Summary
The PATZ1 gene encodes a transcription factor that belongs to the BTB/POZ group of transcriptional regulators and has been implicated as a transcriptional repressor. We cloned cDNA from glioma cell lines and found they expressed transcript variant 2 of PATZ1. We designed a specific siRNA against PATZ1 and showed that this siRNA, but not a control randomized siRNA, reduced PATZ1 expression in glioma cells as determined by quantitative PCR. In a panel of human glioma cell lines incubated with proapoptotic FasL, those transfected with PATZ1 siRNA displayed reduced cell numbers by the MTT colorimetric assay, relative to those transfected with randomized siRNA. Further studies showed that in 10-08-MG, U-251MG, U-87MG, and T98G cells PATZ1 siRNA significantly increased apoptosis in response to incubation with soluble FasL, as shown by a morphologic acridine orange/ethidium bromide apoptotic assay. Using an apoptosis specific cDNA microarray we further demonstrated that down-regulation of PATZ1 by siRNA resulted in the upregulation of death receptor pro-apoptotic genes including caspase 8 and Death Receptor 5 (DR5) in U-373MG cells. Since DR5 is the receptor for TRAIL we tested whether PATZ1 downregulation also sensitized cells to TRAIL-induced apoptosis and found that PATZ1 siRNA, but not control siRNA, sensitized U-251MG and T98G glioma cells to TRAIL-induced apoptosis. Altogether, these data demonstrate a previously unknown role for the transcription factor PATZ1 in conferring resistance to apoptosis and indicate that modulation of PATZ1 expression may be a therapeutic strategy for gliomas.
Keywords: siRNA, astrocytomas, apoptosis, FasL, TRAIL
I. Introduction
In adults, glial neoplasms are the most common primary malignant tumors of the central nervous system. The prognosis of patients with gliomas is discouraging. The mean survival of glioblastoma patients with current therapies, which include surgery, chemotherapy and radiation, is 12-14 months (Stupp et al, 2005; Hutterer et al, 2008). They represent a significant cause of death in young adults (Greenlee et al, 2000; CBTRUS, 2005). Factors contributing to the inefficacy of existing treatments include the locations of primary tumors within the brain as relates to possible resection, the cytogenetic heterogeneity of the cell populations, and the infiltrative nature of the tumor cells. Hence, there is a necessity to explore and develop alternative therapies for this disease.
Apoptosis is an active process of cell death. With roles in both normal and pathological processes, it can be induced by a number of unrelated stimuli. Fas (CD95) and Fas ligand (FasL) are members of the TNF death receptor/ligand family and FasL binding to Fas-expressing cells will trigger their apoptosis. A significant proportion of cancer cell types including many gliomas express Fas and FasL on the cell surface and yet are resistant to apoptosis induced by this death receptor/ligand pair (Muschen et al, 2001; Riffkin et al, 2001; Barnhart et al, 2004). It is thought that expression of FasL is a mechanism by which cancer cells escape immune surveillance (Maecker et al, 2002). Phosphorylated PEA-15/PED and c-FLIP have been shown to contribute to Fas resistance in astrocytomas and gliomas (Yang et al, 2003). Fas associated phosphatase-1 (FAP-1) has been shown to be upregulated in gliomas compared to normal brain. Its role in Fas resistance was validated in tissue culture studies where siRNA against FAP-1 was able to sensitize glioma cells to Fas-induced apoptosis (Foehr et al, 2005). It is not surprising that glioma cells would ultimately provide themselves with multiple compensatory factors to resist Fas/FasL-induced apoptotic death (Gomez and Kruse, 2006). Characterized by genomic instability (Gomez et al, 2004), glioma cells, through epigenetic and genetic alterations, can readily select for numerous resistance mechanisms by possessing what has been termed a severe mutator phenotype (Merlo, 2003). Reversing this process to render cells sensitive to apoptotic stimuli can have important therapeutic implications.
As described in an earlier report, we screened a retroviral ribozyme library for proapoptotic genes that would activate FasL-induced apoptosis in tumor cells resistant to Fas-induced apoptosis (Tritz et al, 2005). One of the identified genes, PATZ1 (also known as ZNF278), is a member of the BTB/POZ (for Poxvirus and zinc finger) transcriptional regulatory protein family (Bardwell and Treisman, 1994; Zollman et al, 1994). Currently 4 different transcript variants with unknown functions have been detailed for the PATZ1 gene (Fedele et al, 2000; Mastrangelo et al, 2000; Mitchelmore et al, 2002). The PATZ1 gene has been associated with the repression of androgen receptor biosynthesis (Pero et al, 2002). Although PATZ1 has not previously been functionally associated with FasL-induced apoptosis or any other part of the known apoptotic pathways, it has recently been shown to be required for spermatogenesis in a mouse knockout model (Fedele et al, 2008). The affected tissue in the testes showed increased apoptosis, indicating that when PATZ1 is absent, cells may become sensitive to apoptotic stimuli.
In this report we describe that down regulation of the PATZ1 gene in glioma cells by a specific PATZ1 siRNA results in pleiotropic anti-tumor effects. Similar to the cycloheximide-mediated sensitization of glioma cells to FasL and TRAIL-induced apoptosis (Knight et al, 2001), we found that actinomycin D (actD) could sensitize glioma cells to FasL-induced apoptosis. We used actD sensitization to select a panel of Fas/FasL-resistant glioma cells for testing a siRNA targeting the PATZ1 gene. In these cell lines we found that the PATZ1 siRNA imparted cell sensitization to death receptor mediated apoptosis. Our data indicate that PATZ1 may be an effective therapeutic target in glioma cells.
II. Materials and methods
A. Cell lines and tissues
Human glioma cell lines 10-08-MG, U-138MG, U-251MG, U-373MG, U-87MG, and T98G were cultured in Dulbecco’s modified essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Two freshly-resected human primary brain specimens were obtained from adult seizure patients undergoing temporal lobectomies. The tissues, collected under IRB-approved guidelines, were without tumors or other major histopathologic abnormalities. The snap-frozen specimens were stored at -80°C until use.
