, an enzyme, Fe(II), and alpha-KG-dependent peak was observed in the chiral radio-HPLC chromatogram (Figure 5a). The peak co-eluted with the L-erythro-3-OH-Asp authentic standard and LC-MS indicated its mass coincided with that of 3-OH-Asp (3-OH-Asp 149.10 [(M+H)+] calculated, 149.02 observed). The generation of the L-erythro product was quite unexpected, since syringomycin contains the L-threo diastereomer. In a similar set of experiments, D-Asp, D-Glu, L-Glu, D-Asn, and L-Asn were tested as substrates for AspH, but no hydroxylation activity was noted for any of these substrates.
FIGURE 5.
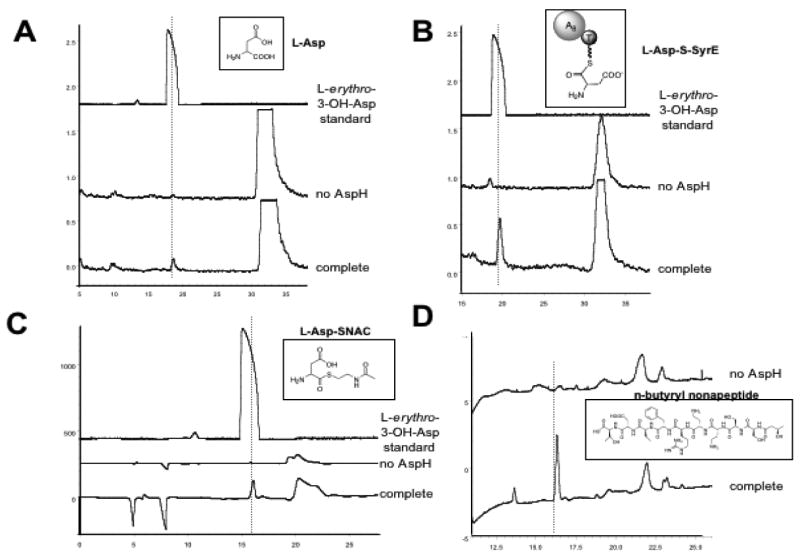
HPLC chromatograms of products from AspH incubation with various substrates. “Complete” denotes reactions containing all components, and “no AspH” denotes control reactions lacking the enzyme. The substrates are shown as insets in each trace. A dashed line is shown passing through the product peaks. (A) Chiral radio-HPLC chromatogram of AspH incubation with L-[14C]Asp, (B) Chiral radio-HPLC chromatogram of AspH incubation with SyrE-A8T8-S-L-[14C]Asp. (C) Chiral HPLC of AspH incubation with L-Asp-SNAC with detection at 220 nm. (D) C18 HPLC of AspH incubation with the linear N-acyl-nonapeptide mimic of the syringomycin lipononapeptide.
Characterization of AspH dioxygenase activity with thioester-linked substrates
The kinetic parameters of AspH with L-[14C]Asp (kcat = 0.15 min-1, Km = 51.9 μM) suggest that the free substrate is sub-optimal, so the question arose as to whether the enzyme requires a thioester-linked substrate as opposed to one with a free carboxylate. Therefore, the SyrE-A8T8-S- L-[14C]Asp substrate was generated by phosphopantetheinylation of SyrE-A8T8 via Sfp followed by auto-aminoacylation of the holo construct with L-[14C]Asp as described above. The SyrE-A8T8-S- L-[14C]Asp substrate was incubated with AspH under standard conditions, and the product was released hydrolytically with LiOH. Chiral radio-HPLC and LC-MS analysis of the reaction products again showed formation of the L-erythro-3-OH-Asp product in an enzyme, Fe(II) and alpha-KG-dependent fashion (3-OH-Asp 149.10 [(M+H)+] calculated, 149.02 observed) (Figure 5b)
Since it is difficult to accurately determine the concentration of the SyrE-A8T8-S- L-[14C]Asp substrate, the kinetic parameters of AspH with this substrate could not be determined. However, an aspartyl-S-N-acetylcysteamine (Asp-SNAC) substrate was generated as a mimic of the PCP-tethered substrate such that kinetic studies of AspH with a thioester-linked substrate could be carried out. The L-Asp-SNAC substrate was incubated with AspH under standard conditions, the thioester linkage was cleaved with LiOH, and the reaction products were analyzed as described. In this case we again noted the formation of the L-erythro-3-OH-Asp product (Figure 5c) and the mass of the 3-OH-Asp product was noted by LC-MS (3-OH-Asp 149.10 [(M+H)+] calculated, 149.02 observed). The kinetic parameters of the hydroxylation of the L-Asp-SNAC (kcat = 42.5 min-1, Km = 43.6 μM) were significantly improved over those for the free L-Asp substrate, with kcat increasing by about 283-fold.
Characterization of AspH dioxygenase activity with peptide substrates
To determine AspH activity on a third class of substrates, a linear N-butyryl-nonapeptide mimic of the syringomycin lipononapeptide was generated by chemical synthesis. The linear nonapeptide was incubated with AspH under standard conditions, and the reaction products were characterized by C18 HPLC and LC-MS. In the HPLC chromatogram, an AspH-dependent peak was observed (Figure 5d), and the mass of the hydroxylated peptide was apparent by LC-MS (1098.53 [(M+H)+] calculated, 1099.04 observed). However, the exact position of hydroxylation in the nonapeptide could not be determined by these methods, and would have to be determined by hydrolysis of the reaction product followed by chiral HPLC. It has thus far not been possible to generate sufficient material for the hydrolytic analysis of the hydroxylated product.
