Summary
Nonsense mediated mRNA decay (NMD) eliminates transcripts carrying premature translation termination codons, but the role of NMD on yeast unspliced pre-mRNA degradation is controversial. Using tiling arrays we show that many unspliced yeast pre-mRNAs accumulate in strains mutated for the NMD component Upf1p and the exonuclease Xrn1p. Intron identity and suboptimal splicing signals resulting in weak splicing were found to be important determinants in NMD targeting. In the absence of functional NMD, unspliced precursors accumulate in the cytoplasm, possibly in P-bodies. NMD can also complement RNase III-mediated nuclear degradation of unspliced RPS22B pre-mRNAs, degrades most unspliced precursors generated by a 5’ splice site mutation in RPS10B, and limits RPS29B unspliced precursors accumulation during amino acid starvation. These results show that NMD has a wider impact than previously thought on the degradation of yeast unspliced transcripts and plays an important role in discarding precursors of regulated or suboptimally spliced transcripts.
Introduction
Nonnsense mediated mRNA decay (NMD) is an RNA surveillance pathway that eliminates transcripts carrying premature translation termination codons (PTCs; reviewed in Behm-Ansmant et al., 2007; Chang et al., 2007; Conti and Izaurralde, 2005; Lejeune and Maquat, 2005). This surveillance mechanism relies on the Upf proteins, which target PTC-containing transcripts for degradation (Chang et al., 2007; Conti and Izaurralde, 2005; Leeds et al., 1992; Lejeune and Maquat, 2005). The physiological functions of NMD have been analyzed in a variety of organisms. In mammalian cells, NMD prevents the accumulation of truncated proteins that would result from mutations found in patients affected by genetic diseases (Behm-Ansmant et al., 2007; Frischmeyer and Dietz, 1999). NMD also plays a role in regulating transcripts that have incorporated PTC-containing exons by alternative splicing (Green et al., 2003; Hillman et al., 2004; Hori and Watanabe, 2005; Lareau et al., 2007; Ni et al., 2007). In yeast and mammalian cells, NMD controls the expression of a large number of natural transcripts (Guan et al., 2006; He et al., 2003; Johansson et al., 2007; Lelivelt and Culbertson, 1999; Mendell et al., 2004). In C. elegans, Arabidopsis and in Paramecium, NMD contributes to the elimination of unproductively spliced mRNAs (Arciga-Reyes et al., 2006; Jaillon et al., 2008; Mitrovich and Anderson, 2000). NMD also controls the expression of C. elegans pseudogenes (Mitrovich and Anderson, 2005).
Due to the great likelihood of encountering intronic PTCs, it seems logical that NMD would target unspliced pre-mRNAs for degradation in all eukaryotes. However the impact of NMD on unspliced pre-mRNA degradation in the model organism yeast S. cerevisiae has been controversial. Unspliced forms of three pre-mRNAs were found to accumulate in the upf1Δ mutant (He et al., 1993; Li et al., 1995). However, the analysis of the splicing and expression of intron-containing transcripts using splicing microarrays in the xrn1Δ, upf1Δ or upf3Δ mutants failed to reveal widespread accumulation of unspliced pre-mRNAs (Burckin et al., 2005; Clark et al., 2002; Pleiss et al., 2007). In addition, several studies have pointed to nuclear or cytoplasmic degradation pathways for yeast unspliced pre-mRNAs that are independent of NMD. Unspliced transcripts generated by a mutation of the Prp2p splicing factor are not stabilized by NMD mutations (Bousquet-Antonelli et al., 2000), but their accumulation is increased by inactivation of the nuclear exosome, suggesting nuclear decay. Inactivation of the nuclear exosome component Rrp6p also increases the accumulation of unspliced transcripts of genes for which the spliceosome is not associated with the sites of transcription (Moore et al., 2006). In support of the hypothesis of a nuclear retention and degradation system is the finding that depletion of the nuclear pore-associated Mlp1p protein increases the leakage of unspliced pre-mRNAs of an inefficiently spliced reporter transcript (Galy et al., 2004). Specialized nuclear degradation systems for unspliced pre-mRNAs have also been described that rely on the nuclear RNase III Rnt1p (Danin-Kreiselman et al., 2003). In contrast to these findings, which suggest nuclear degradation of several yeast unspliced pre-mRNAs, the decay of actin intron-based reporter transcripts containing splicing signals mutations depends on Xrn1p, the cytoplasmic 5’->3’ exonuclease, but is not affected by Upf1p absence of (Hilleren and Parker, 2003). The decay of several cytoplasmic unspliced meiotic transcripts is also independent of Upf1p (Scherrer and Spingola, 2006).
Interestingly, these observations were made on different transcripts, suggesting transcript-specific degradation pathways. These observations led us to revisit the global impact of NMD on yeast intron-containing transcripts (hereby called the intronome). In this study, using tiling array analysis of strains lacking Xrn1p or Upf1p, we show that NMD eliminates a large number of unspliced pre-mRNAs, many of which are not spliced with high efficiency, due in part to suboptimal splicing signals. Taken together with previous studies, these results show that the mechanisms of degradation of unspliced pre-mRNAs are multiple and sometimes complementary, underscoring the importance of various surveillance pathways for unprocessed species.