B. siRNA and primers
The siRNA targeting the PATZ1 gene was designed using the siRNA target finder web site at AMBION.com. Potential target sites of the siRNA were subjected to a homology search with the megablast algorithm against the genome plus transcript database at NCBI to identify siRNA that avoids potential human off target transcripts. siRNA targeting PATZ1 (transcript variants 1, 2, and 3) and - as controls - randomized (rand) siRNA and siRNA targeting luciferase (luc) were synthesized on an ABI3900 DNA synthesizer using standard procedures, purified by AEX HPLC, and annealed in phosphate buffered saline (PBS, Ambion). siRNA sequences with chemical modifications follow: lower case letters indicate 2′-O-methyl modification at that position; S indicates a phosphorothioate linkage. dT indicates deoxythymidine. PATZ1 siRNA: sense 5′-GucuAuGGAAGAAAuAGuuusU-3′; antisense 5′-AACuAUUUCUUCcAuAGACUsU-3′; randomized control siRNA: sense 5′-GuAGuAGuAGuAGuAGuAAusU-3′; antisense 5′-UuACuACuACuACuACuACUsU-3′; luc siRNA: sense 5′-cuuAcGcuGAGuAcuucGAdTsdT-3′; antisense 5′-UCGAAGuACUcAGCGuAAGdTsdT-3′. Primers for quantitative real time-polymerase chain reaction (qPCR) were synthesized by IDT (Coralville, IA). PATZ1 primer set: Fwd 5′ACATCAGGATCCGATTGAGA3′; Rev 5′ATGCACCTTCTGGATGTGTT3′; a GAPDH primer set was obtained from RealTime Primers: Fwd 5′GAGTCAACGGATTTGGTCGT 3′; Rev 5′TTGATTTTGGAGGGATCTCG 3′.
C. Transfection of siRNA into glioma cells
Glioma cells were seeded at 2.5 × 104 cells/well in DMEM with 10% FBS in 24-well plates the day prior to transfection. Oligofectamine/siRNA complexes were formed in serum free DMEM by adding siRNA (50 nM final concentration) to 2 μl of Oligofectamine (Invitrogen, Carlsbad, CA) per well. Complexes were allowed to form at 25°C for 10 min and added to wells (50 μl per well) containing attached glioma cells in the 24-well plates and incubated for 16-18 hr at 37°C in 0.5 ml of DMEM. The transfection medium was then aspirated from the wells and replaced with 0.5 ml DMEM with 10% FBS.
D. Quantitative PCR for PATZ1 mRNA
RNA was isolated according to the manufacturer’s protocol from nontransfected glioma cells or siRNA transfected cells at 48 hr post-transfection using the Absolutely RNA kit (Stratagene, San Diego, CA). The RNA was reverse transcribed into cDNA using the iScript cDNA kit (BioRad, Hercules, CA). The PATZ1 mRNA level was quantitated by the SYBR green method on the iQ5 ribocycler (BioRad) in a 96-well format. Briefly, standard curves were obtained for PATZ1 and the internal control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) with template dilutions from 1:50 to 1:6750. Samples to be quantitated were run at a template dilution of 1:200 along with minus RT and minus template controls. Amplification was continued for 40 cycles as follows: 94°C-10s, 55°C-15s, 65°C-30s.
E. MTT assays to determine reductions in viable cell number
Initial experiments were performed with nontransfected glioma cells to determine their sensitivity to low concentrations of actD (Alexis Biochemical, Lausen, Switzerland), to FasL (SuperFasLigand, Alexis), or both. Glioma cells (2.5 × 104 cells/well) were seeded into DMEM with 10% FBS in 24-well plates the day prior to the assay. Culture medium with actD was added to the cells at a final concentration of 0.01 to 0.1 μg/ml and incubated for 24 hrs at 37°C. FasL (10 to 150 ng/ml) was then added to the medium and incubated overnight before determining viable cell numbers by MTT assay.
Experiments were then similarly conducted with nontransfected or siRNA transfected glioma cells at 48 hr post-transfection when incubated with FasL (10 to 100 ng/ml) or TRAIL (200 to 300 ng/ml, Cell Sciences Inc., Sharon, MA).
Cells were incubated at 37°C for 18 hr, then medium was aspirated and replaced with serum-free DMEM. The yellow tetrazole, 3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide (MTT, EMD Chemicals, Inc., Gibbstown, NJ) was added at a final concentration of 1 mg/ml and incubated at 37°C for 30 min. Blue formazon, solubilized by adding 100% DMSO to the wells, produced by viable cells was quantified from spectrophotometer readings at 570 nm after subtracting the background reading at 650 nm. The data were presented as the percentage reduction in viable cell numbers of the treated cells from those in untreated control wells, calculated as 100 - [(A570-A650 sample/A570-A650NT) × 100].
F. Morphologic identification of apoptotic cells by fluorescence microscopy using acridine orange/ethidium bromide staining
Cells were seeded and transfected as described for the MTT assay. FasL was added 48 hr post-transfection and the cells incubated for 18 hr. Acridine orange/ethidium bromide (32 μl, Sigma) at 100 μg/ml in PBS were added to cells for 15 min at 37°C. The plates were then centrifuged for 5 min. Staining medium was aspirated and replaced with 0.5 ml of PBS. Cells were photographed on an Olympus CKX41 fluorescent microscope. Two high field magnifications were photographed for each treatment. The percentages of apoptotic cells were determined by counting the apoptotic cells from a total of 400 cells (i.e., 200 cells counted in each of two high fields).
G. Apoptosis-pathway specific gene expression microarray analysis
The gene expression profiles of siRNA transfected and nontransfected glioma cells were analyzed using the GEArray Q Series Human Apoptosis Gene Array (SuperArray Bioscience Corp., Frederick, MD). The array contains 96 apoptosis-related gene specific cDNA fragments and 4 housekeeping genes for normalization of the data. Total RNA (5 mg) was reverse transcribed and the resulting cDNA probes amplified (GeneAmp PCR System 9700, Applied Biosystems, Foster City, CA) according to the manufacturer. Prior to hybridization, biotinlabeling efficiency of the probes was determined by manufacturer instructions. Hybridization of the probes to the membranes was detected by chemiluminescence and the hybridization signals captured on X-ray film. Images were scanned and converted to grayscale TIF files. The hybridization signals were converted into numerical data using the ScanAlyze software program (http://rana.lbl.gov/EisenSoftware.htm). The raw intensity signals were corrected for background by subtracting the minimum value of the data set and normalized to the housekeeping gene RPL13A (ribosomal protein L13A). Gene expression changes between the treated or untreated gliomas were determined with the GEArray Analyzer software program.