Although the exact position and stereochemistry of hydroxylation in the nonapeptide remains to be determined, the kinetic parameters of AspH with the linear lipononapeptide substrate were obtained (kcat = 120 min-1, Km = 26.9 μM) and suggest that this substrate is the most kinetically competent substrate of those tested, with an apparent catalytic efficiency (kcat/Km) improved 1500- and 4.6-fold over that of L-Asp and L-Asp-SNAC, respectively.
In combination, these data suggest that AspH preferentially modifies substrates in which the C-terminus of L-Asp is bound in a thioester or amide linkage, which may indicate that AspH is involved in posttranslational modification of peptides and proteins (Table 1). However, in all cases where stereochemistry could be determined, AspH generates the L-erythro-3-OH-Asp diastereomer, while syringomycin contains the L-threo diastereomer. This observation, in combination with the fact that AspH is located far upstream of the syringomycin gene cluster, strongly suggests that AspH is probably not involved in tailoring Asp8 in syringomycin biosynthesis.
Table 1.
Kinetics data collected for AspH with aspartate-containing substrates.
| Substrate | kcat (min-1) | Km (μM) | kcat/Km |
|---|---|---|---|
| L-Asp | 0.15 ± 0.03 | 52.9 ± 4.7 | 0.002 |
| L-Asp-SNAC | 42.5 ± 5.2 | 43.6 ± 2.3 | 0.97 |
| Linear nonapeptide | 120.0 ± 15.4 | 26.9 ± 9.9 | 4.6 |
Overproduction and purification of SyrP
The fact that AspH does not appear to be involved in syringomycin tailoring led us to re-examine the members of the syringomycin gene cluster in search of a putative dioxygenase, since it is most likely that the tailoring enzyme would be in or near the gene cluster itself. A BLAST analysis of SyrP, which was previously assigned as a regulator of syringomycin production, revealed a high degree of homology to TauD, the well-studied taurine dioxygenase (31-33) (Figure 6). Indeed SyrP contained the conserved His2-Asp facial triad responsible for iron-binding, as well as the conserved Arg residues that act as alpha-KG ligands. This strongly suggested that SyrP might be the aspartyl dioxygenase responsible for hydroxylation of Asp8.
FIGURE 6.
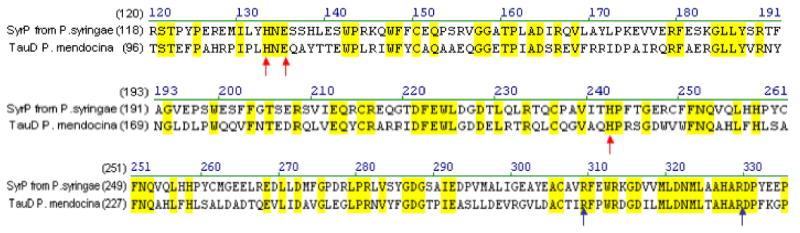
Alignment of the protein sequences of SyrP from P. syringae pv. syringae B728a with that of TauD from P. mendocina. Red arrows indicate the His2/Asp iron-binding motif and blue arrows point to the two conserved Arg residues that bind alpha-ketoglutarate.
A synthetic gene for syrP with codons optimized for expression in E. coli was cloned into N-terminal His6-tagged vector. Then SyrP was expressed in E. coli at 15°C following induction with 0.1 mM IPTG. The soluble 40.4 kDa protein was purified using nickel-affinity chromatography to produce a yield of approximately 10 mg/ L of culture (Figure 2c).
Examination of SyrP dioxygenase activity with Asp-containing substrates
The ability of SyrP to act as an aspartyl hydroxylase was examined with a number of Asp-containing substrates, including free L-[14C]Asp, SyrE-A8T8-S- L-[14C]Asp, L-Asp-SNAC, the linear N-acyl-nonapeptide, the N-acyl-nonapeptidyl-CoA, and the cyclic N-dodecanoyl nonapeptidolactone. Assays were performed as described above, and products were characterized by HPLC and LC-MS. However, SyrP did not generate a hydroxylated product from any of the listed substrates except SyrE-A8T8-S- L-[14C]Asp. Furthermore, with this substrate, the stereochemistry of the 3-OH-Asp product was found to be exclusively in the L-threo configuration (Figure 7). Interestingly, the SyrP-mediated hydroxylation was found to be dependent on alpha-KG, but was independent of exogenous (NH4)2Fe(SO4), leading to the hypothesis that SyrP is purified with Fe(II) bound. The Ferrene S assay (34) revealed that indeed, pure SyrP contains 28% Fe(II).
FIGURE 7.