Results
Tiling microarray analysis of the S. cerevisiae intronome in upf1Δ and xrn1Δ deletion strains
To investigate the role of NMD in intron-containing transcripts expression and unspliced pre-mRNA degradation, we monitored the accumulation of intron-containing genes (ICGs) using Affymetrix S. cerevisiae tiling arrays in the upf1Δ and xrn1Δ deletion strains. Upf1p is one of the main components of the yeast NMD machinery (Leeds et al., 1991), and Xrn1p is the major 5’->3’ exonuclease that degrades transcripts through the NMD pathway (He and Jacobson, 2001). Although Xrn1p also plays a role in constitutive mRNA degradation, we expected bona fide NMD targets to present similar profiles in upf1Δ or xrn1Δ mutant strains, as shown in previous expression microarray studies (He et al., 2003). We chose to use tiling arrays, since these have been previously used to efficiently detect intronic RNAs (Juneau et al., 2007; Zhang et al., 2007), and also because previously described custom splicing arrays did not reveal widespread effects of NMD mutants on unspliced precursor accumulation (Burckin et al., 2005; Clark et al., 2002; Pleiss et al., 2007). Four independent cultures were used for wild-type, xrn1Δ and upf1Δ mutants and probes prepared from the corresponding RNAs were hybridized to Affymetrix tiling arrays. We monitored the expression of probe sets corresponding to intronic and exonic regions of 278 intron-containing genes (ICGs), corresponding to 287 intronic features, since nine ICGs contain two introns. Following bioinformatics analysis (described in the Supplemental Materials), classification of transcripts according to phenotypic classes was confirmed by visual inspection of the tiling array profiles (see examples in supplemental materials).
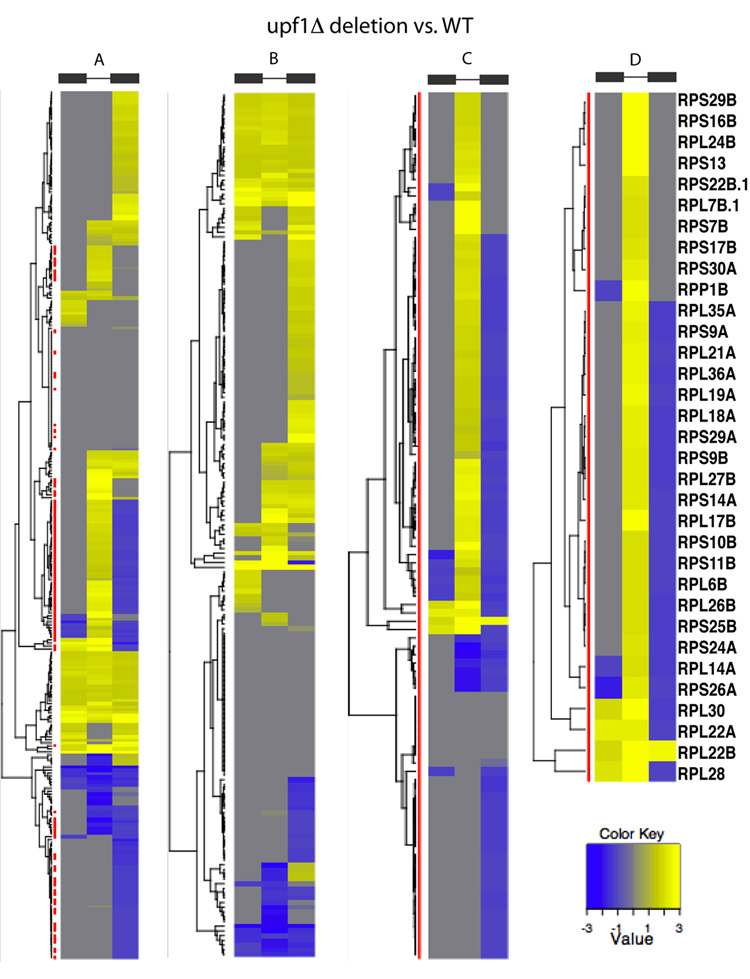
Figure 1 shows the clustering of exon1, intron and exon 2 signals in the upf1Δ mutant strain compared to wild-type for all ICGs. This clustering revealed that a subpopulation of ICGs shows an increase of intronic signal in the upf1Δ strain compared to the wild-type, without major change of signal in exonic sequences (Fig.1A). Strikingly, most of these ICGs corresponded to ribosomal protein genes (RPGs), as represented by red bars on the left side of the dendrogram (Fig. 1). Analysis of the intron-containing RPGs subpopulation (Fig.1C) showed that 31% of RPG introns (33 introns) had a more than two-fold increase of intronic signal in the upf1Δ mutant compared to wild-type. Compared to the number of ICGs showing a more than two-fold increase of intronic levels in the non-RPGs population (19), there is a significant enrichment for RPGs (p-value < 1.6×10−5). The subpopulation of RPGs for which elevated (> two-fold) levels of intronic signals were observed upon Upf1p inactivation (hereby called NMD-sensitive) is shown in detail in Figure 1D. Most of these did not exhibit any change or only a slight decrease in exonic signal. One exception was RPL22B, for which an increase in exonic signal was detected in upf1Δ (Fig.1D; Suppl.Fig.S1). This effect could be explained by the fact that the amount of unspliced RPL22B that accumulates in upf1Δ is so high that it exceeds that of spliced RPL22B in wild-type (see below, Fig. 7). When non-RPG ICGs were clustered separately (Fig.1B), the main cluster was a family of ICGs for which a signal increase in upf1Δ occurred throughout exonic and intronic regions (Fig.1B). Many of these ICGs corresponded to subtelomeric genes encoding helicase-like proteins. The overall signal increase in these regions for the upf1Δ mutant was also observed in neighboring regions outside of these genes, therefore this phenotype might have been the result of a decrease of subtelomeric silencing. Because it is unclear whether or not these genes are actually functional, we did not analyze this subset of genes further.
