Summary
Plasmodium and other apicomplexan parasites are deficient in purine biosynthesis, relying instead on the salvage of purines from their host environment. Therefore interference with the purine salvage pathway is an attractive therapeutic target. The plasmodial enzyme adenosine deaminase (ADA) plays a central role in purine salvage and, unlike mammalian ADA homologs, has a further secondary role in methylthiopurine recycling. For this reason, plasmodial adenosine deaminase accepts a wider range of substrates, as it is responsible for deamination of both adenosine and 5′-methylthioadenosine. The latter substrate is not accepted by mammalian ADA homologs. The structural basis for this natural difference in specificity between plasmodial and mammalian ADA has not been well understood. We now report crystal structures of Plasmodium vivax adenosine deaminase in complex with adenosine, guanosine, and the picomolar inhibitor 2′-deoxycoformycin. These structures highlight a drastic conformational change in plasmodial ADA upon substrate-binding that has not been observed for mammalian ADA enzymes. Further, these complexes illuminate the structural basis for the differential substrate specificity and potential drug selectivity between mammalian and parasite enzymes.
Keywords: Medical Structural Genomics of Pathogenic Protozoa, malaria, Plasmodium, adenosine deaminase, drug selectivity
Introduction
Nearly half of the world's population is at risk of contracting malaria, the devastating disease caused by protozoan parasites of the genus Plasmodium, and several million malaria-related deaths occur each year. Of the four species of Plasmodium that cause human disease, P. falciparum is the most lethal and P. vivax is the most prevalent. The disease burden is greatest on the world's poorest nations in tropical and subtropical regions and is a major contributing factor in the perpetuation of poverty in these areas. Complicating efforts to stem the disease is the emergence and spread of strains that are resistant to the currently available antimalarial drugs. There is an urgent need for novel drugs against these parasites (www.searo.who.int/EN/Section10/Section21.htm). The Medical Structural Genomics of Pathogenic Protozoa Consortium (MSGPP, www.msgpp.org)1 is addressing this need through the structural characterization of proteins that are essential to these eukaryotic pathogens, in particular through the identification of potential pharmacophores.
Plasmodium and other apicomplexan parasites lack enzymes critical for de novo purine synthesis and thus rely on purine salvage to supply precursors for nucleic acid synthesis and energy metabolism. Thus the purine salvage pathway is an attractive drug target. Indeed, disruption of this pathway by inhibition of purine nucleoside phosphorylase (PNP) has been shown to kill malaria parasites cultured in human erythrocytes.2; 3 Adenosine deaminase (ADA; EC 3.5.4.4) is a metallo-dependent hydrolase that precedes PNP in the salvage pathway. It catalyzes the irreversible hydrolytic deamination of adenosine/deoxyadenosine to inosine/deoxyinosine and ammonia. The product of ADA is subsequently converted to hypoxanthine, the fundamental building block for purine nucleotides in Plasmodium. Though there is redundancy in purine salvage pathways of many organisms, Plasmodium apparently lacks a gene that codes for adenosine kinase, suggesting that ADA and PNP are essential enzymes in the lifecycle of the malaria parasite.4 Supporting the notion that ADA is essential to Plasmodium survival, a dramatic decrease of parasitemia has been observed in Plasmodium knowlesi infected primates who have been injected with 2′-deoxycoformycin (DCF), a non-specific, picomolar inhibitor of ADA that mimics the tetrahedral transition state intermediate.5 However, DCF does not inhibit Plasmodium falciparum growth in cultured erythrocytes 6 underscoring the difficulties in drawing parallels between the human disease response and that of other primates and rodents, and also the differences between cultured cells and the more complex environment of a host organism.
The plasmodial ADA homologs show greater than 70% sequence identity with each other while the amino acid sequence of Plasmodium ADA is less than 25% identical to the human protein overall. The active sites show much higher conservation, particularly among the residues that interact with the catalytic zinc ion and the substrate. Unlike the human host's ADA, however, Plasmodium ADA has a secondary function in recycling methylthiopurines from the polyamine biosynthetic pathway. In this role Plasmodium ADA catalyzes the deamination of 5′-methylthioadenosine (MTA) to 5′-methylthioinosine, though less efficiently than it catalyzes the deamination of adenosine.4 Human and other mammalian ADAs do not accept MTA as a substrate. This striking difference in substrate acceptance suggests that an exploitable difference exists at the active site of the plasmodial enzyme that may be used to create selective inhibitors relative to the human enzyme. Indeed, knowledge of this expanded activity has led to the recent design of a series of 5′-substituted deoxycoformycins that selectively inhibit the P. falciparum enzyme but not the human or bovine enzymes.7 Insight into the structural basis of this selectivity has been lacking, however.
We report here the crystal structures of adenosine deaminase from the human malaria parasite Plasmodium vivax (PvADA) in complex with its natural substrate adenosine, the purine guanosine, and the picomolar inhibitor DCF, also known as Pentostatin. This provides the first detailed look at the closed, ligand-bound conformation of the plasmodial enzyme. Comparison to the open, ligand-free ADA homolog from the rodent malaria parasite, P. yoelii (PDB ID 2AMX)8, highlights a dramatic conformational change that occurs upon ligand binding where a Plasmodium-specific loop swings approximately 15 Å over the ligand, effectively sequestering it from the surrounding solvent. Such a large conformational reordering is not observed in available structures of the apo versus ligand-bound mammalian enzyme. Additionally, these structures shed light on the observed differences in substrate acceptance of plasmodial ADA compared to mammalian ADA 4 and in their selective inhibition by 5′-substituted DCFs 7 while providing a more substantial foundation for continued studies in molecular modeling, biochemistry, and structure-based drug design.
Results and Discussion
Structure of Plasmodium vivax adenosine deaminase (PvADA)
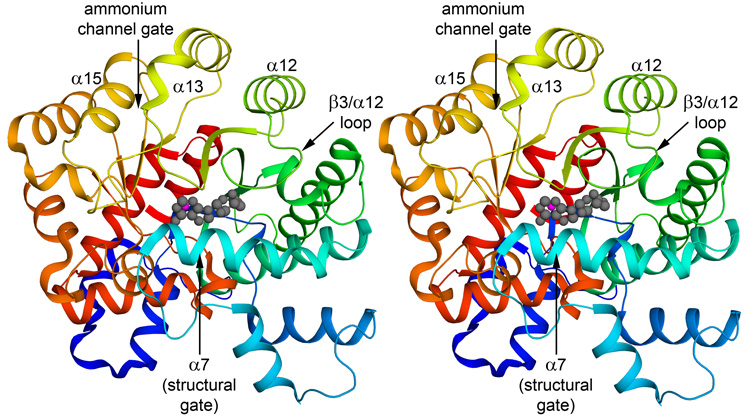
The structure of PvADA has been solved in complex with the natural substrate adenosine (Figure 1), the purine guanosine, and the picomolar inhibitor 2′-deoxycoformycin (DCF, Pentostatin) and refined to final resolution of 1.89 Å, 2.19 Å, and 2.30 Å, respectively. Despite the different ligands bound in the active sites of the three complexes, their structures are essentially identical. The backbones superimpose 9 on one another with a root mean square deviation of less than 0.15 Å for all modeled Cα atoms. PvADA is an α/β protein displaying a TIM-barrel fold and is very similar to previously determined adenosine deaminase (ADA) structures with the exception of a Plasmodium-specific loop that has undergone a major conformational change upon substrate/inhibitor binding. The core is composed of an 8-stranded, parallel β-sheet of strand order 12345678 that closes into a slightly distorted barrel and is surrounded by twenty-four α-helices. Notably in all three models His253, which lies at the C-terminus of strand β6, deviates from typical φ/ψ backbone torsion angles. This histidine is conserved in ADAs from bacteria to mammals and is involved in the catalytic reaction, interacting with the leaving amine on the substrate.10 It is a Ramachandran outlier in nearly all ADA structures currently available in the PDB, suggesting that a strained backbone conformation at this particular histidine is structurally important for catalysis.
Figure 1.