H. Statistical analysis
The MTT assay data and the morphologic apoptotic data were analyzed by a 2 way-ANOVA using the Prism statistics package (Graph Pad Software Inc., La Jolla, CA). Bonferroni post-hoc tests were performed to compare treatment groups and to determine statistical significance.
III. Results
A. Criteria used for selection of human glioma cell panel for siRNA experiments
Human glioma cells had to meet three defined criteria to be included on the panel to be screened for activation of Fas/FasL-induced apoptosis when transfected with PATZ1 siRNA: i) the cells had to be resistant to apoptosis induced by FasL, ii) they had to exhibit increased FasL-induced apoptosis in the presence of actD, and iii) they had to express PATZ1 mRNA.
B. Selection of FasL-resistant glioma cell lines
Initial experiments involved screening human glioma cell lines that met criteria i and ii above. They were tested for their sensitivity to FasL (10-150 ng/ml), to low concentrations of actD (0.02-0.1 μg/ml), or both. Data obtained with the U-87MG cell line, representative of that obtained with the 10-08-MG, U-251MG, U-373MG, and T98G cell lines, showed that they were relatively insensitive to the anti-tumor effects of low concentration actD alone, FasL alone, but were sensitive when exposed to both (Figure 1A). Relative to untreated cells, each of the 5 cell lines displayed small reductions in viable cell numbers in vitro when FasL was added to the supernatants, but the percentages increased (3.5 to 48 fold) upon the simultaneous addition of low concentrations of actD.
Figure 1.
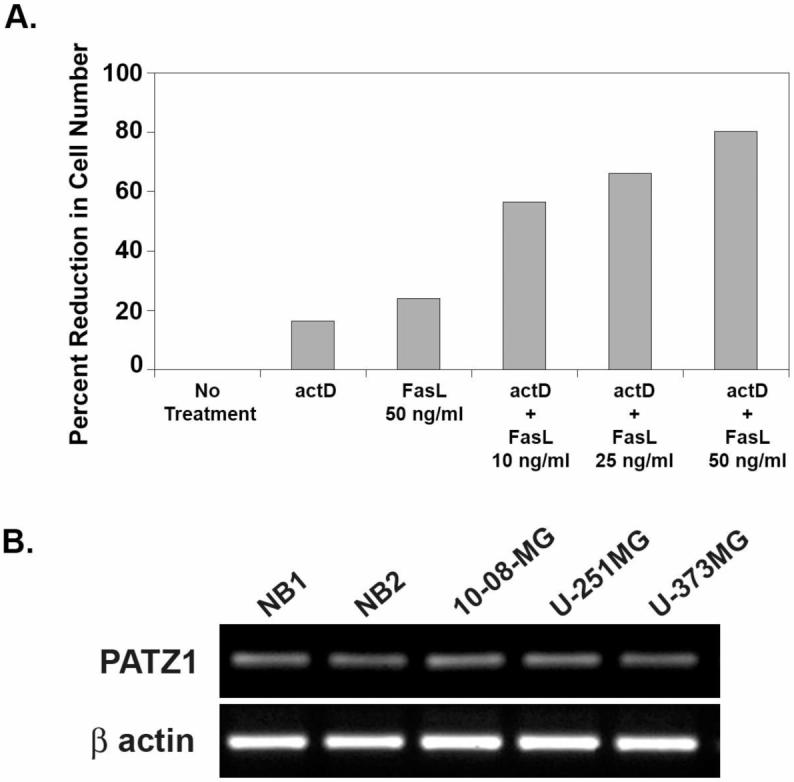
Screening data collected with glioma cells to demonstrate their resistance to FasL-induced apoptosis and the presence of PATZ1 mRNA. (A) Relative to those given no treatment, the percentage reduction in viable cell number by MTT assay is shown for U-87MG glioma cells when treated with actinomycin D (actD), FasL, or both. The percentage reductions in U-87MG viable cell number are given for treatments with actD (0.02 μg/ml), FasL (50 ng/ml), or both with FasL titrated from 10 to 50 ng/ml. (B) P ATZ1 mRNA levels are shown for extracts from two different normal brain (NB) specimens (lanes 1, 2) and three gliomas (10-08-MG, U-251MG, U-373MG in lanes 3-5, respectively). The β actin loading controls are shown at the bottom.
C. Expression of PATZ1 in normal brain and glioma cells
Indicating that glioma cells meet criterion iii above, Figure 1B shows PATZ1 mRNA extracted from three different glioma cell lines, 10-08-MG, U-251MG, and U-373MG, and from two different normal brain (NB) cell specimens. By densitometry, with NB normalized to actin at 1.0, the values for glioma cell lines ranged from 0.949 to 1.05, indicating that PATZ1 mRNA is not overexpressed in gliomas relative to normal brain. With cDNA cloned from two glioma cell lines, we found they expressed only transcript variant 2 of PATZ1 as determined by BLAST search.
D. PATZ1 siRNA decreases PATZ1 expression in glioma cells
Next we generated siRNA to PATZ1. To validate the siRNA we determined its effect on PATZ1 mRNA expression. When glioma cell lines were transfected with PATZ1 siRNA, expression of PATZ1 mRNA was reduced approximately 2-5 fold compared to cells transfected with control siRNA or to nontransfected cells (Figure 2).
Figure 2.
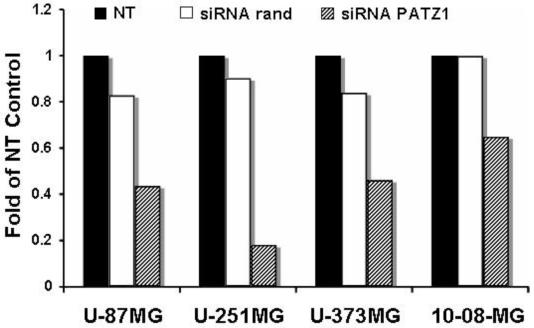
RT-PCR analysis of PATZ1 gene knockdown in a glioma cells after transfection with siRNA. Cells were either not transfected (NT) or transfected with siRNA to either a randomized sequence (rand) or to PATZ1. Total RNA was isolated from the cells 48 hr post transfection and converted to cDNA. Real time PCR was performed using gene specific primers and SYBR green detection on an iQ5 ribocycler (BioRad). All data were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) prior to comparison for fold changes in mRNA levels determined from numerical readings taken from the ribocycler.