Figure 1. Tiling array analysis of exons and intron signals in the upf1Δ strain relative to wild-type.
A. Dendrograms of all ICG. B. Dendrogram of non-RPGs. C. Dendrogram of RPGs; D. dendrogram of RPGs that show an increase of intronic signal. Exons are represented by boxes, intron by a line. Yellow represents increase, blue, decrease. For genes exhibiting two introns, the first intron is indicated by .1.
We also analyzed intronic and exonic signals in the xrn1Δ strain relative to the wild-type strain (Suppl. Fig S2). The increase of intronic signal observed in the upf1Δ strain for many RPGs was also observed in xrn1Δ (Supp. Fig.S2). To visualize the differences between ICGs expression in the xrn1Δ and upf1Δ strains, we compared the array profiles of ICGs between these two strains. Several ICGs exhibited an increase in exonic signals in xrn1Δ compared to upf1Δ (Suppl.Fig.S3), without change in intronic signal (top of the dendrogram, Suppl. Fig.S3), which suggested increased spliced mRNA levels. This increase could be attributed to the role of Xrn1p in general mRNA degradation (Parker and Song, 2004). We also found transcripts for which intronic signals were higher in xrn1Δ compared to upf1Δ, suggesting that some unspliced precursors accumulate to higher levels when the exonuclease component is disrupted. Quantification of intronic and exonic signals is provided in Supplemental Table 3. Some genes also showed a stronger accumulation of both intronic and exonic signals in the xrn1Δ strain compared to upf1Δ strain, consistent with a stronger accumulation of unspliced and spliced species in the xrn1Δ strain compared to upf1Δ. Both RPGs and non-RPGs were found among those, including the YRA1 transcript, which exhibited a strong increase of intronic and exonic signals in the xrn1Δ strain compared to wild-type, but not in the upf1Δ strain (Suppl. Fig.S3). These findings are consistent with previous studies, which involved Xrn1p, but not Upf1p in the degradation of YRA1 unspliced mRNAs (Dong et al., 2007; Preker and Guthrie, 2006). However, the overall clustering patterns were very similar in the strain lacking Xrn1p compared to that lacking Upf1p, strengthening the argument that transcripts for which intronic signal accumulation is observed are targeted by NMD.
Effects detected on tiling arrays can be reproduced by northern blot analysis
Tiling arrays analysis showed an accumulation of intronic signal for many RPG transcripts in the upf1Δ and xrn1Δ mutants. While this accumulation was consistent with an increase of unspliced pre-mRNAs in these mutants (with the level of unspliced largely inferior to those of spliced mRNAs), we could not formally exclude that the observed intronic signal was the result of a lack of degradation of the excised introns. We therefore validated the microarray results by analyzing a set of candidates by northern blot. Figure 2 shows the northern blot analysis of several ICGs in the upf1Δ and xrn1Δ deletion strains. To further test if unspliced pre-mRNA accumulation occurs generally in NMD mutants, we also analyzed unspliced transcripts accumulation in mutants lacking Upf2p and Upf3p (Fig.2C). As a positive control, we used a strain carrying a thermosensitive mutation in the splicing factor Prp2p to detect the presence of unspliced pre-mRNAs. These experiments showed that whenever we detected an increase in intronic signal by tiling arrays, we were also able to detect unspliced pre-mRNAs by northern blot analysis. (Fig.2). This effect was observed mostly for RPGs but also for the REC107 meiotic transcript (Fig. 2A). We also analyzed RPL25 by northern blot, which did not show any intronic signal accumulation in the upf1Δ and xrn1Δ strains on tiling arrays. We were unable to detect an increase of unspliced RPL25 precursors in these mutants, while these precursors were readily detectable in the prp2-1 splicing mutant. Interestingly, the quantitative differences between strains on the tiling arrays could be reproduced by northern blot. For example, the RPL21A intronic signal relative to wild-type was similar for the upf1Δ and xrn1Δ mutant strains as judged by tiling arrays (Suppl.Fig.S5A) or by northern blot (Fig.3B). The RPP1B transcript exhibited higher intronic signal in upf1Δ than in xrn1Δ on the tiling arrays (Suppl.Fig S5B), which was confirmed by northern (Fig.2B). Reciprocally, RPS14B exhibited stronger intronic signal in xrn1Δ than in upf1Δ on the arrays (Suppl.Fig.S5B), which was also detected by northern blot (Fig.2B). In addition, elevated exonic RPS14B signal was detected in the xrn1Δ strain on the arrays (Supp.Fig. S5C), which was confirmed by higher levels of spliced mRNAs on the northern blot (Fig.2B). Therefore, the relative differences detected by the tiling arrays can be used with confidence to predict relative differences in gene expression between strains. However, numbers extrapolated from the tiling array analysis were generally lower than the quantifications of the northern analysis (compare numbers in Fig. 2 and Suppl. Table 3), suggesting that the tiling array analysis underestimates the quantitative effects of NMD. Finally, the observation that unspliced precursors accumulate to similar levels in the upf1Δ, upf2Δ and upf3Δ strains (Fig.3C) emphasizes the fact that these unspliced precursors are NMD targets.