Stereo view of Plasmodium vivax adenosine deaminase in complex with adenosine. The color of the protein ramps from blue at the N-terminus to red at the C-terminus. Since the structure of all the complexes is essentially identical, only the adenosine complex (2PGF) is shown. The bound adenosine is shown as a gray ball and stick model and the catalytic zinc ion is shown as a magenta sphere. Features discussed in the text are labeled. The view is into the active site. Figures were prepared with PyMol.39
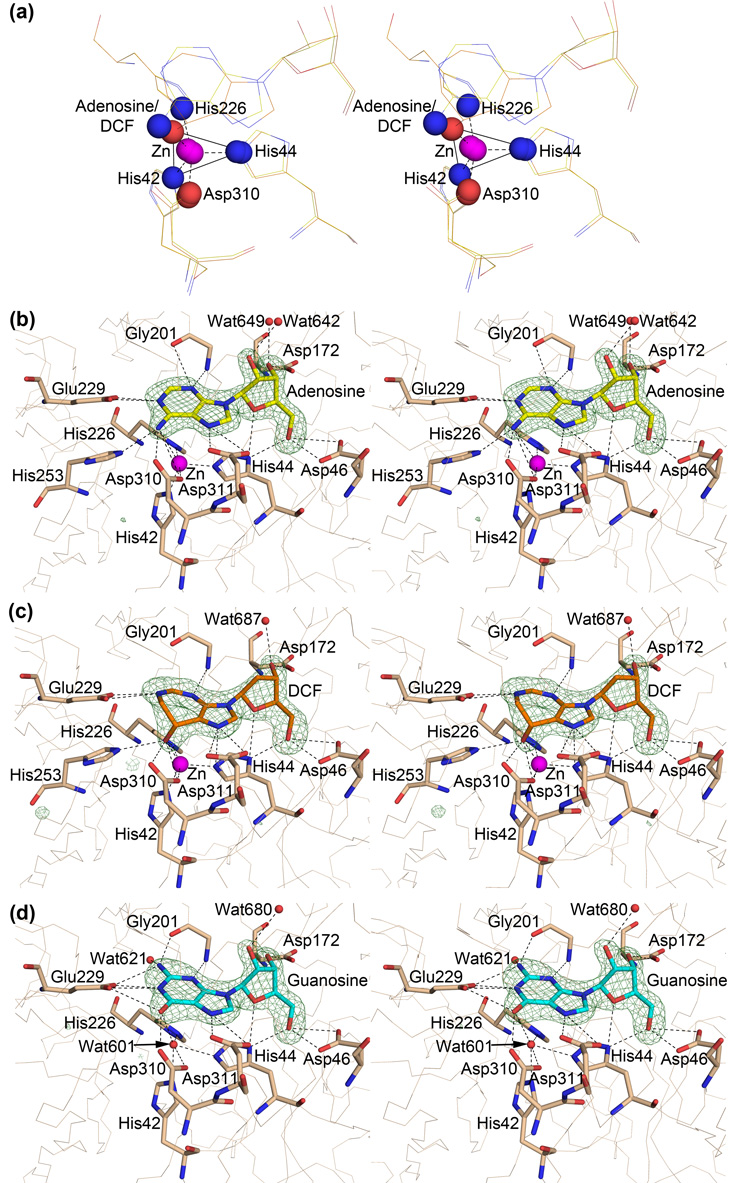
ADA is a metallo-dependent hydrolase that utilizes a catalytic Zn2+. Though Zn2+ was not present in the buffers used for purification or crystallization, a strong anomalous difference density peak exists at the site suggesting the presence of a metal. Therefore, a Zn2+ ion was modeled in the active sites of the adenosine and DCF complexes. The occupancy of the metal, however, was decreased to 0.6 so that the B factors are more comparable to the surrounding residues. The catalytic Zn2+ ion sits roughly in the center of the C-terminal end of the barrel and is coordinated by three histidines (His42 and His44 of β1 and His226 of β5) and one aspartic acid (Asp310 in the β8/α21 loop). The coordination geometry of the bound Zn2+ is roughly trigonal bipyramidal with atom Nε from both His42 and His44 occupying two of the three triangle vertexes and atoms Nε from His226 and Oδ from Asp310 occupying the apex of each pyramid. Atoms N6 from adenosine or O8 from DCF occupy the last vertex of the triangle and the coordination geometry is much less distorted when bound to DCF (Figure 2a). This geometry is identical to that described for the coordination environment for the bound zinc ion in the murine enzyme.11 In the case of the guanosine complex, the Zn2+ site was modeled as a water molecule despite the presence of a very weak anomalous peak. The coordinating residues are shifted slightly yielding donor atom to metal distances greater than would be expected in this coordination environment.
Figure 2.
(a) Stereo view of the coordination environment of the catalytic Zn2+ ion (magenta sphere). Nitrogen atoms involved in coordination are shown as blue spheres and oxygen atoms as red spheres. The view is rotated −60° along the x-axis with respect to figure 1. (b–d) Stereo views of the ligand environments. Residues that make polar contacts within 3.5 Å of the ligand or that coordinate the catalytic zinc ion (magenta sphere; water 601 in the case of the guanosine complex) are shown as sticks. Waters that are in contact with the ligand are shown as red spheres. 2Fo-dFc ligand-omit difference density is shown as green mesh around the ligands at a contour level of 5σ. In all three cases the ribose 2′-endo sugar pucker is clearly defined by the electron density. The view is rotated −90° around the X-axis with respect to figure 1. (b) Adenosine environment (2PGF). (c) Guanosine environment (2QVN). (d) 2'-deoxycoformycin environment (2PGR).
The ligands, adenosine (PDB ID 2PGF), 2′-deoxycoformycin (DCF; PDB ID 2PGR), and guanosine (PDB ID 2QVN), are situated adjacent to the Zn2+ ion in a tight cavity that is almost completely enclosed by the protein (Figure 2 and Figure 3). The purine(-like) rings lie roughly perpendicular to the β-barrel above the Zn2+ ion and the sugar groups are situated between β-strands 2, 3, and 4. Helices α7 and α8 lie above the ligands on one side of the barrel and loops β3/α12 and β4/α13 comprise the other side. The sidechains of residues Glu229, His253, Asp310, and Asp311, and the main chain nitrogen of Gly201 make hydrogen bonds with the adenosine base and stereospecifically position the leaving amine group adjacent to the bound Zn2+ ion (Figure 2b).
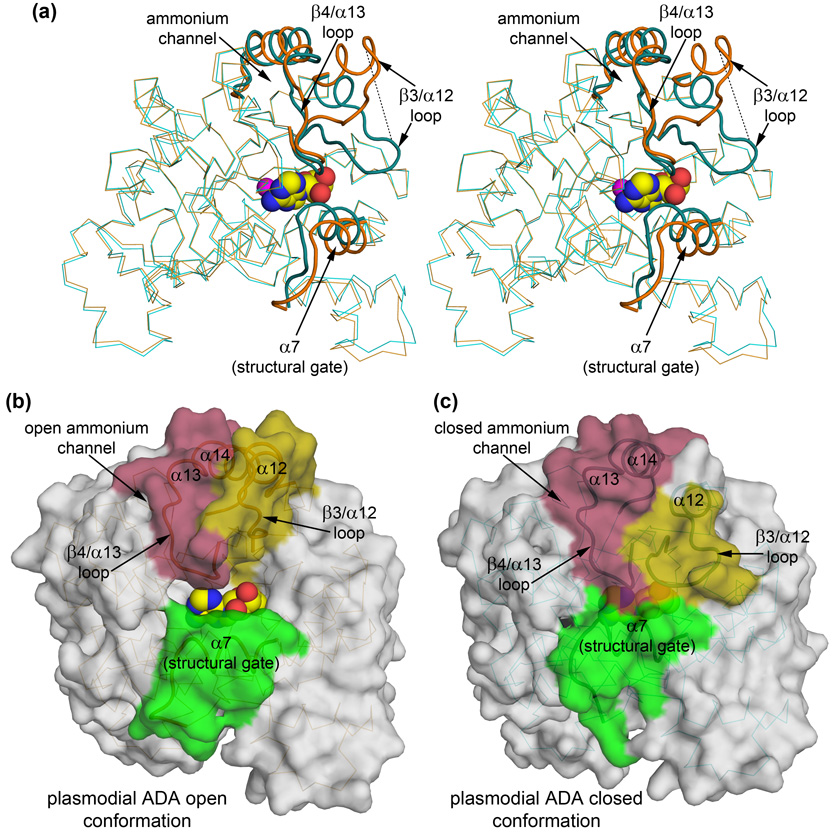
Figure 3.
Ligand-binding induces a large conformational change around the active site of the enzyme. (a) Stereo view highlighting the conformational changes associated with ligand-binding. The P. yoelii structure (2AMX; orange), corresponding to the open form, is superimposed 9 onto the P. vivax structure (2PGF, 2PGR, 2QVN; cyan), corresponding to the closed form. In addition to the relatively small rigid body movements of α7 and β4/α13 loop-α13- α14 toward the substrate associated with the closed state of the mammalian enzymes, the β3/α12 loop of plasmodial ADA undergoes a reordering and is stabilized over the substrate, blocking access to the active site. The dashed line runs between PvADA Ala177 (cyan loop) and PyADA Ala190 (orange loop), which are the equivalent alpha carbons at the maximal displacement of this conformational change, approximately 15.5 Å. The features of greatest change between the apo, open, form and the substrate-bound, closed form are highlighted as tubes of slightly darker color. The view is rotated 45° around the Y-axis in relation to that of figure 1 to emphasize the changes upon substrate-binding. (b) Surface representation of PyADA (2AMX; the plasmodial open form) with adenosine (spheres) modeled in the active site pocket by superposition. (c) Surface representation of the PvADA: adenosine complex (2PGF; the plasmodial closed form). In panels b and c, the surface corresponding to the regions of greatest conformational change between the apo and ligand-bound forms are highlighted. α7, the structural gate, is highlighted green; the β3/α12 loop and the N-terminus of α12 are highlighted yellow; and the β4/α13 loop, α13, and the N-terminus of α14 is highlighted pink. The surface is transparent to allow visualization of the backbone and, in the case of the closed form, the bound substrate. The view is the same as depicted in figure 1.