E. PATZ1 siRNA increases sensitivity to FasL-mediated anti-tumor effects in glioma cells
The concentration of FasL for each particular glioma cell line that resulted in ≥ 30% killing in the presence of actD (listed in the legend to Figure 3) was chosen for use in MTT assays testing the effects of downregulation of PATZ1 with siRNA. Of the 5 glioma cell lines, the 10-08-MG and U-373MG cell lines were the most resistant to the effects of FasL alone (Figure 3). The glioma cell lines transfected with PATZ1 siRNA and treated with FasL showed significant increases in the percentage reductions in cell numbers when compared to those of the most relevant control, cells transfected with randomized control siRNA and treated with FasL. The approximate 20% to 40% reductions in cell number were significantly different (*p<0.05, **p<0.01) as indicated in Figure 3.
Figure 3.
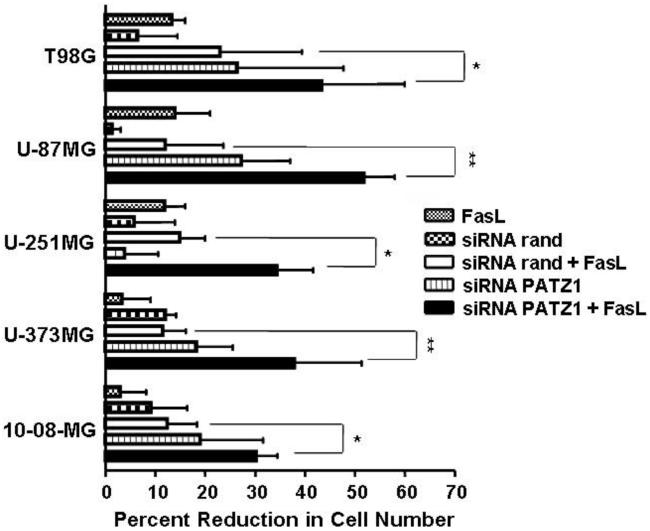
The reductions in viable cell number by MTT assay following siRNA-mediated downregulation of PATZ1 in human glioma cell lines and exposure to FasL. Cells were seeded at 2.5 × 104 cells/well in a 24-well plate the day prior to transfection. Transfections were performed with Oligofectamine with siRNA at a concentration of 50 nM. At 48 hr post-transfection, FasL (at the concentrations listed: T98G & U-87MG, 10 ng/ml; U-251MG & U-373MG, 50 ng/ml; and 10-08-MG, 150 ng/ml) was added and incubated overnight at 37°C. The percentage reductions in viable cell number were estimated by the MTT colorimetric assay. The wells contained either 1) nontransfected cells and FasL, 2) cells transfected with a randomized (rand) siRNA sequence that were without or with FasL, or 3) cells transfected with siRNA against PATZ1 (PATZ1) that were without or with FasL. Experiments were performed in duplicate and repeated at least twice. Representative data are shown giving reductions in cell number with standard error of the mean. Data were analyzed by two-way ANOVA. Statistical significance between the treatment groups, determined by Bonferroni post-hoc tests, at * p≤0.05 and **p≤0.01 are shown.
Glioma cells were then assessed for apoptosis by the acridine orange/ethidium bromide morphologic assay. Figure 4 shows fluorescent photomicrographs demonstrating low basal levels of apoptotic cells within the nontransfected U-251MG cells in the presence or absence of FasL, and in U-251MG cells transfected with the control luc siRNA in the presence or absence of FasL. Similarly U-251MG cells transfected with PATZ1 siRNA alone show very few cells with condensed or disintegrated nuclei. In contrast, PATZ1 siRNA transfected U-251MG cells exposed to FasL demonstrate many apoptotic cells. An even larger number of apoptotic cells is visible in U-87MG cells transfected with PATZ1 siRNA exposed to FasL, whereas U-373MG glioma cells similarly transfected with PATZ1 siRNA and exposed to FasL are seemingly resistant to apoptosis.
Figure 4.
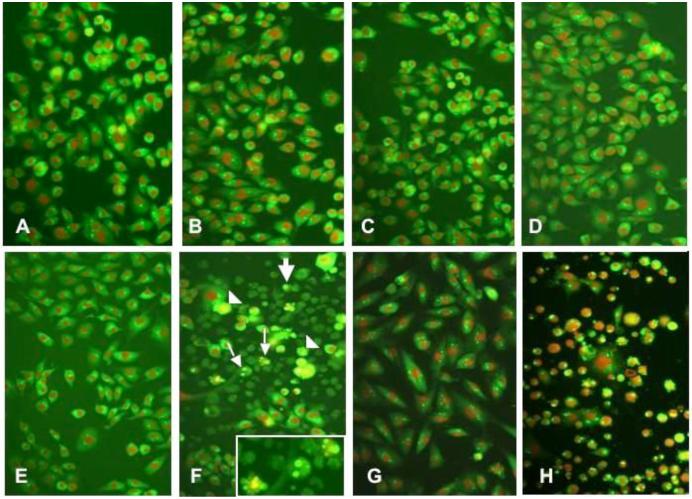
Glioma cells shown by fluorescence microscopy after undergoing a morphologic assay for apoptosis by the acrid ine orange/ethidium bromide staining. The cells shown were incubated under the following control or experimental conditions: (A) nontransfected U-251MG cells, (B) nontransfected U-251MG cells + FasL (C) U-251MG cells transfected with siRNA luc, (D) U-251MG cells transfected with siRNA luc + FasL, (E) U-251MG cells transfected with siRNA PATZ1, (F) U-251MG cells transfected with PATZ1 siRNA + FasL, (G) U-373MG cells transfected with PATZ1 siRNA + FasL, and (H) U-87MG cells transfected with PATZ1 siRNA + FasL. In panels F and H, a large number of glioma cells are seen with morphologic changes indicative of a poptosis that include cells with nuclear fragmentation (small arrows), membrane blebbing (arrowheads), and small cells with condensed nuclei (large arrows). In F, the inset shows three cells at a higher magnification where nuclear fragmentation is occurring.