Figure 2. Northern blot analysis of ICGs in wild-type and NMD mutant strains.
A. and B. Detection of unspliced ICGs in wild-type, upf1Δ and xrn1Δ strains. Shown are the signals obtained by hybridization of northern blots with the indicated probes, which covered exon1 (E1), intron (I), and exon2 (E2), unless indicated otherwise. US, Unspliced pre-mRNA. S, Spliced mRNA. Numbers indicate the fold increase accumulation of unspliced relative to wild-type. SCR1 or G3PDH were used as loading controls. C. Detection of unspliced RPGs in wild-type, upf1Δ, upf2Δ, upf3 Δ strains. The prp2-ts strain was used as a positive control for the detection and migration of unspliced precursors.
Figure 3. Effects of NMD mutations combined with inactivation of Rrp6p, Mlp1 or of Rnt1 stem-loops on the accumulation of unspliced pre-mRNAs.
A. Expression of RPGs and detection of unspliced RPGs in wild-type, upf1Δ and rrp6Δ and upf1Δrrp6Δ strains for NMD-sensitive transcripts. For some transcripts, different exposures are shown to properly visualize mRNAs. Legends as in Fig. 2. B. Expression of RPGs and detection of unspliced RPGs in wild-type, upf1Δ and mlp1Δ and upf1Δmlp1Δ strains for NMD-sensitive transcripts. Legends as in Fig. 2. C. Expression of RPGs and lack of detection of unspliced RPGs in wild-type, upf1Δ, mlp1Δ, rrp6Δ, upf1Δmlp1Δ and upf1Δrrp6Δ strains for NMD-insensitive transcripts. Legends as in Fig. 2. D. Inactivation of NMD and of Rrp6 shows synergistic effects on the accumulation of two unspliced pre-mRNAs. Legends as in A. E. Analysis of RPS22B in strains carrying a deletion of the Rnt1p-target stem-loops (SL1,2Δ) and/or carrying mutations of NMD components. Shown are the signals obtained by hybridization of the same membrane with the indicated probes.
Effects of the combined inactivation of NMD and of the nuclear exosome component Rrp6p or of the nuclear retention protein Mlp1p on the accumulation of unspliced transcripts
To test whether additional nuclear degradation or retention systems contributed to the degradation of unspliced NMD targets, we combined the upf1Δ deletion with a deletion of either RRP6 or MLP1. Rrp6p is a component of the nuclear exosome, and some unspliced pre-mRNAs have been shown to accumulate in rrp6 mutants (Bousquet-Antonelli et al., 2000; Moore et al., 2006). The accumulation of unspliced pre-mRNAs was first analyzed for the NMD-sensitive RPGs RPL19A, RPL21A, RPL36A and RPP1B (Fig. 3A). The inactivation of Rrp6p did not result in an increase of these unspliced pre-mRNAs, nor did it exacerbate the accumulation of unspliced pre-mRNAs when combined with the upf1Δ deletion (Fig. 3A). This result suggests that these pre-mRNAs are not subjected to nuclear surveillance, but that their principal mode of degradation relies on NMD.
Mlp1p is required for nuclear retention of an inefficiently spliced RP51B-based pre-mRNA reporter transcript (Galy et al., 2004). We analyzed the accumulation of the NMD-sensitive pre-mRNAs RPL21A, RPP1B and RPS11B in upf1Δ mlp1Δ and upf1Δ m1p1Δ strains (Fig.3B). Mlp1p depletion in the upf1Δ strain did not exacerbate the accumulation of these unspliced pre-mRNAs (Fig.3B), suggesting that the Mlp1p-mediated nuclear retention system is not active on these pre-mRNAs. We conclude that these transcripts are not retained in the nucleus and that they can escape to the cytoplasm where they are targeted by NMD (see below, Fig.6).