DCF is a transition state analog of adenosine deamination in that it mimics the tetrahedral intermediate at the C6 position of adenine, from which the amine is abstracted. The hydroxyl group of DCF occupies the transition state position of the leaving amine, lying at a near-optimal distance to fill the sixth coordination spot on the Zn2+ ion. It also forms an additional hydrogen bond with the sidechain of His226, which is also involved in Zn2+ coordination. These additional interactions are responsible for the extremely high affinity of this picomolar inhibitor (Figure 2c). Guanosine is not able to form as favorable of a hydrogen-bonding network due to the presence of the additional chemical group attached to the purine ring (Figure 2d). In all three cases, the sugar group is hydrogen bonded by the sidechains of His44, Asp46, and Asp172. In addition to coordinating the Zn2+, His44 hydrogen bonds with the ether oxygen of the ribose ring and with the 5′-hydroxyl group. Asp46 also hydrogen bonds with the 5′-hydroxyl group suggesting that its positioning within the substrate pocket is important. Asp172 hydrogen bonds with the ribose 3′-hydroxyl group and, as discussed later, this interaction induces a different sugar ring pucker than previously observed in complexes with mammalian ADAs. There are no interactions between the protein and the ribose 2′-hydroxyl group, which makes sense because the enzyme acts on both adenosine and deoxyadenosine.
Apo versus substrate-bound conformations of plasmodial ADA
Prior to this work, the structure of apo adenosine deaminase from the rodent malaria parasite, Plasmodium yoelii (PyADA), was reported (PDB ID 2AMX).8 In the PyADA structure, two copies are present in the asymmetric unit and residues Thr186- Ile193, which make up a large part of an extended surface loop, are only modeled in the A chain due to conformational flexibility. As would be expected from protein sequences that are 71% identical, the structures of the enzymes from the two closely related parasites are nearly identical with a backbone root mean squared deviation (RMSD)9 of approximately 1 Å along the entire length of the protein. This global value, however, masks pronounced conformational changes localized around the active site that occur in the enzyme when it is bound to substrate, particularly at the previously mentioned flexible loop in the PyADA structure. The equivalent loop in the ligand-bound P. vivax structures, the β3/α12 loop, is well-ordered and adopts a significantly different conformation from the completed loop in chain A of the PyADA structure, shifting approximately 15 Å (Figure 3a). SSM superposition9 of only the regions of greatest change around the active site (α7, the β3/α12 loop, the β4/α13 loop, and α13 into α14; emphasized in Figure 3) yields an RMSD of 2.4 Å for 42 of 49 alpha carbons whereas the remainder of the proteins superimpose with an RMSD of 0.66 Å for 307 of 308 alpha carbons. Structures of ADAs from other organisms show conformational changes between the apo and bound states so these structural differences between PyADA and PvADA are likewise the result of closure of the substrate pocket upon substrate binding rather than being due to the minute differences in primary sequence. Therefore the P. yoelii structure represents an open form of the plasmodial enzyme (Figure 3b) while the P. vivax structure represents a closed form (Figure 3c).
Closure of ADA upon substrate binding has been previously described for structures of the murine and bovine enzymes (MmADA and BtADA, respectively).10; 12; 13; 14 The mammalian enzyme has been described as being in an "open" conformation in the ligand-free state and as a "closed" form when bound by a ligand. However, neither the open nor the closed states observed for the mammalian structures correspond in detail to those observed for the plasmodial structures. Ligand binding in the mammalian enzymes induces shifts in a leucine-containing helix called the “structural gate,”14 equivalent to PvADA α7 (colored green in Figures 3b and 3c), and in a loop and helix on the opposite wall of the pocket, equivalent to the β4/α13 loop, α13, and much of α14 in PvADA (colored pink in Figures 3b and 3c). This produces a tighter fit to the base moiety of the substrate, the site of catalysis, but the sugar moiety remains largely accessible to the solvent. In contrast, the entire substrate is completely sequestered in the binding pocket of the P. vivax enzyme (Figure 3c). In addition to the conformational changes observed in the mammalian enzymes, substrate binding by the plasmodial enzyme is accompanied by a partial unwinding at the N-terminus of helix α12 and a dramatic shift of approximately 15 Å in the loop leading from the β-barrel to this helix, the β3/α12 loop (Figure 3a). These additional structural elements that undergo conformational change in the plasmodial enzyme are colored yellow in Figures 3b and 3c. Given that the β3/α12 loop is well-ordered in only one copy of P. yoelii crystal structure, the “open” conformation of this loop may not constitute a distinct stable state of the plasmodial enzyme. Therefore, the dramatic conformational change of this loop between the P. yoelii apo structure and the ligand-bound P. vivax structure may more properly be considered as a transition from disorder to order rather than “open” to “closed.” The result of the ordering of these additional structural elements in the plasmodial enzyme is the closure of the active site pocket into a cavity that is nearly completely filled by the substrate (Figures 3b and 3c), in contrast to the “closed” conformation of the mammalian enzyme where the bound substrate is still largely accessible by the solvent.
The active site cavity and ammonium channel
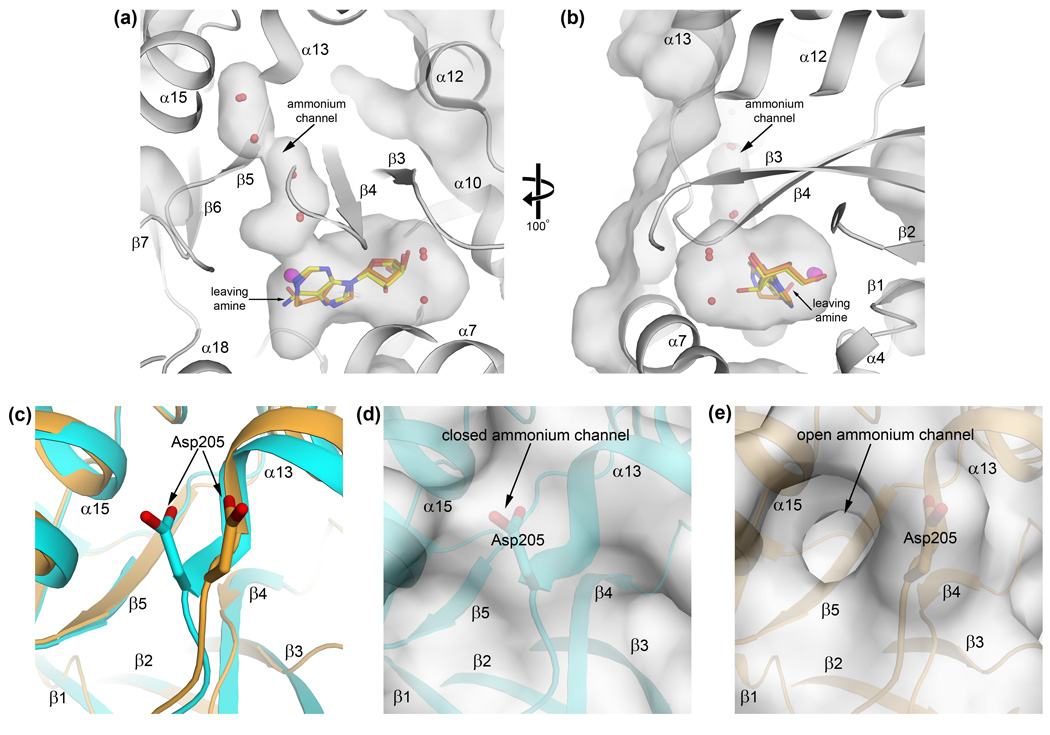
Upon substrate binding, plasmodial ADA assumes a closed conformation in which a boot-shaped cavity is created that is sealed off from the surrounding solvent and nearly completely filled by the substrate (Figures 4a and 4b). The base of the adenosine/inhibitor occupies the "heel" adjacent to the catalytic zinc ion while the sugar moiety occupies much of the "toe" area (Figure 4a). Considering only the substrate, there appears to be a significant volume of free space in the cavity off the 2′-position of the ribose. Two well-ordered water molecules form hydrogen bonds with the substrate and occupy this region, however, leaving very little unoccupied space (Figure 4b). As previously mentioned, the conformational changes associated with substrate-binding in the Plasmodium structures are much more dramatic than those seen in the mammalian structures, where a wide channel extends away from the sugar moiety of the substrate to the bulk solvent. Thus, whereas the substrate is completely sequestered in the closed Plasmodium enzyme (Figure 3c), it is still largely solvent accessible in the closed mammalian structures.
Figure 4.
The boot-shaped active site cavity and putative ammonium channel gate of the active conformation of plasmodial ADA. (a) Side view of the cavity, looking into the side opposite the catalytic zinc. The enclosed adenosine and DCF (yellow and orange sticks, respectively) and waters (red spheres) that occupy the cavity are displayed. The catalytic zinc (magenta spheres) makes up one wall of the "heel" of the boot. (b) The view has been rotated −100° along the Y-axis and is now into the "toe" of the boot. Note that the hydroxyl group of DCF that is equivalent to the leaving amine group of adenosine is oriented toward the putative ammonium channel. (c) The ammonia channel gate. Conformational changes in α13 and in the side chain of Asp205 exist between the closed, substrate-bound (d) and open, apo (e) forms of ADA. In the closed form, the solvent-filled channel leading to the surface from the active site is blocked by the side chain of Asp205. When the enzyme is not bound to ligand, the Asp205 side chain adopts an alternate conformation that allows the channel access to the surrounding solvent, presumably facilitating the release of the ammonia product.