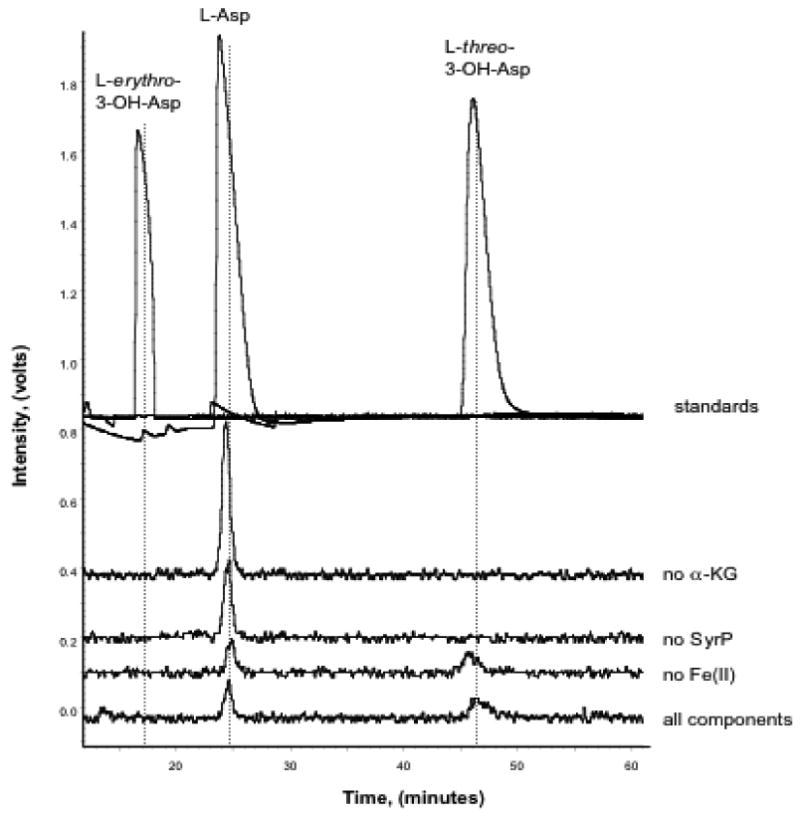
Chiral radio-HPLC traces of products from the reaction of SyrP with the SyrE-A8T8-S-L-[14C]Asp substrate and products of the control reactions. In the reaction containing all components, a peak at ∼49 min is evident in the B-RAM chromatogram, corresponding to the retention time of the L-threo-3-OH-Asp authentic standard. A peak with similar retention time is evident in the control reaction lacking Fe(II), suggesting that SyrP may have been purified with Fe(II) already bound. The mass of the L-threo-3-OH-Asp product was verified by LC-MS.
These data suggest that SyrP is likely to be the Fe(II)- and alpha-KG-dependent aspartyl hydroxylase responsible for hydroxylating Asp8 in syringomycin biosynthesis. In addition, the fact that SyrP acts exclusively on the PCP-tethered substrate indicates that hydroxylation occurs while the substrate is on the assembly line, corresponding with the results of the ATP-PPi exchange assay.
Properties of the SyrP-mediated hydroxylation reaction
Due to the difficulty of determining the exact concentration of the SyrE-A8T8-S- L-[14C]Asp substrate because of small variations in the loading of A8T8 with L-[14C]Asp, the kinetic parameters of SyrP dioxygenase activity with this substrate could not be determined. Despite this obstacle, the catalytic properties of the SyrP mediated hydroxylation could be characterized by approximately calculating the amount of SyrE-A8T8-S- L-[14C]Asp present using the TCA precipitation assay.
In order to determine whether product formation catalyzed by SyrP is proportional to the amount of substrate present, the hydroxylation reaction was analyzed at increasing concentrations of SyrE-A8T8-S- L-[14C]Asp. This experiment revealed the linear correlation of the amount of SyrE-A8T8-S- L-[14C]Asp to the amount of L-threo-3-OH-Asp product (Figure 8a). To further examine the catalytic role of SyrP, an experiment was carried out in which SyrE-A8T8-S- L-[14C]Asp was held constant at 4 nmol, and SyrP was titrated from 25 to 500 pmol. In this case, SyrP was found to catalyze multiple turnovers at low SyrP:A8T8 ratios, indicating that it does indeed function catalytically (Figure 8b).
FIGURE 8.