We also investigated whether Rrp6p or Mlp1p depletion, alone or in combination with Upf1p depletion, could result in the accumulation of unspliced pre-mRNAs for NMD-insensitive ICGs. Northern blot analysis of RPS18A, RPL39 and GOT1, which are unaffected by the upf1Δ or xrn1Δ mutations did not reveal any accumulation of unspliced pre-mRNA in the rrp6Δ or rrp6Δ upf1Δ mutants (Fig.3C), indicating that these transcripts either do not produce significant levels of unspliced pre-mRNAs, or that their degradation relies on other pathways. Similarly, MLP1 deletion in combination with the UPF1 deletion did not result in accumulation of unspliced RPL25,RPL16A/B or GOT1 mRNAs (Fig.3C), suggesting that these transcripts are probably not retained in the nucleus, which would have explained their insensitivity to NMD.
Finally RPS11B and RPL16A were the only transcripts for which inactivation of both Rrp6p and of Upf1p resulted in a minor increase in unspliced transcripts compared to the single mutants (Fig.3D). These results show that in some particular cases, the nuclear exosome can functionally complement NMD in the degradation of unspliced transcripts.
NMD complements Rnt1p-mediated degradation of unspliced RPS22B species
RPS22B contains two introns, the second of which contains the box H/ACA snoRNA snR44. We previously showed that Rnt1p, a nuclear double-stranded RNA endonuclease, cleaves a stem-loop structure in the first intron, triggering degradation of the unspliced or partially spliced precursors containing intron 1 (Danin-Kreiselman et al., 2003). Since RPS22B was found among NMD targets (Fig. 1), we tested whether NMD could complement Rnt1p-mediated degradation for this transcript by combining the upf1Δ deletion with a deletion of both stem-loops in the first intron of RPS22B (ΔSL1+2; see (Danin-Kreiselman et al., 2003). We then monitored the accumulation of unspliced RPS22B species in these mutants (Fig. 3E). While the deletion of the stem-loops or of UPF1 resulted in only a modest increase of unspliced pre-mRNAs, the double mutant showed a dramatic increase of partially spliced pre-mRNAs that retain intron 1 (Fig. 3E). An increase in partially spliced RPS22B transcripts that retain intron 1 was also observed when the stem-loop deletion was combined to Upf2p or Upf3p inactivation (Fig. 3E). These results show that the NMD pathway complements Rnt1p-mediated degradation in preventing the accumulation of unspliced RPS22B species. We also found that the amount of RPS22B spliced mRNAs were increased in the upf1Δ, ΔSL1+2 double mutant, compared to wild-type or to the corresponding single mutants. This result was also observed when Upf2p or Upf3p were inactivated in addition to the stem-loop deletion (Fig. 3D). In contrast, the amount of snR44 was not increased in any of the double mutants (Fig. 3D, snR44 probe). This result shows that both Rnt1p-mediated degradation, and more surprisingly the NMD pathway compete with splicing. These two degradation pathways thereby titrate a significant fraction of precursors away from the splicing pathway, rendering precursor degradation rate-limiting for the production of spliced RPS22B mRNAs.
Intron identity is a key determinant for targeting of unspliced pre-mRNAs to NMD
We next investigated the determinants of NMD targeting for unspliced transcripts. Experiments described in Supplemental Materials showed that UTR sequences, promoter or chromatin environment were not likely to be major determinants for NMD targeting. Therefore we focused our investigations on the role of intronic elements using an intron replacement strategy. We replaced the small intron (80nt) of the GOT1 gene, which is NMD-insensitive with those of RPL19A or RPS11B, which are NMD-sensitive. The lack of sensitivity of GOT1 to NMD is not due to the lack of PTCs, as these were found in unspliced GOT1. Intron replacement was performed by homologous recombination at the GOT1 chromosomal locus using the delitto perfetto technique (Storici et al., 2001), followed by UPF1 knockout. When the GOT1 intron was replaced with that of RPL19A, unspliced chimeric GOT1-RPL19A precursors were found to accumulate, and this accumulation was strongly exacerbated by Upf1p inactivation (Fig. 4A). The same result was observed when the GOT1 intron was replaced by that of RPS11B (Fig.4B). A doublet of bands was observed for unspliced chimeric GOT1-RPS11B precursors, which might indicate, in addition to the bona fide unspliced transcripts, a cryptic splice site or cryptic transcription initiation event triggered by the insertion of the RPS11B intron into the GOT1 gene. To rule out that modifying GOT1 intronic structure perturbs its splicing efficiency, we replaced the GOT1 intron by those of ACT1 or RPL25, which are NMD-insensitive. In contrast to the result observed with RPL19A or RPS11B, we did not detect unspliced precursor accumulation when the GOT1 intron was replaced with ACT1 or RPL25, even in the absence of Upf1p (Fig. 4C). This was not due to the lack of PTCs, as many could be found in these chimeric precursors (not shown). Finally, the intron sizes of RPL19A (506nt), RPS11B (511nt), ACT1 (305nt) or RPL25 (414nt) are comparable, ruling out a size-specific effect. This experiment unambiguously shows that intron identity is a major determinant of NMD targeting, and that substituting the intron of an NMD-insensitive transcript (GOT1) by one of an NMD-sensitive one (RPL19A or RPS11B) is sufficient to target this transcript to NMD.