Leading out of the heel and extending towards the surface from the zinc ion and the leaving amine group of the substrate is a narrow solvent-filled channel (Figure 4a). This channel runs between the C-termini of β-strands 4 and 5 of the TIM barrel, along the β4/α13 loop, and reaches the surface between helices α13 and α15. At the surface, the sidechain of Asp205 at the N-terminus of α13 closes off the channel, serving as a gate. In the open conformation, this residue adopts a slightly different conformation and, in conjunction with a slight backbone shift, the channel extends from the active site through the surface (Figures 4c, 4d, and 4e). Interestingly, a similar channel exists in mammalian enzymes in the same vicinity but it is much shorter than seen in the Plasmodium structures. The position of this channel in relation to the catalytic site suggests that it is the likely path followed by the ammonium product but, like the inosine product, a conformational change is necessary to allow its release. In support of this notion, the hydroxyl group on the DCF inhibitor equivalent to the leaving amine group of the adenosine is oriented toward this putative channel (Figures 4a and 4b).
Structural basis for differing substrate acceptance by human and plasmodial ADA
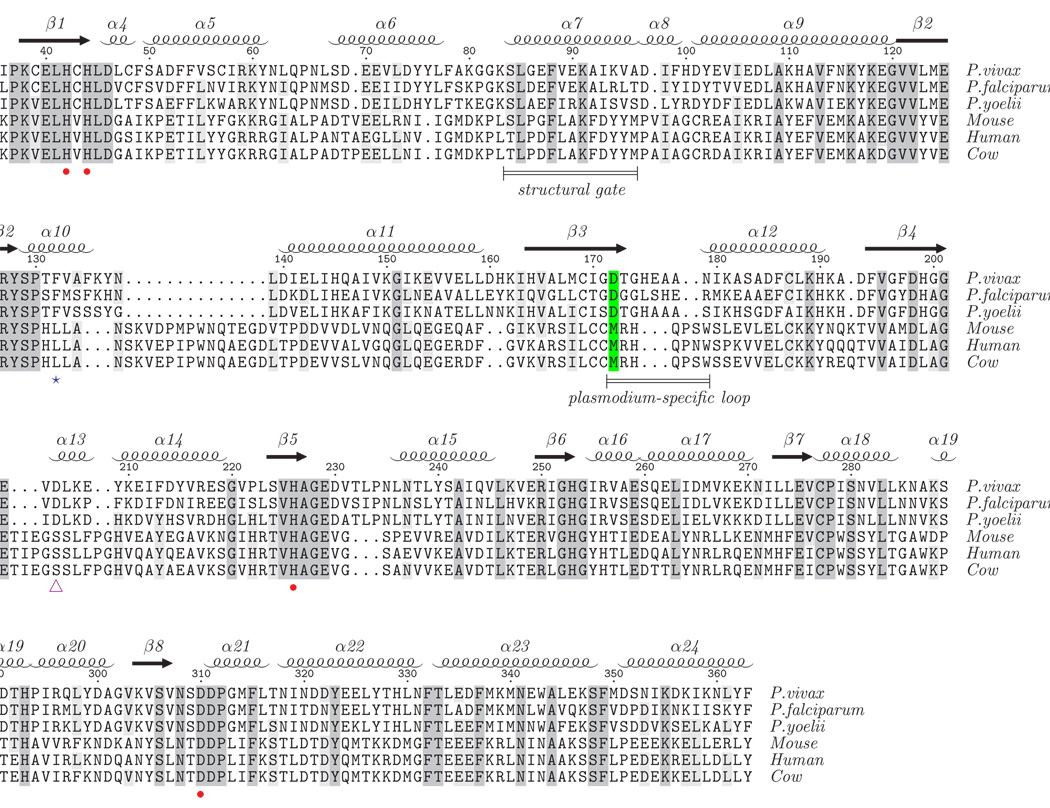
Sequence and especially structure conservation among ADAs across species, particularly in the region of the active site, is apparent from available sequences and structures (Figure 5). The structure of the closed Plasmodium ADA (PDB ID 2PGF) superimposes 9 on the closed MmADA (PDB ID 1ADD) with an RMSD of 1.74 Å for 308 aligned Cα atoms and on the closed BtADA (PDB ID 1KRM) with an RMSD of 1.75 Å for 308 aligned Cα atoms. It has not been clear from the available structures how the alternate substrate, MTA,4 or the specific substituted inhibitors, 5′-MeS-DCF, 5′-PrS-DCF, and 5′-PhS-DCF 7 bind selectively to the plasmodial enzyme over the mammalian counterparts.
Figure 5.
Structure-based sequence alignment of the "closed" forms of plasmodial and mammalian ADAs. The structure-based sequence alignment was created using CE-MC 40 with the structures of ADA enzymes reported to be in the closed, inhibitor-bound form; P. vivax (2PGR), cow (1KRM), and mouse (1A4L). The sequences of P. falciparum, P. yoelii, and human ADA, which lack structures in the closed form, were then manually aligned to the structure-based sequence alignment. For conciseness, residues at the termini that do not structurally align are not shown. Absolutely conserved residues are shaded dark gray and residues conserved in at least four of the sequences are shaded light gray. Secondary structural elements corresponding to the P. vivax structures are presented at the top of the alignment. Zn2+-coordinating residues and the structural gate, which are common to mammalian and plasmodial ADAs, are indicated by red dots and a labeled bar. The loop that makes a large, Plasmodium-specific conformational change upon substrate/inhibitor binding is indicated by a labeled bar. The critical amino acid difference between plasmodial and mammalian ADA that facilitates the broader substrate range for plasmodial ADA and its selective inhibition (D172M) is highlighted green. The putative ammonium channel gate is indicated by the purple triangle and the phenylalanine that adopts an alternate rotamer in plasmodial ADA to facilitate binding to 5′-functionalized substrates/inhibitors is indicated by a blue star.
Structural differences in the mammalian and plasmodial active sites in the vicinity of the 5′-position of the ribose of the bound substrate have been suggested to create a larger pocket that may accommodate 5′-functionalized compounds.7 In particular, the structural changes implicated are a 1 Å shift in the backbone away from the substrate at conserved Tyr128 and an additional backbone shift in the vicinity of Ile170 and Asp172 of plasmodial ADA 7 (residue numbering is from PvADA). The shift at this second location was attributed to substitutions of Cys153 to Ile170 and Met155 to Asp172 between mammalian and plasmodial ADA sequences (Figure 6). However, this hypothesis was based on a comparison of the open, ligand-free form of the plasmodial enzyme, represented by the P. yoelii structure (PDB ID 2AMX), to the closed, ligand-bound form of the mammalian enzyme (PDB ID 1ADD), which do not represent structurally equivalent states of the enzyme.
Figure 6.
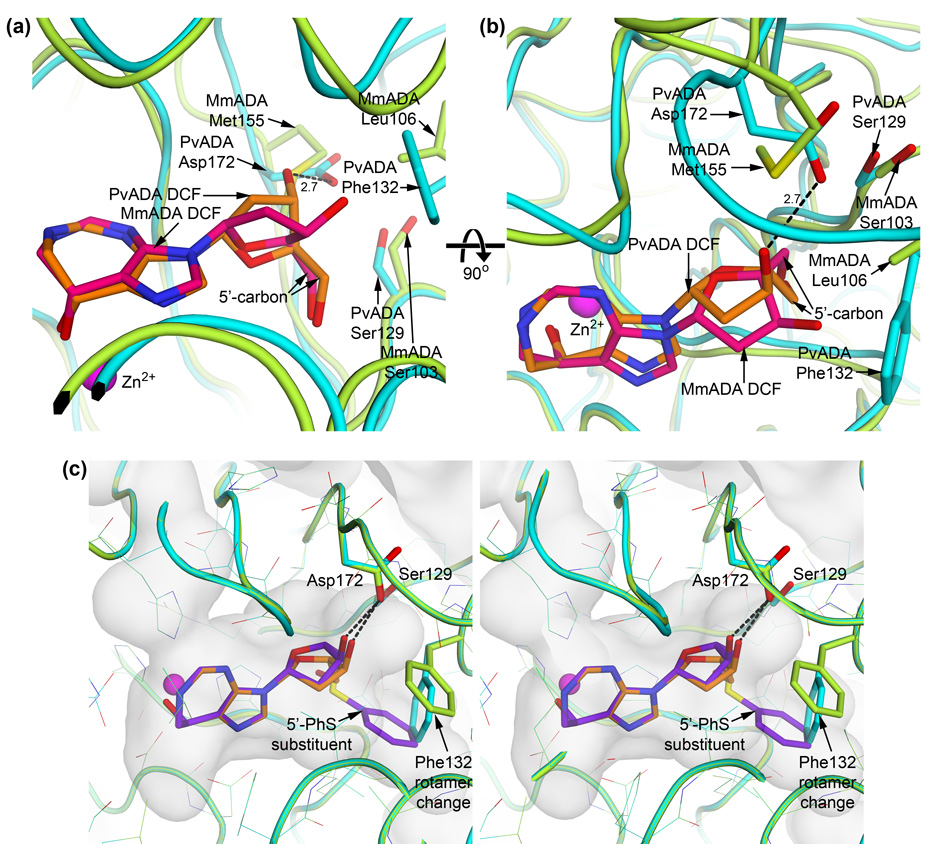
(a) and (b) Alternate sugar pucker of substrate/inhibitor induced by the plasmodial ADA Asp172 : mammalian ADA Met155 sequence difference. Plasmodial ADA is cyan and its bound DCF in orange while mammalian ADA is green and its bound DCF in pink. Plasmodial ADA Asp172 hydrogen bonds with the ribose 3′-hydroxyl group, an interaction that mammalian Met155 is incapable of making. This causes the plasmodial ADA-bound inhibitor to adopt a C2′-endo sugar pucker while the mammalian ADA-bound inhibitor adopts a C4′-exo pucker. The result is that the 5′-carbon of the two riboses are oriented significantly differently with respect to the ribose ring although the 5′-hydroxyl groups occupy nearly the same location and are less than 0.4 Å apart. The different orientations of the 5′-carbon, however, has a great affect on the space that additions at this position may occupy while maintaining a biologically relevant glycosidic linkage with the purine ring. (c) Stereo view of 5′-PhS-DCF (purple sticks) docked into the active site cavity of plasmodial ADA and superimposed on the crystallographically observed DCF (orange sticks). The plasmodial ADA crystal structure is cyan, while the protein following docking is green. The most significant change in the structure of plasmodial ADA in order to accommodate the 5′-thiophenyl addition is an alternate rotamer adopted by Phe132, which both enlarges the cavity and stabilizes the 5′-addition.