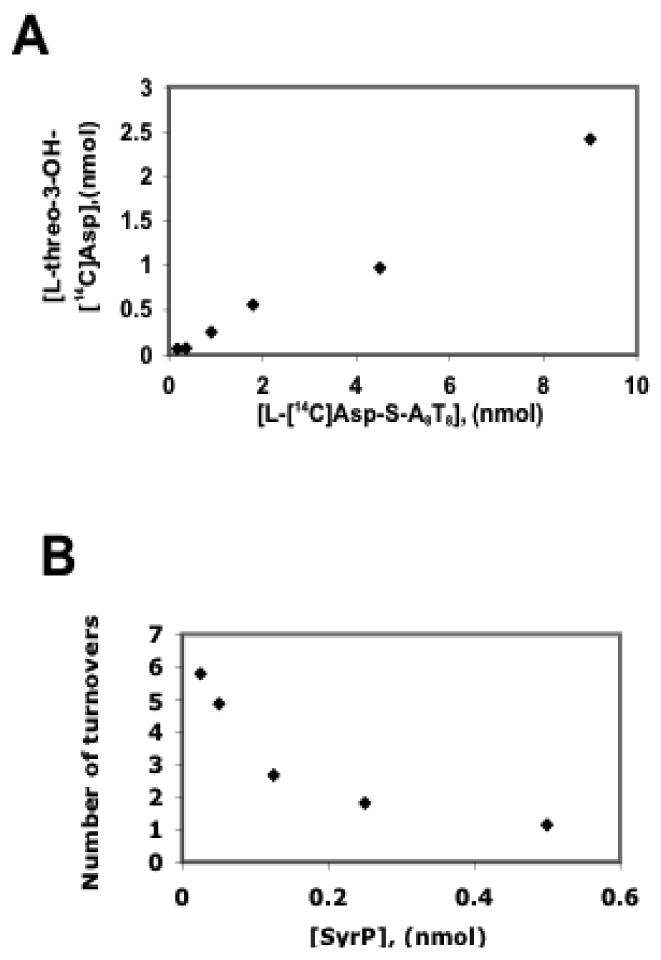
(A) In a SyrP-mediated reaction, formation of the L-threo-3-OH-[14C]Asp product is proportional to the amount of SyrE-A8T8-S-L-[14C]-Asp substrate. (B) When SyrE-A8T8-S-L-[14C]-Asp is held constant at 4 nmol and SyrP is titrated, multiple turnovers are observed at low SyrP:A8T8 ratios, indicating that SyrP functions catalytically.
Generation and characterization of syrP and aspH knockout strains of P. syringae pv. syringae
The ΔaspH and ΔsyrP strains of P. syringae pv. syringae B301D were generated by the introduction of gene-replacement vectors containing the FRT-flanked, cat-disrupted aspH or syrP genes into the parent strain as described in the methods. The deletions were returned to the P. syringae pv. syringae B301D chromosome through successive selection steps for each of two homologous recombination events. The resulting aspH∷cat and syrP∷cat strains were then electroporated with the plasmid pFLP2. Subsequent expression of the Flp recombinase, which acts at the FRT sites flanking the cat gene in the P. syringae chromosome, resulted in Flp-catalyzed excision of the cat elements from the host chromosome and produced the unmarked ΔaspH and ΔsyrP strains. PCR was performed on genomic DNA isolated from the aspH∷cat, syrP∷cat, ΔaspH, and ΔsyrP strains to confirm the presence of the desired insertions and deletions.
The syrP∷cat, ΔsyrP, and wild type strains were grown in PDB media in still culture at 25°C for 6 days, and the ΔaspH strain was grown under identical conditions for an additional two days due to slow growth. Cultures were extracted in water-saturated n-butanol and were assayed for syringomycin production by HPLC and LC-MS (Figure 9). This analysis revealed that the ΔaspH strain produced roughly equivalent amounts of syringomycin (1225.6 [(M+H)+] calc., 1225.6 obs.) as the wild type strain, despite its reduced growth rate, supporting the hypothesis that AspH is not involved in syringomycin biosynthesis. However, analysis of the syrP∷cat strain indicated that syringomycin production was entirely abolished. This led us to suspect that the insertion of the cat gene in the P. syringae chromosome was causing polar effects on downstream genes in the syringomycin gene cluster. Subsequent HPLC analysis of the unmarked ΔsyrP strain suggested that it, too, did not produce any syringomycin, but instead produced a new compound with a retention time that is 0.3 minutes later than that of syringomycin. The LC-MS analysis of material eluting in this peak showed it to have m/z of 1216.6 rather than the 1209.6 m/z expected for the des-hydroxy species.
FIGURE 9.
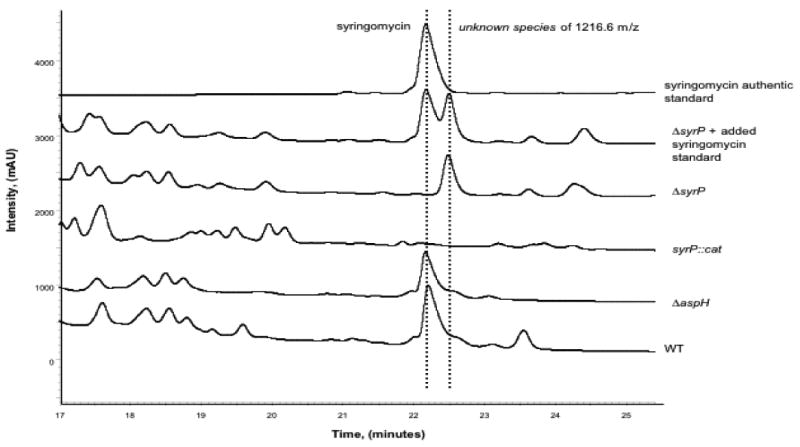
HPLC analysis of n-butanol extracts of wild type, ΔaspH, syrP∷cat, ΔsyrP, ΔsyrP spiked with syringomycin authentic standard, and syringomycin authentic standard. The traces show a new product formed in the ΔsyrP culture with a retention time that is 0.3 minutes later than syringomycin. LC-MS analysis of this product shows it to have a mass of 1216.6 m/z, and NMR studies indicate that it is an immature form of syringomycin, missing the β-OH group on Asp8. Insufficient amounts of the 13C-, 15N-labeled compound were obtained for determination of the other structural variations in this compound.