Figure 4. NMD targeting is influenced by intron identity and suboptimal splicing signals.
A. Detection of spliced and unspliced GOT1 transcripts (GOT1 riboprobe) in wild-type, prp2-ts, upf1Δ, and in a strains where the natural GOT1 intron was replaced by that of RPL19A, and in the same strain where Upf1p was inactivated. B. Similar as A, except that the GOT1 intron was replaced by that of RPS11B. C. Similar as A, except that the GOT1 intron was replaced by that of actin (ACT1) or RPL25. D. Legends as in A. Strains indicated as N contain the natural RPL19A branchpoint (BP) UAACUAAC; Strains indicated as CS contain the consensus sequence UACUAAC.
Introns with suboptimal splicing signals are enriched among NMD targets, and the accumulation of unspliced RPL19A in the upf1Δ mutant can be prevented by changing its suboptimal branchpoint to the consensus sequence
The previous experiments showed that intron identity is a major determinant of NMD targeting. This effect was not likely due to PTC position of the within the intron (which would mimic the faux UTR model; Amrani et al., 2004), as transcripts with different intron sizes, 3’ exon sizes and PTC positioning are found in NMD-sensitive and NMD-insensitive clusters (not shown). Thus, we investigated whether NMD-sensitive ICGs are enriched for introns containing suboptimal splicing signals. When we analyzed the proportion of introns containing suboptimal splicing signals (i.e. that contain either a 5’ splice site that differs from the canonical GUAUGU, and/or a branchpoint that deviates from the canonical UACUAAC), we found that they comprise 46% of NMD sensitive introns (45/98) compared to 35% for NMD-insensitive introns (67/189). This significant (P<0.05) enrichment for suboptimal splicing signals in NMD-sensitive introns suggested that unspliced precursors might accumulate in NMD mutants because of their intrinsically inefficient splicing. To test the hypothesis of a contribution of suboptimal splicing signals to NMD targeting, we mutated the branchpoint of the RPL19A intron that was previously transposed in GOT1. The suboptimal UAACUAAC sequence was changed to the consensus UACUAAC by delitto perfetto, followed by Upf1 inactivation. Although this mutation had no effect on GOT1 expression in the context of active NMD (Fig. 4D, lanes 1and 2), it was sufficient to prevent unspliced GOT1 pre-mRNAs accumulation when Upf1p was inactivated (Fig.4D). This experiment demonstrates that NMD targeting resides, at least for RPL19A and for a fraction of NMD-sensitive ICGs, in their suboptimal splicing signals that drive production of unspliced precursors.
Unspliced RPS10B transcripts resulting from a 5’ splice site mutation are stabilized by NMD inactivation
Unspliced precursors resulting from splice site or branchpoint mutations of ACT1 or RP51 are typically detectable without NMD perturbation (Chanfreau et al., 1994; Hilleren and Parker, 2003; Vijayraghavan et al., 1986). However, the previous results suggested that NMD might be involved in degrading unspliced transcripts generated by splice site mutations. We tested this hypothesis by introducing a mutation in the 5’ splice site (SS) of RPS10B. This gene was chosen because inactivation of Upf1p resulted in increased RPS10B intronic signal by tiling arrays (Suppl. Fig. S4). The 5’SS was mutated from GUAUGU to GcAUaU at the RPS10B chromosomal locus by delitto perfetto. As expected, this mutation resulted in a strong decrease of spliced RPS10B (Fig. 5A). While this mutation is expected to completely block RPS10B splicing, the residual spliced mRNA detected might be due to cross-hybridization of the riboprobe with the RPS10A paralogue. More importantly, and in contrast to results observed with ACT1 or RP51 – based reporters (Chanfreau et al., 1994; Hilleren and Parker, 2003; Vijayraghavan et al., 1986), unspliced precursors did not accumulate to high levels in this 5’SS mutant strain. However when this 5’SS mutation was combined with deletion of UPF1, UPF2 or UPF3, all double-mutants showed a dramatic stabilization of unspliced RPS10B pre-mRNAs (Fig. 5A). This result shows that NMD targets unspliced mutant transcripts of the RPS10B gene for degradation, and that these unspliced species do not accumulate to high levels unless NMD is inactivated. We also tested whether inactivation of the nuclear exosome component Rrp6p would exacerbate RPS10B precursors accumulation in the RPS10B 5’SS mutant. Strikingly, Rrp6p inactivation in this mutant resulted only in a slight increase of unspliced precursors (Fig.5B), indicating that these transcripts are in majority targeted by NMD, and that the nuclear exosome only plays a modest role in their degradation. When the same 5’SS mutation was introduced into the ACT1 intron transposed into the GOT1 gene (see Fig.4), unspliced precursors levels were not exacerbated by NMD inactivation (Fig.5C). This result is consistent with previous results showing that unspliced ACT1 precursors are immune to NMD (Hilleren and Parker, 2003) and confirms that different precursor transcripts obey different modes of degradation, as the ACT1 precursor and several other unspliced transcripts resulting from a prp2 mutation are discarded through degradation pathways other than NMD (Bousquet-Antonelli et al., 2000; Hilleren and Parker, 2003).