Comparison of the now available ligand-bound form of plasmodial ADA, represented by the P. vivax structures, to the ligand-bound mammalian structures, however, indicates that the structural differences suggested by Tyler, et al.7 do not actually have a significant effect on the pocket size in the bound state. The indicated mammalian Cys to plasmodial Ile substitution is located at the beginning of the β3/α12 loop that undergoes a 15 Å shift upon substrate binding. Due to this large conformational change, the Cα of Asp172 in the plasmodial ADA moves approximately 4 Å to end up almost 1 Å closer to the substrate than the corresponding Cα of Met155 in the mammalian ADA. Indeed, the structures of the ligand-bound plasmodial enzyme show that its active site pocket is even more tightly constrained than that of the mammalian enzyme in their substrate/inhibitor-bound states. Obviously, some other property of the protein is responsible for the observed differences in substrate/inhibitor acceptance between Plasmodium and mammalian ADAs. The large, Plasmodium-specific conformational change in the β3/α12 loop may be responsible for facilitating a favorable interaction with a 5′-functionalized substrate. It creates a surface that can favorably interact with the additional chemical groups upon rotation of the 5′-carbon of the sugar. The additional chemical substituent would primarily remain solvent-exposed in the current closed mammalian enzyme structures. As a side note, it is plausible that the plasmodial ADA cannot reach its fully closed active conformation when acting on the larger substrate, MTA, and this may explain why it is not as efficient as when acting on adenosine (Km adenosine ~ 5× Km MTA, kcat adenosine ~ 5× kcat MTA).4
Molecular modeling of Plasmodium ADA-specific inhibitors
In the interest of designing drugs that selectively inhibit Plasmodium ADA, an understanding of the binding mode of the substituted deoxycoformycins, 5′-MeS-DCF, 5′-PrS-DCF, and 5′-PhS-DCF 7 would be invaluable. It is important to understand how the plasmodial ADA active site pocket is able to accommodate the largest of these three inhibitors, 5′-PhS-DCF, while mammalian ADA is not able to form a favorable interaction even with the smallest inhibitor, 5′-MeS-DCF. Thus, these compounds were modeled into the closed active site cavities of the PvADA:DCF complex (PDB ID 2PGR) and the MmADA:DCF complex (PDB ID 1A4L)11 with the crystallographically determined conformation of the bound DCF as the starting point for a direct comparison. Residues in the active site adjacent to the 5′-position of the bound inhibitors were allowed conformational freedom throughout the docking, while the remainder of the protein was fixed (see Methods section). The catalytic zinc ion and the base moiety of the substituted DCFs were also fixed because coordination of the Zn2+ by the 8R-hydroxyl of the base greatly contributes to the high potency of this transition state intermediate analog.11
Role of ribose sugar pucker in ADA substrate range and inhibitor selectivity
These modeling experiments help to illuminate why 5′-substituted DCF inhibitors selectively bind to the plasmodial enzyme. To a small extent, the Plasmodium-specific conformational change, whereby the β3/α12 loop swings down over the ribose, creates a surface for additional chemical groups to interact with. In the absence of this conformational change in mammalian ADAs, the additional, hydrophobic chemical groups are left largely exposed to the solvent. More importantly, the Met to Asp172 substitution between the mammalian and plasmodial enzymes (Figure 5 and Figure 6) plays a key role in the expanded substrate range by altering the preferred ribose sugar pucker of bound nucleoside(-like) ligands.15 The ribose ring in all complexes of mammalian ADA with adenosine-like compounds 10; 11; 13; 16; 17 is observed to adopt a C4′-exo or a nearly superimposable C3′-endo pucker, which allows the sugar O3′ to avoid interaction with the bulky hydrophobic Met155 sidechain. These puckers cause the sugar 5′-carbon to be nearly equatorial to the ribose ring. In plasmodial ADA, the hydrogen-bond between the sugar O3′ and Asp172 instead stabilizes a C2′-endo sugar pucker, which concomitantly results in the sugar C5′ being more axial to the ribose ring. C5′-substituents on a C2′-endo ribose are thereby positioned to interact with the binding site at a different angle than would be possible for substituents on a C4′-exo or C3′-endo ribose (Figures 6a and 6b).
Models based on the sugar conformation observed in the present P. vivax ADA:ligand complexes indicate that the plasmodial enzyme can accommodate addition even of the large thiophenyl substituent at the 5′-position with only minor conformational changes to the protein and the ligand (Figure 6c). The glycosidic bond anti-conformation observed crystallographically is preserved in the models, and the primary conformational change is a rotation about the C4′-C5′ bond. The 5′-sulfur of the thio-substituted compounds, which takes the place of the 5′-hydroxyl group of ribose, is oriented more towards the purine-like ring so that the phenyl group may occupy the opposite side of the pocket from the ribose 5′-hydroxyl group in the crystal structures. To accommodate the phenyl group, the sidechain of Phe132 must adopt an alternate rotamer that both enlarges the toe area of the boot-shaped cavity and helps stabilize the inhibitor by π-stacking with the phenyl substituent (Figure 6c). To the contrary, the C4′-exo/C3′-endo conformation favored by the mammalian enzymes would cause even the smallest chemical substituent at the 5′-position, a thiomethyl group, to clash with the backbone in the vicinity of Ser103 unless the conformation of the glycosidic bond is altered to the syn-conformation. The biological relevance of such syn-docking poses is questionable, however, because ADA activity is dependent on the substrate being in the anti- conformation.11; 18 Further, in all structures of ADA that contain a nucleoside or nucleoside-like substrate or an inhibitor available in the protein data bank (www.pdb.org), the glycosidic bond is in an approximately anti- conformation with the 5′-position of the sugar oriented towards the same side of the pocket as seen in the structures described here, including the mouse ADA:ligand structures. It should be noted that PvADA Ser129 is equivalent to MmADA Ser103 and that the backbones of the mouse and P. vivax structures superimpose to within 1 Å of each other in this region; so the clashes observed in modeling the 5′-substituted DCF molecules in the mammalian active site while maintaining an anti-conformation of the glycosidic bond are predominantly due to differences in the sugar pucker adopted by the bound substrates/inhibitors.
The results described above lead to a prediction that substitution of Met155 in mammalian ADA by any residue capable of interacting with the 3′-hydroxyl of the substrate ribose moiety may permit the broader substrate range observed for the plasmodial enzyme. Further, the resultant enzyme would likewise be inhibited by this series of 5′-substituted inhibitors. The converse should also hold true for the substitution of Asp172 in the plasmodial enzyme by a bulky hydrophobic residue. To test the plausibility of these hypotheses, the reciprocal in silico mutations were made in PvADA and MmADA, specifically Asp172Met in PvADA and Met155Asp in MmADA, and the docking calculations repeated on the mutant enzymes. The mutant enzymes did show altered binding energetics from the native proteins. As anticipated, the predicted binding modes of the 5′-functionalized DCF inhibitors in the Asp172Met mutant plasmodial enzyme are very unfavorable energetically and more closely resemble those of the docked native mouse enzyme. Conversely, the binding modes of the thio-substituted inhibitors in the Met155Asp mutant mouse enzyme are now much more favorable and resemble those of the docked native plasmodial enzyme. Though the energies associated with these docking studies are less favorable overall than those observed with docking in the native enzymes, they do suggest that Met155Asp MmADA is able to bind 5′-PhS-DCF while Asp172Met PvADA is not likely to bind even 5′-MeS-DCF in a biologically relevant manner. These results strengthen the suggestion that the selective inhibition and expanded substrate range of plasmodial ADA in comparison to mammalian ADA is largely a result of the altered sugar pucker induced by the change of a hydrophobic residue to one capable of forming an additional hydrogen bond with the sugar O3′.