The syrP knockout strain was grown in the presence of 13C-Asp and 15N-NH4Cl in order to obtain 13C and 15N-labeled syringomycin derivatives. NMR studies of the compound produced by the ΔsyrP species indicated that it is, in fact, an immature form of syringomycin lacking the β-OH form of Asp8. However, due to difficulties in extracting sufficient amounts of the labeled compound, these NMR studies were unable to fully elucidate the structure of this species.
Discussion
Syringomycin and related nonribosomal peptides from pseudomonads are pore-forming N-acylpeptidolactones. The long-chain acyl group is added at the N-terminal residue at the start while the lactone/depsipeptide linkage is formed in the terminal chain release/cyclization step. The full-length nonapeptidyl chain, tethered as a pantetheinyl-S-peptide on the ninth and last module of the megasynthetase SyrE enzyme, is released by intramolecular capture of the thioester carbonyl of Cl-Thr9 by the OH-side chain of Ser1. In the pseudomonal lipodepsipeptide family the residues at positions 7-9 are conserved as dehydrobutyrine7-3-OH-Asp8-4-Cl-Thr9, arising respectively from the proteinogenic monomers, Thr, Asp, and Thr in a set of enzyme-mediated tailoring steps.
In principle, the tailoring of those three residues could happen (a) prior to NRPS assembly line action, (b) during assembly line action, or (c) after release of the nonapeptidolactones from the assembly lines (35). Our prior studies on 4-Cl-Thr9 have established that it is chlorinated on the assembly line, as a Thr-S-pantetheinyl-SyrB1 covalent intermediate, and then transferred by the agency of the shuttle enzyme SyrC to the T9 thiolation domain on SyrE. Nothing is known on the timing of dehydration of Thr7 to dehydrobutyrine although it most likely occurs after Thr7 has been incorporated into the growing chain to avoid enolization and adventitious hydrolysis to 2-ketobutyrate. In this study we have investigated the timing of Asp8 enzymatic tailoring.
The adenylation-thiolation didomain pair, A8T8, expected to activate and load Asp8 onto the syringomycin synthetase assembly line is embedded as part of the eighth module in the megadalton protein SyrE. We have been able to express A8T8 as a free-standing fragment and posttranslationally prime T8 with phosphopantetheine via the enzyme Sfp (36) to yield the holo form of A8T8 as a soluble, well-folded 70.1 kDa protein. The A domain shows a preference for activating L-Asp by 60-fold over the 3-OH erythro diastereomer and 30 fold over the 3-OH-threo diastereomer, consistent with the view that hydroxylation occurs after selection, activation, and installation of Asp as the S-pantetheinyl thioester on T8. By use of [14C]-L-Asp, we could generate A8T8-S-L-[14C]Asp as a potential substrate for the two candidate hydroxylases, AspH and SyrP, examined in this work.
Based on precedent, we expected the conversion of the L-Asp residue into the L-3-OH-Asp residue in syringomycin would be carried out by an iron-based hydroxylase. Such an oxygenase could possibly be a heme protein but more likely would be a nonheme mononuclear iron with a two-His, one-carboxylate facial triad as the active site ligands (22, 33). Initial inspection of the syringomycin biosynthetic gene cluster annotations (15) did not reveal either type of iron hydroxylase within the cluster. Reasoning that such a hydroxylase might be encoded elsewhere in the genome or even be borrowed from another pathway, we examined all the orfs in the producing Pseudomonas syringae genome. The most likely candidate was an ORF at the primary locus Psyr_1584. Residues 149-229 of AspH align well with Asp nonheme iron hydroxylases from Alcanivorax and from Ralstonia as well as LpxO from S. typhimurium. In particular, the putative iron ligands, two histidines and a conserved aspartate in an HXD array, as well as the two arginine side chains observed to coordinate α-ketoglutarate in this superfamily (22, 33) are all conserved, strongly suggesting AspH would be an Asp-selective α-ketoglutarate-requiring nonheme iron hydroxylase.
Indeed, AspH is such a catalyst. Our evidence shows that free aspartate is a detectable but poor substrate (kcat = 0.15 min-1) for hydroxylation in an O2- and FeII-dependent manner. Asp-SNAC, a soluble mimic of the Asp-S-pantetheinyl tether for covalent intermediates on NRPS assembly lines, has a 280 fold higher kcat at 42 min-1. While this might point to oxygenative tailoring on module eight of SyrE, we also examined the ability of AspH to hydroxylate a linear N-acylnonapeptide surrogate of syringomycin and found an even higher kcat of 120 min-1. This latter finding, that an Asp side chain in the peptide is hydroxylated 800 fold faster than the free Asp amino acid would argue that the function of AspH may be to hydroxylate Asp side chains in a peptide scaffold, but that has yet to be resolved.
However, in each of the above three contexts, free Asp, Asp-SNAC, and the linear nonapeptide, the AspH enzyme generated the L-erythro 3-OH diastereomer, with no detectable formation of the L-threo diasteromer. Since it is the L-threo form that is found in syringomycin, we were forced to the following conclusion. AspH, while indeed a mononuclear iron aspartyl β-hydroxylase with robust activity toward the aspartyl side chain when the carboxyl was derivatized, is not likely to be involved in a syringomycin tailoring step.