By morphologic assessments, the percentages of apoptotic cells were determined for the entire panel of glioma cells undergoing the acridine orange assay (Figure 5). The percentages were determined by counting the number of condensed or disintegrated nuclei for 200 cells in each of two different high power fields. With exception of the U-373MG cells, significant increases in apoptosis (*p<0.001) were obtained with glioma cells exposed to FasL, which were transfected with PATZ1 siRNA compared to those transfected with control luc siRNA. The apoptotic percentages for 4 of the 5 glioma cell lines were similar and roughly corresponded to about two thirds of the effect seen in the MTT assays (Figure 3). In contrast to 10-08-MG, U-251MG, U-87MG, and T98G, the U-373MG cell line appears resistant to FasL-induced apoptosis after transfection with the PATZ1 siRNA, even though it showed a significant reduction in cell number by MTT assay. This result suggests that the downregulation of PATZ1 and treatment with FasL in the U-373MG cell line is anti-proliferative. To determine whether downregulation of PATZ1 had any pro-apoptotic effects in U-373MG cells, we determined the expression levels of genes involved with death receptor-mediated apoptosis.
Figure 5.
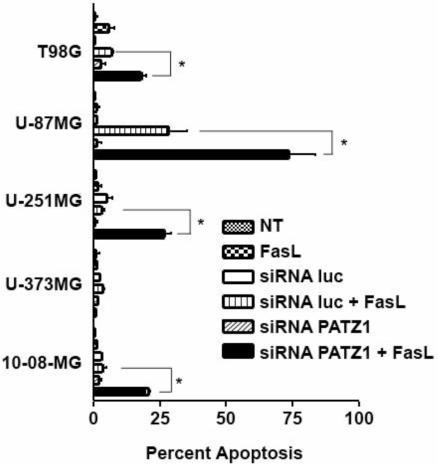
Percentages of apoptotic glioma cells from nontransfected (NT) or siRNA-transfected glioma cells (luc or PATZ1), treated or untreated with FasL. With the exception of U-373MG cells, the glioma cells transfected with siRNA against PATZ1 and incubated with FasL generally demonstrate enhanced apoptosis; U-87MG showed the most remarkable effect. Apoptotic cells (bright green or orange) visualized by fluorescence microscopy were counted from a total of 400 cells. The percentages of apoptotic cells calculated are shown with error bars indicating the standard deviations from duplicate experiments. Data were analyzed by a 2 way-ANOVA. Bonferroni post-hoc tests to compare treatment groups demonstrated statistical significance at *p≤0.001.
F. Downregulation of PATZ1 increases the expression of several genes associated with the extrinsic apoptosis pathway
To test whether PATZ1 gene downregulation could affect apoptosis pathways other than FasL-induced apoptosis we used an apoptosis pathway-specific cDNA microarray and probed with total RNA isolated from U-373MG cells transfected with either siRNA against PATZ1 or to the randomized control sequence (Figure 6). The microarray data indicate six genes normally linked to the death receptor apoptosis pathway were up-regulated in the PATZ1 siRNA-transfected cells compared to the control. The six genes include RIP, DAP-kinase 2, FADD, Fas receptor, caspase 8, and of particular interest relative to another report (Knight et al, 2001), DR5, which is the TRAIL receptor. None of the members of the Bcl-2 or inhibitor of apoptosis (IAP) gene families had changes in their expression levels.
Figure 6.
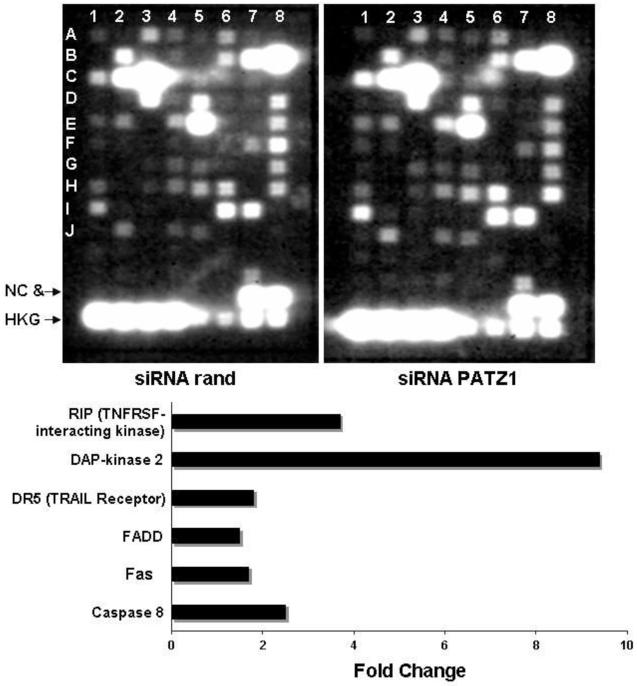
Apoptotic-pathway specific microarrays of U-373MG gliomas cells transfected with a randomized siRNA or siRNA to PATZ1. The cells transfected with siRNA to PATZ1 generally demonstrate upregulation of proapoptotic genes. The location by column, row are: caspase 8 (1,E); Fas (2,J); FADD (4,F); TRAIL receptor (1,I); Dap-kinase 2 (1,F); RIP (3,H).
G. Downregulation of PATZ1 decreases viable glioma cell numbers and/or increases apoptosis by TRAIL
We next tested whether exposure to TRAIL caused a reduction in viable glioma cell number when PATZ1 was downregulated in them by siRNA-transfection. The gliomas from the cell panel were nontransfected, or transfected with the siRNA to PATZ1 or with the siRNA to the control luc sequence. TRAIL was then added or not added before assay by MTT. The concentration of TRAIL for each particular glioma cell line (listed in the legend to Figure 7) that resulted in ≥ 30% killing in the presence of actD was chosen for testing the sensitivity to TRAIL exposure after downregulation of PATZ1 with siRNA. In these experiments, only two of the five PATZ1 siRNA transfected glioma cell lines, T98G and U-251MG, tended to have reduced viable cell numbers when TRAIL was added to them, compared to the luc siRNA transfected cell counterparts (Figure 7). Interestingly, the PATZ1 siRNA transfected U-87MG, U-373MG and 10-08-MG cell lines alone showed similar viable cell reductions to those also exposed to TRAIL.
Figure 7.