Conclusion
The ADA structures from P. vivax presented here provide the first look at the substrate-bound conformation of the plasmodial enzyme in the presence of two different purines and a picomolar inhibitor. The overall fold of the enzyme and the ligand-filled active site pocket are virtually identical in these three complexes. The structures are reminiscent of the closed form of the enzyme described for substrate-bound mammalian homologs; 10; 12; 13; 14 however, a significant Plasmodium-specific conformational change is observed in comparison to the previously solved apo structure of the plasmodial enzyme8 that is not observed in comparison of mammalian apo and substrate complex structures. The unique conformational change involves a 15 Å shift in the β3α12 loop whereby it closes over the ribose moiety of the substrate and results in an active site cavity that is completely sealed off from the surrounding solvent. This is in sharp contrast to the mammalian enzyme whose substrate remains largely solvent accessible.
Inspection of the substrate-filled cavity of the closed plasmodial structures reveals a sealed, solvent-filled channel extending from the active site adjacent to the leaving amine group of the substrate towards the surface of the protein. In the plasmodial apo structure, this channel is open to the surrounding solvent due to a slight conformational change in the protein at the N-terminus of helix α13. The location of this channel suggests that it is the path followed by the reaction product ammonium.
These structures of distinct ligand complexes of Plasmodium ADA also provide an explanation for the ability of this enzyme to deaminate the alternate substrate methylthioadenosine, an activity not observed in mammalian enzymes, and for the selectivity of 5′-functionalized 2′-deoxycoformycin inhibitors for the plasmodial enzyme over the mammalian enzyme. Two specific differences between the Plasmodium and mammalian enzymes are suggested to be responsible for these observations. Firstly, the Plasmodium-specific loop that closes over the active site provides a favorable interaction surface for the additional 5′ hydrophobic chemical groups that would otherwise remain solvent exposed. Secondly, and probably more importantly, the substitution of a Met in the mammalian enzyme to an Asp in the plasmodial enzyme at a key location within the active site adjacent to the 3′-position of the bound substrate modulates the sugar pucker adopted by the ribose moiety. The C2′-endo sugar pucker observed in the plasmodial structures allows substituents at the 5'-position of the substrate or inhibitor to sample a favorable environment within the active site. This favorable orientation is not accessible to 5′-substituents of the C4′-exo or C3′-endo sugar puckers observed in complexes of mammalian ADA. As the ligand-bound structures reported here shed light on observed exploitable differences between plasmodial and mammalian ADA, they will be invaluable to ongoing structure-based drug design efforts that target this key enzyme in the life cycle of the deadly malaria parasite.
Materials and Methods
Target selection, protein expression and purification
Plasmodium adenosine deaminase (TargetDB: Pfal005676AAA and Pviv005676AAA) was selected as a medically relevant target because it is a key enzyme in the essential purine salvage pathway of these purine auxotrophs. The protein sequences for P. falciparum and P. vivax adenosine deaminase are 72% identical. An established research base exists in inhibition of the activity of this enzyme class (EC 3.5.4.4) and potent inhibitors, both nucleoside-like (e.g. 2′-deoxycoformycin) and non-nucleoside-like [e.g. erythro-9-(2-hydroxy-3-nonyl)adenine], are well-characterized, providing the opportunity to piggyback on these molecules to aid in the drug design process. Further, several inhibitor-bound structures of murine 10; 11 and bovine 13; 14; 19; 20; 21; 22 ADAs are available for comparison of the active site environments between mammalian and parasite enzymes.
The full-length ADA genes from P. falciparum and P. vivax were cloned and expressed, and protein from the P. vivax construct yielded diffraction-quality crystals. The gene sequence (PlasmoDB: Pv111245)23 was PCR-amplified from genomic DNA and cloned into expression construct BG1861, a modified version of pET14b that includes a noncleavable hexahistidine tag.24 Protein was expressed in Escherichia coli BL21 [DE3] and purified using immobilized metal affinity chromatography on a Ni-NTA column followed by size exclusion chromatography on a HiLoad Superdex 200 column (Amersham Pharmacia Biotech). Protein was eluted in a standard buffer (0.5M NaCl, 2mM DTT, 0.025% NaN3, 5% glycerol, 20 mM HEPES at pH 7.5), concentrated to greater than 10 mg/mL, flash frozen in liquid nitrogen, and stored at 80° C.25 Selenomethionyl-derivative protein was produced according to the protocols of Studier 26 and SGPP 27 and purified as described for the native protein.
Protein crystallization
Purified protein was screened at the high-throughput facility at the Hauptman Woodward Institute to identify initial crystallization conditions.28 Crystallization leads identified in the high-throughput screen were optimized in-house using sitting-drop vapor diffusion to produce crystals suitable for data collection. Frozen selenomethionyl protein stocks were thawed and diluted with adenosine or guanosine stock solutions in standard buffer to a final concentration of 7 mg/ml protein and 5 mM adenosine or guanosine for cocrystallization experiments. The adenosine cocrystallization drop consisted of 1 µl protein + adenosine mixed with 1 µl reservoir solution (33% PEG 20K, 0.1 M NaH2PO4, 0.1 M TAPS at pH 9.0) and a final concentration of 16% acetonitrile. The guanosine cocrystallization drop consisted of 1 µl protein + guanosine mixed with 1 µl reservoir solution (27.3% PEG 20K, 0.1 M NaH2PO4, 0.1 M CHES at pH 9.5). In both cases, the drop was equilibrated over 100 µl reservoir solution at 25° C and needle-shaped crystals appeared within two days. For the picomolar inhibitor 2′-deoxycoformycin (DCF, Pentostatin) complex, DCF stock solution was added to a drop containing crystals from an adenosine cocrystallization experiment to a final concentration of 10 mM. Crystals were soaked in this solution for 75 minutes. Crystals were directly mounted in cryoloops and frozen directly in liquid nitrogen in preparation for diffraction experiments.
Data collection and structure determination
Crystals of PvADA were screened at the Stanford Synchrotron Research Laboratory (SSRL) on beamlines 9-2 and 11-1 using the SSRL automated mounting (SAM) system.29 Single wavelength data from single crystals were collected using the Blu-Ice software package30 and processed using HKL2000.31 The crystals belong to space group C2221 with unit cell dimensions of 144.0 Å × 146.6 Å × 50.4 Å. The Matthews coefficient is approximately 3.1 Å3/Da and one protein molecule is present in the asymmetric unit. The structure of the P. vivax ADA (PvADA) – adenosine complex was solved by molecular replacement with a resolution limit of 3.0 Å and a search model consisting of each separate chain from the Plasmodium yoelii ADA (PyADA) structure 8 (PDB ID 2AMX, 71% sequence identity) using MOLREP.32 Though both chains present in 2AMX successfully led to a solution of the P. vivax structure, the B chain produced a slightly better score likely owing to it lacking residues equivalent to 173–180 in the P. vivax structures. These residues adopt a significantly different conformation upon ligand binding. The resulting MOLREP model was rigid body refined using Refmac5 33 and then manually edited using Coot.34 All steps after the initial molecular replacement phase assignment utilized the full dataset to a resolution of 1.89 Å. The presence of adenosine in the active site was immediately obvious from a very strong difference density peak displaying the characteristic shape of adenosine. Adenosine and waters were placed and refinement continued by iteration of manual editing in Coot followed by restrained refinement in Refmac5. In the final cycles of refinement, perturbational displacement of the protein chain was described by twenty TLS groups identified by the TLSMD server 35; 36 and TLS parameters were refined for each group prior to restrained refinement in Refmac5. Model quality was validated using Coot and MolProbity.37 The final model consists of residues 5–363, the adenosine substrate, the catalytic divalent zinc ion, and 270 water molecules. Additionally, two acetonitrile molecules were placed in the final model corresponding to distinct difference density peaks that did not appear to be waters since the crystallization conditions included 16% acetonitrile. The N-terminal His-tag and the first four residues of the protein sequence are not visible in the model, presumably due to disorder in this region of the protein.
The structures of the guanosine and 2′-deoxycoformycin (DCF; Pentostatin) complexes were solved by placing a protein-only model from the adenosine complex into the isomorphous unit cells by rigid body refinement in Refmac5. The presence of the respective ligands in the active sites was immediately obvious from large difference density peaks of characteristic shape, and clearly distinct from the shape of adenosine. Again, the models were completed by cycles of manual editing in Coot followed by restrained refinement with Refmac5, with the final rounds using TLS groups identified by the TLSMD server (ten groups for the guanosine complex and nine groups for the DCF complex). The final model of the DCF complex also contains two acetonitrile molecules, though they are in different locations than those in the adenosine complex. The final model of the guanosine complex includes residue 4 and also contains an N-Cyclohexyl-2-aminoethanesulfonic acid (CHES) molecule from the crystallization conditions. The resolution of the final, refined structure is 2.19 Å for PvADA:guanosine complex and 2.30 Å for the PvADA:DCF complex. Data collection and model refinement statistics are presented in Table 1 and Table 2, respectively.
Table 1.