Figure 5. NMD inactivation exacerbates accumulation of unspliced precursors generated by a 5’ splice site mutation of RPS10B.
A. Detection of RPS10B transcripts in strains carrying a wild-type RPS10B gene or with a 5’ splice site (5’SS) mutation (M) in RPS10B, alone or in combination with deletion of UPF1, UPF2 or UPF3. Legends as in Figure 2. B. Legends as in A, with the effects of the inactivation of Rrp6p. C. Legends as A, except that the 5’SS mutation was introduced in the ACT1 intron that was transposed in the GOT1 gene (see Fig. 4B).
Unspliced RPS10B precursors are degraded by NMD in the cytoplasm and maybe targeted to P-bodies
A yeast NMD reporter substrate was previously shown to be targeted to cytosolic P-bodies (Sheth and Parker, 2006). If unspliced precursors are NMD targets, we expect that they should accumulate in P-bodies in the absence of functional NMD. In addition, we would expect them to accumulate in diffuse cytoplasmic signal in the absence of Upf1p, which is required for P-body formation and substrate targeting (Sheth and Parker, 2006). To test this hypothesis, we investigated the localization of unspliced RPS10B precursors using fluorescence in situ hybridization. In the context of functional NMD, unspliced RPS10B transcripts that accumulate naturally are detected exclusively in the nucleus (Fig. 6A), adjacent or overlapping with the DAPI staining (80% of cells, n=60), possibly because unspliced transcripts are degraded quickly once they reach the cytoplasm. Strikingly, in strains lacking NMD components Upf1p or Upf2p, unspliced RPS10B were found mostly in the cytoplasm (Fig.6A; upf1Δ: 95.2 % of cells, n=63; upf2Δ: 72.3% of cells, n=36). In upf2Δ cells, this cytoplasmic signal was concentrated in multiple foci that we suspect to be P-bodies (Fig.6A). In contrast to unspliced RPS10B pre-mRNAs, the nuclear localization of the control U14 snoRNA transcripts was unaffected in NMD mutants (Fig.6B). These results are consistent with previous studies describing the localization of a reporter NMD substrate in P-bodies in the absence of Upf2p (Sheth and Parker, 2006). We could not demonstrate formally that those are P-bodies, but given their cytoplasmic localization and previous demonstration that yeast NMD substrates are targeted to P-bodies (Sheth and Parker, 2006), this makes this conclusion very likely.
Figure 6. Localization of unspliced RPS10B precursors by Fluoresencent In Situ Hybridization (FISH).
A. Shown are the DAPI staining (blue) for nuclear DNA, intron RPS10B FISH (red) and merged images for the wild-type, upf1Δ and upf2Δ strains. B. U14 snoRNA FISH (red) is shown as a control in the same strains.
NMD limits the accumulation of unspliced precursors during amino acid starvation
Unspliced precursors of many RPGs accumulate during amino acid starvation (Pleiss et al., 2007). These results have been interpreted as a selective inhibition of splicing of these RPGs during this stress. We noticed a large overlap of NMD targets detected in this study and of RPGs affected by amino acid starvation (Fig. 7A). Furthermore, it has been shown that amino acid starvation can inhibit NMD (Mendell et al., 2004). Therefore an accumulation of unspliced RPGs in amino acid starvation could be interpreted by a possible inactivation of NMD and by the inhibition of the degradation of some of these unspliced precursors in these conditions. We therefore investigated whether NMD inactivation would be epistatic to the unspliced precursor accumulation phenotype observed during amino acid starvation. Wild-type and upf1Δ strains were shifted to a medium containing 50 mM aminotriazole (3AT) to induce amino acid starvation. The level of unspliced RPS29B and RPL22B, which were found to be both NMD-sensitive and affected by amino acid starvation, was then assessed by northern blot. In wild-type cells, unspliced precursors of RPS29B and RPL22B were detected transiently after 10 minutes in 3AT-medium, after which their levels reverted to close to normal levels (Fig. 7). We observed the same increase of unspliced RPS29B precursors in the upf1Δ strain, showing that this increase was not likely due to NMD inhibition since NMD is defective in this mutant. The accumulation of unspliced transcripts was however higher in the upf1Δ strain than in wild-type, indicating that NMD is used to limit the accumulation of unspliced RPS29B precursors generated during amino acid starvation. We could not detect an increase of unspliced precursors for RPL22B in the upf1Δ strain, possibly because the very high levels of unspliced precursors observed before the shift could mask the small increase of unspliced transcripts observed in the wild-type (1.8 fold). As a negative control, RPL25, which is not an NMD target and was not found to be affected by amino acids starvation did not exhibit unspliced transcripts accumulation during this treatment (Fig. 7). In conclusion, the results observed with RPS29B show that NMD is used to limit the amount of some unspliced transcripts resulting from splicing inhibition during amino acid starvation.