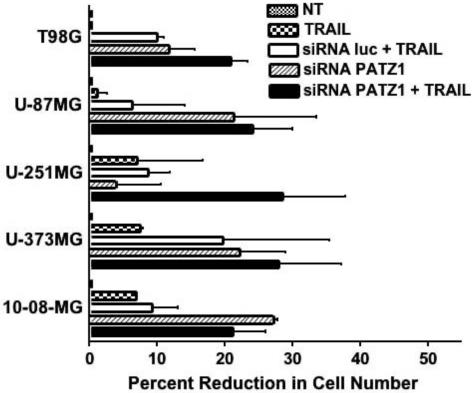
The reductions in viable glioma cell numbers by MTT assay following downregulation of PATZ1 in human glioma cell lines and exposure to TRAIL. Cells were either nontransfected (NT) or transfected with a luciferase (luc) control siRNA or siRNA targeting the PATZ1 gene. At 48 hr post transfection TRAIL was added at 200 ng/ml for T98G and 300 ng/ml for U-373MG, U-251MG, U-87MG and 10-08-MG. The percentage reductions in cell number are given plus or minus the standard error of the mean. Data were analyzed by a 2 way-ANOVA. Although trends in reduced cell numbers were noted, statistical significance was not reached by Bonferroni post-hoc tests between the groups transfected with siRNA to PATZ1 and luc that were treated with TRAIL.
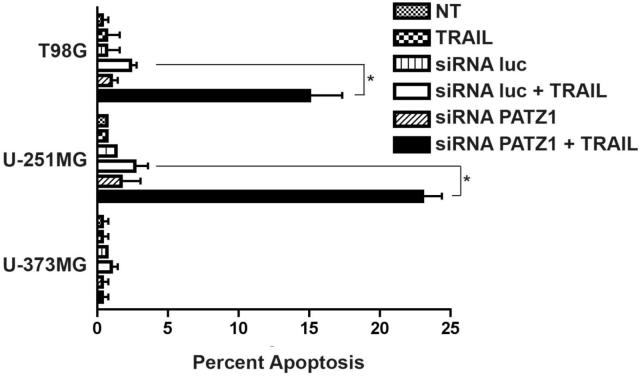
The acridine orange/ethidium bromide morphologic apoptosis assay similarly confirmed the findings of the MTT assay with three of the cell lines tested (Figure 8). As expected the U-373MG cell line remained resistant to TRAIL induced apoptosis. Consistent with data from the MTT assay, U-251MG and T98G glioma cells were sensitive to TRAIL induced apoptosis when the PATZ1 gene was downregulated. Both cell lines showed statistically significant increases in apoptosis (*p<0.001) when TRAIL was added to PATZ1 siRNA transfected cells compared to those given TRAIL that were transfected with luc siRNA.
Figure 8.
Percentages of apoptotic glioma cells from nontransfected (NT) or siRNA-transfected glioma cells (luc or PATZ1), treated or untreated with TRAIL as determined from apoptotic cells counted by fluorescence microscopy after staining with acridine orange/ethidiium bromide. Nontransfected T98G, U-251MG and U-373MG glioma cells were or were not exposed to TRAIL, or the cells were transfected with a luciferase (luc) control siRNA or siRNA targeting the PATZ1 gene that were or were not exposed to TRAIL. At 48 hr post transfection TRAIL ligand was added at 200 ng/ml for T98G or 300 ng/ml for U-373MG and U-251MG. The percentage of apoptotic cells are given plus or minus the standard error of the mean. Statistical differences at *p≤0.001 were obtained between TRAIL-treated siRNA transfected PATZ1 cells vs the TRAIL-treated siRNA luc transfected cells where shown, after the data were analyzed by a 2 way-ANOVA and Bonferroni post-hoc tests.
Altogether, these data indicate that down regulation of PATZ1 in glioma cell lines decreases their resistance to both FasL- and TRAIL-induced apoptosis.
IV. Discussion
Previous studies indicate that glioma cell lines vary in their resistance to apoptosis induced by FasL or TRAIL, especially in the presence of other drugs or inhibitors to the PI3K pathway (Knight et al, 2001; Opel et al, 2008). Our data confirm those findings. We showed first that Fas/FasL-resistant glioma cells can be activated to undergo apoptosis after cotreatment with actD and FasL. This finding was critical to our study because it meant we could identify a panel of glioma cells that were resistant to apoptosis upon addition of FasL, but still had the ability to undergo apoptosis after Fas ligation in the presence of low concentration actD. It also indicated that the cells were likely resistant to apoptosis due to the synthesis of inhibitors. Such inhibitors could be targeted.
We identified a novel target gene, that when downregulated, sensitized Fas/FasL-resistant glioma cells to killing by FasL. This gene is the BOB/POZ AT hook zinc finger protein 1 (PATZ1) and was identified after screening a ribozyme library for targets that when downregulated converted Fas-resistant to Fas-sensitive cells (Tritz et al, 2005). All of the glioma cell lines we tested were found to express transcript variant 2 of the PATZ1 gene (NM_032050), but the gene was not over expressed compared to normal human brain tissue (Figure 1B). Studies for protein expression were not possible because antibodies to human PATZ1 are not available. Although it is possible that polyclonal antibodies to mouse zinc finger protein (PATZ/ZNF278) might conceivably cross-react with the human protein, we did not test this.
The downregulation of PATZ1 by siRNA exhibited anti-tumor effects on a panel of glioma cell lines. Cells transfected with PATZ1 siRNA and treated with FasL showed a substantial reduction in viable cell number by MTT assay. The activation of apoptosis by FasL was observed in cells transfected with PATZ1 siRNA for 4 out of 5 cell lines. The random siRNA transfected cells with or without FasL showed a much lower level of apoptosis by the acridine orange morphologic assay compared to the reduction in numbers seen in the MTT assay. This could be due to background in the MTT assay caused by an acute transfection toxicity that is not detected in the subsequent apoptosis assay. We further used luciferase siRNA as a negative control for the morphology and TRAIL studies so negativity was not exclusive to one siRNA control and because the luciferase siRNA could be used in future animal studies. We found that the background activity was comparable between the randomized and the luciferase siRNA controls and, therefore, did not alter the results significantly.