Data collection statistics
| Dataset | Adenosine complex | DCF complex | Guanosine complex |
|---|---|---|---|
| Beamline | SSRL 9-2 | SSRL 9-2 | SSRL 11-1 |
| Spacegroup | C2221 | C2221 | C2221 |
| Unit cell parameters | a=144.0, b=146.6, | a=143.5, b=146.4, | a=144.6, b=146.9, |
| (Å; α=β=γ=90°) | c=50.40 | c=50.03 | c=50.45 |
| Wavelength (Å) | 0.980 | 0.917 | 0.918 |
| Resolution (Å) | 34.2-1.89 (1.97-1.89) | 36.6-2.30 (2.38-2.30) | 34.4-2.19 (2.28-2.19) |
| Unique reflections | 42,835 (4,009) | 23,772 (2,375) | 27,968 (2,662) |
| Completeness (%) | 99.2 (94.6) | 99.6 (99.8) | 99.6 (97.1) |
| Rsym | 0.073 (0.473) | 0.103 (0.427) | 0.146 (0.678) |
| I/σ(I) | 10.3 (2.4) | 6.6 (3.1) | 5.6 (1.7) |
| Redundancy | 7.6 (5.6) | 4.1 (4.1) | 6.6 (5.0) |
| Wilson B factor (Å2) | 24.8 | 32.4 | 30.2 |
Values in parenthesis are for the highest resolution shell.
Table 2.
Model refinement statistics
| Dataset | Adenosine complex | DCF complex | Guanosine complex |
|---|---|---|---|
| Resolution (Å) | 34.2-1.89 | 36.6-2.30 | 34.3-2.19 |
| Rwork | 0.157 | 0.168 | 0.172 |
| Rfree | 0.201 | 0.220 | 0.232 |
| RMSD bonds (Å) | 0.008 | 0.006 | 0.008 |
| RMSD angles (°) | 1.058 | 0.883 | 1.043 |
| Protein atoms | 2,998 | 2,948 | 2,971 |
| Nonprotein atoms | 274 | 136 | 162 |
| Residues in favored regions (%)a | 99.0 | 98.9 | 98.9 |
| Residues in allowed regions (%)a | 99.7 | 99.7 | 99.7 |
| Ramachandran outlier a | His253 | His253 | His253 |
| Unmodeled residues | −7 to 4 | −7 to 4 | −7 to 3 |
| TLS groups (residues) | 5–14, 15–28, 29–47, 48–65, 66–75, 76–88, 89–100, 101–111, 112–153, 154–166, 167–189, 190–203, 204–218, 219–278, 279–288, 289–318, 319–329, 330–337, 338– 351, 352–363 | 5–28, 29–64, 65–99, 100–167, 168–194, 195–258, 259–269, 270–322, 323–363 | 4–15, 16–38, 39–79, 80–99, 100–110, 111–172, 173–210, 211–270, 271–324, 325–363 |
| Mean Biso + BTLS protein atoms (Å2) | 37.0 | 31.2 | 46.6 |
| Mean Biso non-protein or slovent atoms (Å2) | 35.9 | 28.7 | 52.5 |
| Mean Biso solvent atoms (Å2) | 43.2 | 29.0 | 43.2 |
| PDB entry ID | 2PGF | 2PGR | 2QVN |
Determined using the MolProbity Server.37
Molecular modeling of Plasmodium-specific inhibitors
Inhibitors derived from 2′-deoxycoformycin (DCF) with various chemical substituents at the 5′ position of the deoxyribose moiety selectively inhibit plasmodial ADA but not the mammalian homolog.7 To gain insight into the observed selectivity, molecular docking studies were carried out using QXP/FLO 38 and the crystal structures of the DCF-bound ADA from Plasmodium vivax (PDB ID 2pgr) and mouse (MmADA; PDB ID 1a4l).11 The specific inhibitors 5′-methylthio- (5′-MeS-), 5′-propylthio-(5′-PrS-), and 5′-phenylthio- (5′-PhS-) DCF were modeled by building the additional chemical groups onto the crystallographically observed conformation of DCF in each enzyme. Polar hydrogens were added to protein and ligand atoms and water molecules were removed. The active sites were defined as residues within 12.0 Å of DCF. Because the QXP force field is not parameterized for transition state metals and the interaction between the hydroxyl group on the purine-like ring of DCF with the zinc ion coordinated by the protein is critical for the high affinity interaction, the positions of the zinc ion and the atoms in the purine ring of the modeled ligands were fixed during the docking calculations. Protein atoms were fixed except for the sidechains of Asp46, Phe132, Ile170, and Asp172 adjacent to the substituted deoxyribose in PvADA and the corresponding structurally aligned residues Asp19, Leu106, Cys153, and Met155 in MmADA. Docking of each substituted DCF into the plasmodial and mammalian enzymes was carried out using 1000 cycles of Metropolis Monte Carlo conformational searching followed by energy minimization. The 25 lowest energy ligand poses from each run were visually inspected.
The docking simulations were also performed allowing for more conformational freedom in the binding pocket. The sidechains of every residue of the protein in contact with the sugar and its 5' substituent were given full conformational freedom. In addition, flexibility of the protein backbone atoms was permitted for His44, Asp46, Leu47, Val89, Tyr128, Ser129, Phe132, Val133, Asp172, Thr173, Gly174, His175, His202 of PvADA and for His17, Asp19, Leu62, His65, Tyr102, Ser103, Leu106, Leu107, Met157, Asp185 of MmADA. The results of these more elaborate simulations were essentially identical to the earlier ones where only a restricted set of side chains in the immediate vicinity of the 5' substituent were treated as flexible.
To investigate the effect of the substitution of Asp172 in PvADA for Met155 in MmADA on the ability to bind the 5'-substituted DCF inhibitors, these mutations were made in silico and the docking experiments repeated using QXP/FLO as described above for the wild type proteins.
Acknowledgements
We thank Tracy Arakaki and Isolde Le Trong for assistance in crystal screening and data collection and Christine Stewart for bioinformatics and database expertise. Financial support from the Protein Structure Initiative award NIGMS GM64655 and from NIAID award AI067921 is gratefully acknowledged. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
PDB references: 2pgf, 2pgr, and 2qvn
Accession Codes
Atomic coordinates and structure factors are on deposit in the Protein Data Bank (http://www.pdb.org) under accession codes 2PGF, 2PGR, and 2QVN for the adenosine, DCF, and guanosine complexes, respectively.
References
- 1.Fan E, Baker D, Gelb MH, Buckner FS, Van Voorhis WC, Phizicky E, Dumont M, Mehlin C, Grayhack EJ, Sullivan M, Verlinde CL, DeTitta G, Meldrum D, Merritt EA, Earnest TN, Soltis M, Zucker F, Myler P, Schoenfeld L, Kim D, Worthey EA, LaCount D, Vignali M, Li J, Mondal S, Massey A, Carroll B, Gulde S, Luft JR, DeSoto L, Holl M, Caruthers JM, Bosch J, Robien MA, Arakaki T, Holmes MA, Le Trong I, Hol WG. Structural Genomics of Pathogenic Protozoa: An Overview. In: Bostjan K, Mitchell G, Huber T, editors. Methods in Molecular Biology. Vol. 426. 2008. pp. 497–513. [DOI] [PubMed] [Google Scholar]
- 2.Kicska GA, Tyler PC, Evans GB, Furneaux RH, Kim K, Schramm VL. Transition State Analogue Inhibitors of Purine Nucleoside Phosphorylase from Plasmodium falciparum. J. Biol. Chem. 2002;277:3219–3225. doi: 10.1074/jbc.M105905200. [DOI] [PubMed] [Google Scholar]
- 3.Kicska GA, Tyler PC, Evans GB, Furneaux RH, Schramm VL, Kim K. Purine-less Death in Plasmodium falciparum Induced by Immucillin-H, a Transition State Analogue of Purine Nucleoside Phosphorylase. J. Biol. Chem. 2002;277:3226–3231. doi: 10.1074/jbc.M105906200. [DOI] [PubMed] [Google Scholar]
- 4.Ting L-M, Shi W, Lewandowicz A, Singh V, Mwakingwe A, Birck MR, Ringia EAT, Bench G, Madrid DC, Tyler PC, Evans GB, Furneaux RH, Schramm VL, Kim K. Targeting a Novel Plasmodium falciparum Purine Recycling Pathway with Specific Immucillins. J. Biol. Chem. 2005;280:9547–9554. doi: 10.1074/jbc.M412693200. [DOI] [PubMed] [Google Scholar]
- 5.Webster HK, Wiesmann WP, Pavia CS. Adenosine deaminase in malaria infection: effect of 2'-deoxycoformycin in vivo. Advances in Experimental Medicine and Biology. 1984;165(Pt A):225–229. doi: 10.1007/978-1-4684-4553-4_44. [DOI] [PubMed] [Google Scholar]
- 6.Roth E, Jr, Ogasawara N, Schulman S. The deamination of adenosine and adenosine monophosphate in Plasmodium falciparum-infected human erythrocytes: in vitro use of 2'deoxycoformycin and AMP deaminase-deficient red cells. Blood. 1989;74:1121–1125. [PubMed] [Google Scholar]