At this juncture we returned to a bioinformatics approach by re-examining the Syr gene cluster and made an unanticipated observation about SyrP. The publication in 1997 (15) assigning a regulatory function to the 40 kDa SyrP protein noted similarity to phospho-transfer and phospho-receiver domains found in the E. coli chemotaxis protein CheA although SyrP lacked the domains associated with autophosphorylation activity. Despite this predicted lack of autophosphorylation capability, Zhang et al. suggested, but did not directly prove, that SyrP might have a regulatory phosphotransfer role in control of Syr gene cluster expression (5).
However, a contemporary BLAST analysis shows a high degree of homology between SyrP and the P. mendocina TauD protein rather than to chemotaxis homologs. TauD is the nonheme iron enzyme taurine hydroxylase, a paradigm for mechanistic and structural knowledge about the mononuclear FeII hydroxylase family (31-33). Most notably, SyrP contains the conserved HXE motif that comprises two of the three facial triad ligands to the FeII in the TauD active site. In addition, SyrP also has the third conserved His and the two Arg residues that function in this enzyme family to position the cosubstrate α-ketoglutarate.
To evaluate whether SyrP could indeed be the sought after syringomycin Asp hydroxylase, we procured a synthetic gene, expressed it in E. coli, and obtained a soluble, active hydroxylase. It was fortunate that we had prepared the variety of substrates used to profile AspH activity because SyrP showed no hydroxylase activity toward free Asp or the linear nonapeptide analog of syringomycin. However, SyrP was active when presented with Asp-S-pantetheinyl T8 in the context of the didomain A8T8 fragment excised from the nonamodular SyrE. Most reassuringly, chirality analysis after hydrolytic release of product from thioester linkage showed the product to be the desired L-threo-3-OH-aspartate. Kinetic characterization of SyrP with the incompletely loaded Asp-S-A8T8 70 kDa protein substrate was not feasible, but the hydroxylation reaction is linearly dependent on SyrP concentrations and multiple turnovers are detected.
Several conclusions can be drawn. SyrP is indeed a mononuclear iron-containing Asp hydroxylase, but not one that acts on the free amino acid. It works on L-Asp tethered in pantetheinyl thioester tethered to the T8 carrier protein domain of SyrE. This accords with our observation that A8 activates Asp about 30 times faster than L-threo-3-OH-Asp, consistent with oxygenative tailoring after the A8 adenylation domain has activated loaded Asp on the T8 domain. The generation of a ΔsyrP strain of P. syringae pv. syringae B301D indicated that mature syringomycin is no longer produced but instead an immature form of the species, which lacks the β-OH moiety on Asp8, is generated. NMR and MS data indicate that this immature form of syringomycin also lacks other structural features of the mature species, but insufficient amounts of this compound were obtained for complete structural characterization.
At this juncture SyrP joins the SyrB1/B2 couple as a third enzyme in the Syr gene cluster acting in tandem to tailor Asp8 and Thr9 on the NRPS assembly line. SyrC has been assigned by us (13) as the 4-Cl-Thr aminoacyl shuttle enzyme. SyrD is annotated as an efflux pump. As there are no other genes in the cluster, the enzymatic dehydration of Thr7 to dehydrobutyrine7 during maturation of syringomycin has yet to be determined.
Finally, this work reveals that the P. syringae genome encodes two aspartyl hydroxylases, both nonheme mononuclear iron catalysts decarboxylating cosubstrate α-ketoglutarate during oxygen transfer. AspH and SyrP, respectively, are stereospecific for generating the erythro- or the threo-3-OH aspartyl hydroxylated products. While SyrP acts on residue eight in the assembly of the nonapeptidolactone syringomycin, the substrate and product from AspH action is not known. Unlike SyrP, AspH is not in an operon context that is revealing or predictive of function. Given that 3-OH-Asp residues are formed posttranslationally in proteins (16, 17), it may be that a comparative proteomics search of 3-OH-Asp-containing proteins in wild type and AspH knockout strains would produce mass spectrometric evidence of proteins generated by AspH action.
Acknowledgments
We thank Prof. Jon Takemoto (Utah State University, Logan, Utah) for the generous gift of a syringomycin authentic standard. This work was supported in part by National Institutes of Health Grant GM20011 (C.T.W.) and National Science Foundation Predoctoral Fellowship (G.M.S).