The apoptotic index calculated for four of the cell lines could account for approximately 65% of the effect detected in the MTT assay. Interestingly, the PATZ1 transfected cells alone show a greater effect in the MTT assay than the control + FasL and yet the apoptotic index is very low. This indicates that the downregulation of PATZ1 may also have an anti-proliferative component. This was further reinforced by the results obtained with the U-373MG cell line. The U-373MG cell line showed a significant reduction in cell number and viability in the MTT assay but showed little to no apoptosis in the acridine orange/ethidium bromide morphology assay. It is possible that the U-373MG cell number is being reduced by a different mechanism for cell death. Triggering the Fas receptor in Fas-resistant cells has also been shown to cause cell death by necrosis (Hetz et al, 2002). However, we did not see any increase in cell death by trypan blue dye staining (data not shown). It has also recently been shown that engaging the Fas receptor in cells that are resistant to Fas-induced apoptosis can enhance cellular proliferation (Barnhart et al, 2004; Mitsiades et al, 2006) and perhaps activate invasion in vivo (Kleber et al, 2008). To our knowledge our data are the first observation that triggering the Fas receptor can potentially contribute to an anti-proliferative response.
Our preliminary examination of an apoptosis gene specific cDNA microarray suggested that downregulating the PATZ1 gene can cause the upregulation of genes involved in death receptor mediated apoptosis. We were intrigued by the potential upregulation of the TRAIL receptor and the possibility that the downregulation of PATZ1 could also sensitize cells to TRAIL-induced apoptosis. The results from the TRAIL experiments were encouraging and consistent with what we obtained for FasL. They indicate that downregulating the PATZ1 gene can generally sensitize cells to death receptor mediated apoptosis.
Our results provide in vitro support that a siRNA directed against a novel target gene, PATZ1, could be a therapeutic agent for treating gliomas. Downregulation of this gene in a panel of glioma cells exhibited anti-tumor cellular effects including the activation of apoptosis by FasL or by TRAIL as well as an anti-proliferative response. It is possible that PATZ1 siRNA treatment causes inactivation of an inhibitory protein(s), therefore allowing the cells to die by Fas/FasL-induced apoptosis. This same concept may hold true for TRAIL as well. Deciphering how the PATZ1 gene functions in a pathway to confer Fas or TRAIL resistance to glioma cells will increase our understanding of how tumor cells become resistant to death receptor triggered apoptosis. The continued evaluation of this pathway, as well as other novel genes for activating death receptor mediated apoptosis (Xia et al, 2007) could ultimately provide attractive alternative adjuvant therapies to the current small molecule armamentarium for gliomas.
Acknowledgements
Supported in part by NIH NS056300 to CAK and by the R. Herbert and Alma S. Manweiler Memorial Research Fund. Dr. Michelle Hickey is supported by the Joan S. Holmes Fellowship at the Sidney Kimmel Cancer Center. We thank Dr. L.E. Gerschenson for carefully reading the manuscript.
Abbreviations
- MTT
3-(4,5-dimethylthiazole -2-yl)-2,5-diphenyltetrazolium bromide
- actD
actinomycin D
- PATZ1
BOB/POZ AT hook zinc finger protein 1, aka ZNF278
- BOB/POZ
Bric-a brac/Tramtrack/Broad/Pox virus zinc finger
- Fas
Cluster of Differentiation 95, aka CD95
- cDNA
complementary deoxyribonucleic acid
- DAP-kinase 2
Death associated protein kinase 2
- DR5
death receptor 5, aka TRAIL receptor
- dT
deoxythymidine
- DMSO
dimethyl sulfoxide
- DMEM
Dulbecco’s modified essential medium
- FADD
Fas associated death domain protein
- FAP-1
Fas-associated phosphatase 1
- FasL
Fas Ligand
- FBS
fetal bovine serum
- FLIP
FLICE inhibitory protein
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- luc
luciferase
- NT
non-transfected
- NB
normal brain
- PEA-15/PED
phosphoprotein enriched in astrocytes 15
- PCR
polymerase chain reaction
- rand
randomized
- RNA
ribonucleic acid
- RIP
Receptor interacting protein
- siRNA
small interfering RNA
- TRAIL
TNF related apoptosis inducing ligand
References
- Bardwell VJ, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–77. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- Barnhart BC, Legembre P, Pietras E, Bubici C, Franzoso G, Peter ME. CD95 ligand induces motility and invasiveness of apoptosis-resistant tumor cells. EMBO J. 2004;23:3175–85. doi: 10.1038/sj.emboj.7600325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CBTRUS . Statistical Report: Primary Brain Tumors in the United States. Central Brain Tumor Registry of the United States, 2005; 1998-2002. [Google Scholar]
- Fedele M, Benvenuto G, Pero R, Majello B, Battista S, Lembo F, Vollono E, Day PM, Santoro M, Lania L, Bruni CB, Fusco A, Chiariotti L. A novel member of the BTB/POZ family, PATZ, associates with the RNF4 RING finger protein and acts as a transcriptional repressor. J Biol Chem. 2000;275:7894–901. doi: 10.1074/jbc.275.11.7894. [DOI] [PubMed] [Google Scholar]
- Fedele M, Franco R, Salvatore G, Paronetto M, Barbagallo F, Pero R, Chiariotti L, Sette C, Tramontano D, Chieffi G, Fusco A, Chieffi P. PATZ1 gene has a critical role in the spermatogenesis and testicular tumours. J Pathol. 2008;2154:39–47. doi: 10.1002/path.2323. [DOI] [PubMed] [Google Scholar]
- Foehr ED, Lorente G, Vincent V, Nikolich K, Urfer R. FAS associated phosphatase (FAP-1) blocks apoptosis of astrocytomas through dephosphorylation of FAS. J Neurooncol. 2005;74:241–8. doi: 10.1007/s11060-004-7202-x. [DOI] [PubMed] [Google Scholar]