- 7.Tyler PC, Taylor EA, Frohlich RFG, Schramm VL. Synthesis of 5'-Methylthio Coformycins: Specific Inhibitors for Malarial Adenosine Deaminase. J. Am. Chem. Soc. 2007;129:6872–6879. doi: 10.1021/ja0708363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vedadi M, Lew J, Artz J, Amani M, Zhao Y, Dong A, Wasney GA, Gao M, Hills T, Brokx S, Qiu W, Sharma S, Diassiti A, Alam Z, Melone M, Mulichak A, Wernimont A, Bray J, Loppnau P, Plotnikova O, Newberry K, Sundararajan E, Houston S, Walker J, Tempel W, Bochkarev A, Kozieradzki I, Edwards A, Arrowsmith C, Roos D, Kain K, Hui R. Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms. Molecular and Biochemical Parasitology. 2007;151:100–110. doi: 10.1016/j.molbiopara.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallographica Section D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 10.Wilson DK, Rudolph FB, Quiocho FA. Atomic structure of adenosine deaminase complexed with a transition-state analog: understanding catalysis and immunodeficiency mutations. Science. 1991;252:1278–1284. doi: 10.1126/science.1925539. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Quiocho FA. Complexes of Adenosine Deaminase with Two Potent Inhibitors: X-ray Structures in Four Independent Molecules at pH of Maximum Activity. Biochemistry. 1998;37:8314–8324. doi: 10.1021/bi980324o. [DOI] [PubMed] [Google Scholar]
- 12.Cristalli G, Costanzi S, Lambertucci C, Lupidi G, Vittori S, Volpini R, Camaioni E. Adenosine deaminase: Functional implications and different classes of inhibitors. Medicinal Research Reviews. 2001;21:105–128. doi: 10.1002/1098-1128(200103)21:2<105::aid-med1002>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 13.Kinoshita T, Nishio N, Nakanishi I, Sato A, Fujii T. Structure of bovine adenosine deaminase complexed with 6-hydroxy-1,6-dihydropurine riboside. Acta Crystallographica Section D. 2003;59:299–303. doi: 10.1107/s090744490202190x. [DOI] [PubMed] [Google Scholar]
- 14.Kinoshita T, Nakanishi I, Terasaka T, Kuno M, Seki N, Warizaya M, Matsumura H, Inoue T, Takano K, Adachi H, Mori Y, Fujii T. Structural Basis of Compound Recognition by Adenosine Deaminase. Biochemistry. 2005;44:10562–10569. doi: 10.1021/bi050529e. [DOI] [PubMed] [Google Scholar]
- 15.Sun G, Voigt JH, Filippov IV, Marquez VE, Nicklaus MC. PROSIT: Pseudo-Rotational Online Service and Interactive Tool, Applied to a Conformational Survey of Nucleosides and Nucleotides. J. Chem. Inf. Comput. Sci. 2004;44:1752–1762. doi: 10.1021/ci049881+. [DOI] [PubMed] [Google Scholar]
- 16.Sideraki V, Mohamedali KA, Wilson DK, Chang Z, Kellems RE, Quiocho FA, Rudolph FB. Probing the Functional Role of Two Conserved Active Site Aspartates in Mouse Adenosine Deaminase. Biochemistry. 1996;35:7862–7872. doi: 10.1021/bi952920d. [DOI] [PubMed] [Google Scholar]
- 17.Wilson DK, Quiocho FA. A pre-transition-state mimic of an enzyme: x-ray structure of adenosine deaminase with bound 1-deazaadenosine and zinc-activated water. Biochemistry. 1993;32:1689–1694. doi: 10.1021/bi00058a001. [DOI] [PubMed] [Google Scholar]
- 18.Zemlicka J. Formycin anhydronucleosides. Conformation of formycin and conformational specificity of adenosine deaminase. J. Am. Chem. Soc. 1975;97:5896–5903. doi: 10.1021/ja00853a041. [DOI] [PubMed] [Google Scholar]
- 19.Terasaka T, Kinoshita T, Kuno M, Nakanishi I. A Highly Potent Non-Nucleoside Adenosine Deaminase Inhibitor: Efficient Drug Discovery by Intentional Lead Hybridization. J. Am. Chem. Soc. 2004;126:34–35. doi: 10.1021/ja038606l. [DOI] [PubMed] [Google Scholar]
- 20.Terasaka T, Kinoshita T, Kuno M, Seki N, Tanaka K, Nakanishi I. Structure-Based Design, Synthesis, and Structure-Activity Relationship Studies of Novel Non-nucleoside Adenosine Deaminase Inhibitors. J. Med. Chem. 2004;47:3730–3743. doi: 10.1021/jm0306374. [DOI] [PubMed] [Google Scholar]
- 21.Terasaka T, Okumura H, Tsuji K, Kato T, Nakanishi I, Kinoshita T, Kato Y, Kuno M, Seki N, Naoe Y, Inoue T, Tanaka K, Nakamura K. Structure-Based Design and Synthesis of Non-Nucleoside, Potent, and Orally Bioavailable Adenosine Deaminase Inhibitors. J. Med. Chem. 2004;47:2728–2731. doi: 10.1021/jm0499559. [DOI] [PubMed] [Google Scholar]
- 22.Terasaka T, Tsuji K, Kato T, Nakanishi I, Kinoshita T, Kato Y, Kuno M, Inoue T, Tanaka K, Nakamura K. Rational Design of Non-Nucleoside, Potent, and Orally Bioavailable Adenosine Deaminase Inhibitors: Predicting Enzyme Conformational Change and Metabolism. J. Med. Chem. 2005;48:4750–4753. doi: 10.1021/jm050413g. [DOI] [PubMed] [Google Scholar]
- 23.Bahl A, Brunk B, Crabtree J, Fraunholz MJ, Gajria B, Grant GR, Ginsburg H, Gupta D, Kissinger JC, Labo P, Li L, Mailman MD, Milgram AJ, Pearson DS, Roos DS, Schug J, Stoeckert CJ, Jr, Whetzel P. PlasmoDB: the Plasmodium genome resource. A database integrating experimental and computational data. Nucl. Acids Res. 2003;31:212–215. doi: 10.1093/nar/gkg081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexandrov A, Vignali M, LaCount DJ, Quartley E, de Vries C, De Rosa D, Babulski J, Mitchell SF, Schoenfeld LW, Fields S, Hol WG, Dumont ME, Phizicky EM, Grayhack EJ. A Facile Method for High-throughput Co-expression of Protein Pairs. Mol Cell Proteomics. 2004;3:934–938. doi: 10.1074/mcp.T400008-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Deng J, Davies DR, Wisedchaisri G, Wu M, Hol WGJ, Mehlin C. An improved protocol for rapid freezing of protein samples for long-term storage. Acta Crystallographica Section D. 2004;60:203–204. doi: 10.1107/s0907444903024491. [DOI] [PubMed] [Google Scholar]
- 26.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expression and Purification. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 27.Mehlin C, Boni E, Buckner FS, Engel L, Feist T, Gelb MH, Haji L, Kim D, Liu C, Mueller N, Myler PJ, Reddy JT, Sampson JN, Subramanian E, Van Voorhis WC, Worthey E, Zucker F, Hol WGJ. Heterologous expression of proteins from Plasmodium falciparum: Results from 1000 genes. Mol Biochem Parasitol. 2006;148:144–160. doi: 10.1016/j.molbiopara.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 28.Luft JR, Collins RJ, Fehrman NA, Lauricella AM, Veatch CK, DeTitta GT. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. Journal of Structural Biology. 2003;142:170–179. doi: 10.1016/s1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 29.Cohen AE, Ellis PJ, Miller MD, Deacon AM, Phizackerley RP. An automated system to mount cryo-cooled protein crystals on a synchrotron beamline, using compact sample cassettes and a small-scale robot. Journal of Applied Crystallography. 2002;35:720–726. doi: 10.1107/s0021889802016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McPhillips TM, McPhillips SE, Chiu H-J, Cohen AE, Deacon AM, Ellis PJ, Garman E, Gonzalez A, Sauter NK, Phizackerley RP, Soltis SM, Kuhn P. Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. Journal of Synchrotron Radiation. 2002;9:401–406. doi: 10.1107/s0909049502015170. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter C, Sweet R, editors. Macromolecular Crystallography, Part A. Vol. 276. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 32.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallographica Section D. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method D. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallographica Section D. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 36.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. Journal of Applied Crystallography. 2006;39:109–111. [Google Scholar]
- 37.Lovell SC, Davis IW, Arendall WB, III, de Bakker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi, psi, and Cbeta deviation. Proteins: Structure, Function, and Genetics. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 38.McMartin C, Bohacek RS. QXP: Powerful, rapid computer algorithms for structure-based drug design. Journal of Computer-Aided Molecular Design. 1997;11:333–344. doi: 10.1023/a:1007907728892. [DOI] [PubMed] [Google Scholar]
- 39.DeLano WL. The PyMOL Molecular Graphics System. 2002 http://www.pymol.org.
- 40.Guda C, Lu S, Scheeff ED, Bourne PE, Shindyalov IN. CE-MC: a multiple protein structure alignment server. Nucl. Acids Res. 2004;32:W100–W103. doi: 10.1093/nar/gkh464. [DOI] [PMC free article] [PubMed] [Google Scholar]