Funding Information: This work was supported by grants from the National Science Foundation and the National Institutes of Health
Abbreviations
- A
adenylation domain
- α-KG
alpha-ketoglutarate
- Ala
alanine
- Arg
arginine
- Asn
asparagine
- Asp
aspartate
- C
condensation domain
- CoA
Coenzyme A thioester
- Cys
cysteine
- Da
Dalton
- Dap
2,3-diaminopropionic acid
- DMSO
dimethyl sulfoxide
- DMF
dimethylformamide
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- Glu
glutamate
- HPLC
high pressure liquid chromatography
- IPTG
isopropyl thiogalactopyranoside
- LC-MS
liquid chromatography-mass spectrometry
- MS-MS
tandem mass spectrometry
- NMR
nuclear magnetic resonance
- Ser
serine
- SNAC
N-acetylcysteamine
- T
thiolation domain
- TCA
trichloroacetic acid
- TFA
trifluoroacetic acid
- TFE
trifluoroethanol
- TE
thioesterase domain
References
- 1.Bender CL, Palmer DA, Penaloza-Vazquez A, Rangaswamy V, Llrich M. Biosynthesis and regulation of coronatine, a non-host-specific phytotoxin produced by Pseudomonas syringae. Sub-cellular biochemistry. 1998;29:321–341. doi: 10.1007/978-1-4899-1707-2_10. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell RE, Bieleski RL. Involvement of Phaseolotoxin in Halo Blight of Beans: Transport and Conversion to Functional Toxin. Plant Physiol. 1977;60:723–729. doi: 10.1104/pp.60.5.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segre A, Bachmann RC, Ballio A, Bossa F, Grgurina I, Iacobellis NS, Marino G, Pucci P, Simmaco M, Takemoto JY. The structure of syringomycins A1, E and G. FEBS letters. 1989;255:27–31. doi: 10.1016/0014-5793(89)81054-3. [DOI] [PubMed] [Google Scholar]
- 4.Mathews DE, Durbin RD. Mechanistic aspects of tagetitoxin inhibition of RNA polymerase from Escherichia coli. Biochemistry. 1994;33:11987–11992. doi: 10.1021/bi00205a038. [DOI] [PubMed] [Google Scholar]
- 5.Thomas MD, Langston-Unkefer PJ, Uchytil TF, Durbin RD. Inhibition of Glutamine Synthetase from Pea by Tabtoxinine-beta-lactam. Plant Physiol. 1983;71:912–915. doi: 10.1104/pp.71.4.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scaloni A, Dalla Serra M, Amodeo P, Mannina L, Vitale RM, Segre AL, Cruciani O, Lodovichetti F, Greco ML, Fiore A, Gallo M, D'Ambrosio C, Coraiola M, Menestrina G, Graniti A, Fogliano V. Structure, conformation and biological activity of a novel lipodepsipeptide from Pseudomonas corrugata: cormycin A. The Biochemical journal. 2004;384:25–36. doi: 10.1042/BJ20040422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ballio A, Bossa F, Di Giorgio D, Ferranti P, Paci M, Pucci P, Scaloni A, Segre A, Strobel GA. Novel bioactive lipodepsipeptides from Pseudomonas syringae: the pseudomycins. FEBS letters. 1994;355:96–100. doi: 10.1016/0014-5793(94)01179-6. [DOI] [PubMed] [Google Scholar]
- 8.Fukuchi N, Isogai A, Nakayama J, Suzuki A. Structure of syringotoxin B, a phytotoxin produced by citrus isolates of Pseudomonas syringae pv. syringae. Agricultural and biological chemistry. 1990;54:3377–3379. [PubMed] [Google Scholar]
- 9.Sorensen KN, Kim KH, Takemoto JY. In vitro antifungal and fungicidal activities and erythrocyte toxicities of cyclic lipodepsinonapeptides produced by Pseudomonas syringae pv. syringae. Antimicrobial agents and chemotherapy. 1996;40:2710–2713. doi: 10.1128/aac.40.12.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scholz-Schroeder BK, SoμLe JD, Lu SE, Grgurina I, Gross DC. A physical map of the syringomycin and syringopeptin gene clusters localized to an approximately 145-kb DNA region of Pseudomonas syringae pv. syringae strain B301D. Mol Plant Microbe Interact. 2001;14:1426–1435. doi: 10.1094/MPMI.2001.14.12.1426. [DOI] [PubMed] [Google Scholar]
- 11.Vaillancourt FH, Yin J, Walsh CT. SyrB2 in syringomycin E biosynthesis is a nonheme FeII alpha-ketoglutarate- and O2-dependent halogenase. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10111–10116. doi: 10.1073/pnas.0504412102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blasiak LC, Vaillancourt FH, Walsh CT, Drennan CL. Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis. Nature. 2006;440:368–371. doi: 10.1038/nature04544. [DOI] [PubMed] [Google Scholar]
- 13.Singh GM, Vaillancourt FH, Yin J, Walsh CT. Characterization of SyrC, an aminoacyltransferase shuttling threonyl and chlorothreonyl residues in the syringomycin biosynthetic assembly line. Chemistry & biology. 2007;14:31–40. doi: 10.1016/j.chembiol.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Quigley NB, Mo YY, Gross DC. SyrD is required for syringomycin production by Pseudomonas syringae pathovar syringae and is related to a family of ATP-binding secretion proteins. Molecular microbiology. 1993;9:787–801. doi: 10.1111/j.1365-2958.1993.tb01738.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhang JH, Quigley NB, Gross DC. Analysis of the syrP gene, which regulates syringomycin synthesis by Pseudomonas syringae pv. syringae. Applied and environmental microbiology. 1997;63:2771–2778. doi: 10.1128/aem.63.7.2771-2778.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lancaster DE, McDonough MA, Schofield CJ. Factor inhibiting hypoxia-inducible factor (FIH) and other asparaginyl hydroxylases. Biochemical Society transactions. 2004;32:943–945. doi: 10.1042/BST0320943. [DOI] [PubMed] [Google Scholar]