- Gomez GG, Kruse CA. Mechanisms of malignant glioma immune resistance and sources of immunosuppression. Gene Ther Mol Biol. 2006;10:133–46. [PMC free article] [PubMed] [Google Scholar]
- Gomez GG, Read SB, Gerschenson LE, Santoli D, Zweifach A, Kruse CA. Interactions of the allogeneic effector leukemic T cell line, TALL-104, with human malignant brain tumors. Neuro-Oncol. 2004;6:83–95. doi: 10.1215/S1152851703000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenlee R, Murray T, Bolden S, Wingo P. Cancer statistics, 2000. CA: Cancer J Clin. 2000;50:7–33. doi: 10.3322/canjclin.50.1.7. [DOI] [PubMed] [Google Scholar]
- Hetz CA, Hunn M, Rojas P, Torres V, Leyton L, Quest AF. Caspase-dependent initiation of apoptosis and necrosis by the Fas receptor in lymphoid cells: onset of necrosis is associated with delayed ceramide increase. J Cell Sci. 2002;115:4671–83. doi: 10.1242/jcs.00153. [DOI] [PubMed] [Google Scholar]
- Hutterer M, Knyazev P, Abate A, Reschke M, Maier H, Stefanova N, Knyazeva T, Barbieri V, Reindl M, Muigg A, Kostron H, Stockhammer G, Ullrich A. Axl and growth arrest-specific gene 6 are frequently overexpressed in human gliomas and predict poor prognosis in patients with glioblastoma multiforme. Clin Cancer Res. 2008;14:130–8. doi: 10.1158/1078-0432.CCR-07-0862. [DOI] [PubMed] [Google Scholar]
- Kleber S, Sancho-Martinez I, Wiestler B, Beisel A, Gieffers C, Hill O, Thiemann M, Mueller W, Sykora J, Kuhn A, Schreglmann N, Letellier E, Zuliani C, Klussmann S, Teodorczyk M, Grone HJ, Ganten TM, Sultmann H, Tuttenberg J, von Deimling A, Regnier-Vigouroux A, Herold-Mende C, Martin-Villalba A. Yes and PI3K bind CD95 to signal invasion of glioblastoma. Cancer Cell. 2008;13:235–48. doi: 10.1016/j.ccr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Knight MJ, Riffkin CD, Muscat AM, Ashley DM, Hawkins CJ. Analysis of FasL and TRAIL induced apoptosis pathways in glioma cells. Oncogene. 2001;20:5789–98. doi: 10.1038/sj.onc.1204810. [DOI] [PubMed] [Google Scholar]
- Maecker HL, Yun Z, Maecker HT, Giaccia AJ. Epigenetic changes in tumor Fas levels determine immune escape and response to therapy. Cancer Cell. 2002;2:139–48. doi: 10.1016/s1535-6108(02)00095-8. [DOI] [PubMed] [Google Scholar]
- Mastrangelo T, Modena P, Tornielli S, Bullrich F, Testi MA, Mezzelani A, Radice P, Azzarelli A, Pilotti S, Croce CM, Pierotti MA, Sozzi G. A novel zinc finger gene is fused to EWS in small round cell tumor. Oncogene. 2000;19:3799–804. doi: 10.1038/sj.onc.1203762. [DOI] [PubMed] [Google Scholar]
- Merlo A. Genes and pathways driving glioblastomas in humans and murine disease models. Neurosurg Rev. 2003;26:145–58. doi: 10.1007/s10143-003-0267-8. [DOI] [PubMed] [Google Scholar]
- Mitchelmore C, Kjaerulff KM, Pedersen HC, Nielsen JV, Rasmussen TE, Fisker MF, Finsen B, Pedersen KM, Jensen NA. Characterization of two novel nuclear BTB/POZ domain zinc finger isoforms. Association with differentiation of hippocampal neurons, cerebellar granule cells, and macroglia. J Biol Chem. 2002;277:7598–609. doi: 10.1074/jbc.M110023200. [DOI] [PubMed] [Google Scholar]
- Mitsiades CS, Poulaki V, Fanourakis G, Sozopoulos E, McMillin D, Wen Z, Voutsinas G, Tseleni-Balafouta S, Mitsiades N. Fas signaling in thyroid carcinomas is diverted from apoptosis to proliferation. Clin Cancer Res. 2006;12:3705–12. doi: 10.1158/1078-0432.CCR-05-2493. [DOI] [PubMed] [Google Scholar]
- Muschen M, Re D, Betz B, Moers C, Wolf J, Niederacher D, Diehl V, Beckmann MW. Resistance to CD95-mediated apoptosis in breast cancer is not due to somatic mutation of the CD95 gene. Int J Cancer. 2001;92:309–10. doi: 10.1002/1097-0215(200102)9999:9999<::aid-ijc1188>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Opel D, Westhoff MA, Bender A, Braun V, Debatin KM, Fulda S. Phosphatidylinositol 3-kinase inhibition broadly sensitizes glioblastoma cells to death receptor- and drug-induced apoptosis. Cancer Res. 2008;68:6271–80. doi: 10.1158/0008-5472.CAN-07-6769. [DOI] [PubMed] [Google Scholar]
- Pero R, Lembo F, Palmieri EA, Vitiello C, Fedele M, Fusco A, Bruni CB, Chiariotti L. PATZ attenuates the RNF4-mediated enhancement of androgen receptor-dependent transcription. J Biol Chem. 2002;277:3280–5. doi: 10.1074/jbc.M109491200. [DOI] [PubMed] [Google Scholar]
- Riffkin CD, Gray AZ, Hawkins CJ, Chow CW, Ashley DM. Ex vivo pediatric brain tumors express Fas (CD95) and FasL (CD95L) and are resistant to apoptosis induction. Neuro-oncol. 2001;3:229–40. doi: 10.1093/neuonc/3.4.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Tritz R, Habita C, Robbins JM, Gomez GG, Kruse CA. Catalytic nucleic acid enzymes for the study and development of therapies in the central nervous system: Review Article. Gene Ther Mol Biol. 2005;9A:89–106. [PMC free article] [PubMed] [Google Scholar]
- Xia S, Li Y, Rosen EM, Laterra J. Ribotoxic stress sensitizes glioblastoma cells to death receptor induced apoptosis: requirements for c-Jun NH2-terminal kinase and Bim. Mol Cancer Res. 2007;5:783–92. doi: 10.1158/1541-7786.MCR-06-0433. [DOI] [PubMed] [Google Scholar]
- Yang B, Lin H, Hor W, Hwang J, Lin Y, Liu M, Wang Y. Mediation of enhanced transcription of the IL-10 gene in T cells, upon contact with human glioma cells, by Fas signaling through a protein kinase A-independent pathway. J Immunol. 2003;171:3947–54. doi: 10.4049/jimmunol.171.8.3947. [DOI] [PubMed] [Google Scholar]
- Zollman S, Godt D, Prive GG, Couderc JL, Laski FA. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc Natl Acad Sci USA. 1994;91:10717–21. doi: 10.1073/pnas.91.22.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]