- 17.Korioth F, Gieffers C, Frey J. Cloning and characterization of the human gene encoding aspartyl beta-hydroxylase. Gene. 1994;150:395–399. doi: 10.1016/0378-1119(94)90460-x. [DOI] [PubMed] [Google Scholar]
- 18.Chen H, Thomas MG, O'Connor SE, Hubbard BK, Burkart MD, Walsh CT. Aminoacyl-S-enzyme intermediates in beta-hydroxylations and alpha,beta-desaturations of amino acids in peptide antibiotics. Biochemistry. 2001;40:11651–11659. doi: 10.1021/bi0115434. [DOI] [PubMed] [Google Scholar]
- 19.Chen H, Walsh CT. Coumarin formation in novobiocin biosynthesis: beta-hydroxylation of the aminoacyl enzyme tyrosyl-S-NovH by a cytochrome P450 NovI. Chemistry & biology. 2001;8:301–312. doi: 10.1016/s1074-5521(01)00009-6. [DOI] [PubMed] [Google Scholar]
- 20.von Dohren H, Keller U, Vater J, Zocher R. Multifunctional Peptide Synthetases. Chem Rev. 1997;97:2675–2706. doi: 10.1021/cr9600262. [DOI] [PubMed] [Google Scholar]
- 21.Chen H, Hubbard BK, O'Connor SE, Walsh CT. Formation of beta-hydroxy histidine in the biosynthesis of nikkomycin antibiotics. Chemistry & biology. 2002;9:103–112. doi: 10.1016/s1074-5521(02)00090-x. [DOI] [PubMed] [Google Scholar]
- 22.Strieker M, Kopp F, Mahlert C, Essen LO, Marahiel MA. Mechanistic and structural basis of stereospecific Cbeta-hydroxylation in calcium-dependent antibiotic, a daptomycin-type lipopeptide. ACS chemical biology. 2007;2:187–196. doi: 10.1021/cb700012y. [DOI] [PubMed] [Google Scholar]
- 23.Feil H, Feil WS, Chain P, Larimer F, DiBartolo G, Copeland A, Lykidis A, Trong S, Nolan M, Goltsman E, Thiel J, Malfatti S, Loper JE, Lapidus A, Detter JC, Land M, Richardson PM, Kyrpides NC, Ivanova N, Lindow SE. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11064–11069. doi: 10.1073/pnas.0504930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehmann DE, Trauger JW, Stachelhaus T, Walsh CT. Aminoacyl-SNACs as small-molecule substrates for the condensation domains of nonribosomal peptide synthetases. Chemistry & biology. 2000;7:765–772. doi: 10.1016/s1074-5521(00)00022-3. [DOI] [PubMed] [Google Scholar]
- 25.Hoang T, Karkhoff-Schwizer R, Kutchma A, Schwizer H. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 26.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Lorenzo V, Fernandez S, Herrero M, Jakubzik U, Timmis KN. Engineering of alkyl- and haloaromatic-responsive gene expression with mini-transposons containing regulated promoters of biodegradative pathways of Pseudomonas. Gene. 1993;130:41–46. doi: 10.1016/0378-1119(93)90344-3. [DOI] [PubMed] [Google Scholar]
- 28.Nickoloff JA. Electroporation Protocols for Microorganisms. Vol. 47. Humans Press; Totowa, N. J.: 1995. [Google Scholar]
- 29.de Lorenzo V, Timmis KN. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods in Enzymology. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 30.Raetz CR. Regulated covalent modifications of lipid A. Journal of endotoxin research. 2001;7:73–78. [PubMed] [Google Scholar]
- 31.Eichhorn E, van der Ploeg JR, Kertesz MA, Leisinger T. Characterization of alpha-ketoglutarate-dependent taurine dioxygenase from Escherichia coli. The Journal of biological chemistry. 1997;272:23031–23036. doi: 10.1074/jbc.272.37.23031. [DOI] [PubMed] [Google Scholar]
- 32.Ryle MJ, Padmakumar R, Hausinger RP. Stopped-flow kinetic analysis of Escherichia coli taurine/alpha-ketoglutarate dioxygenase: interactions with alpha-ketoglutarate, taurine, and oxygen. Biochemistry. 1999;38:15278–15286. doi: 10.1021/bi9912746. [DOI] [PubMed] [Google Scholar]
- 33.Elkins JM, Ryle MJ, Clifton IJ, Dunning Hotopp JC, Lloyd JS, Burzlaff NI, Baldwin JE, Hausinger RP, Roach PL. X-ray crystal structure of Escherichia coli taurine/alpha-ketoglutarate dioxygenase complexed to ferrous iron and substrates. Biochemistry. 2002;41:5185–5192. doi: 10.1021/bi016014e. [DOI] [PubMed] [Google Scholar]
- 34.Zabinski R, Munck E, Champion PM, Wood JM. Kinetic and Mossbauer studies on the mechanism of protocatechuic acid 4,5-oxygenase. Biochemistry. 1972;11:3212–3219. doi: 10.1021/bi00767a012. [DOI] [PubMed] [Google Scholar]
- 35.Walsh CT, Chen H, Keating TA, Hubbard BK, Losey HC, Luo L, Marshall CG, Miller DA, Patel HM. Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines. Current opinion in chemical biology. 2001;5:525–534. doi: 10.1016/s1367-5931(00)00235-0. [DOI] [PubMed] [Google Scholar]
- 36.Quadri LE, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]