Figure 7. Detection of unspliced precursors in wild-type and upf1 Δ strains during Camino acid starvation.
A. Venn diagram of the overlap between Intron-containing RPGs, ICGs affected by Upf1p inactivation, and ICGs for which splicing is inhibited by amino-acids starvation (Pleiss et al. 2007). B. Indicated strains were shifted for 10 or 20 minutes in a medium containing 50mM 3AT or untreated (mock), and probed for the indicated RPGs. Relative amount of unspliced precursors in the different conditions was standardized to the mock value showing the lowest amount of unspliced precursors.
Discussion
In this study we revisited the effect of the inactivation of the Upf1p and Xrn1p components of the NMD system on unspliced pre-mRNA accumulation in yeast. We show using tiling arrays that a significant fraction of the yeast intronome population is affected by NMD inactivation, as 31% of RPGs and 33% of all introns show an accumulation of unspliced precursors when Upf1p is inactivated. The phenotypes observed in the upf1Δ or xrn1Δ strains are most of the time similar (Fig. 1C). Some exceptions were found, including YRA1, which has been shown to rely on a sequence-specific degradation system to degrade its inefficiently spliced precursors (Dong et al., 2007; Preker and Guthrie, 2006). Previous splicing microarray analysis of the upf1Δ, xrn1Δ and upf3Δ mutants failed to reveal a widespread effect of NMD mutations on unspliced precursor accumulation (Burckin et al., 2005; Clark et al., 2002; Pleiss et al., 20 2007), and in particular on RPGs (Pleiss et al., 2007). While these results may seem contradictory to those presented here, it should be noted that these studies were focused on detecting the effects of splicing mutations or inhibition during stress. Since the accumulation of unspliced precursors is usually higher in splicing mutants than in NMD mutants (Fig.2), it is possible that these previous studies did not consider significant the increase of unspliced precursors observed in NMD mutants. Another possibility to explain this apparent discrepancy may be that using tiling arrays, which cover a large fraction of the intronic sequences increased the detection of smaller effects, leading to a higher sensitivity. Regardless of these differences, our results, combined with studies showing intronic detection by tiling arrays in the debranching enzyme mutant (Juneau et al., 2007; Zhang et al., 2007) demonstrate that these arrays are efficient to detect changes in ICGs transcripts accumulation, as differences predicted by tiling array profiles were confirmed by northern blot analysis (Fig.2B).
What are the determinants of NMD targeting for unspliced yeast pre-mRNAs?
In the case of transcripts for which we did not detect any unspliced pre-mRNAs accumulation in upf1Δ or xrn1Δ strains, we do not know whether this reflects very efficient splicing, or whether some precursors escape splicing but are degraded by alternative degradation pathways. In the first hypothesis, these transcripts might represent a subpopulation with a high level of cotranscriptional splicing (Moore et al., 2006; Tardiff et al., 2006), minimizing the production of unspliced pre-mRNAs. However, many ICGs which do not show co-transcriptional spliceosome recruitment do not show any unspliced precursor accumulation in NMD mutants either (data not shown). Results from Moore et al. (2006) are consistent with the hypothesis that these transcripts are retained in the nucleus by Mlp1p and degraded by the nuclear exosome. Therefore these results would be inconsistent with the idea that a lack of co-transcriptional splicing favors NMD targeting. Instead, we prefer the idea that unspliced precursors that accumulate in upf1Δ or xrn1Δ strains correspond to inefficiently spliced transcripts, not because of a lack of co-transcriptional splicing but because of intronic features. In favor of this hypothesis is the observation that introns containing suboptimal splicing signals are enriched in NMD targets, and that restoring the suboptimal branchpoint of the RPL19A intron back to the canonical sequence is sufficient to prevent unspliced precursors accumulation in the upf1Δ strain (Fig.4D). This strongly suggests that NMD targeting is determined by intronic features, which ultimately drives splicing efficiency. The suboptimal splicing of the other fraction of NMD-sensitive transcripts that present canonical splice site signals could result from competing secondary structures (Goguel et al., 1993), or trans-acting effects. This model cannot completely explain why certain precursors resulting from splicing mutations, including the historical ACT1 model are immune to NMD (Fig. 5C). These transcripts are degraded by other degradation systems such as the nuclear exosome for some meiotic pre-mRNAs (Moore et al., 2006) or by Xrn1p in the cytoplasm (Hilleren and Parker, 2003; Scherrer and Spingola, 2006). The determinants of this differential targeting remain to be investigated.
Significance of NMD-dependent degradation of unspliced pre-mRNAs
The involvement of NMD in the degradation of a large number of unspliced transcript is not unprecedented and seems to be conserved throughout eukaryotic evolution. In C. elegans, NMD degrades inefficiently spliced ribosomal protein precursor transcripts (Mitrovich and Anderson, 2000) and in Paramecia, NMD also limits the accumulation of unspliced precursors (Jaillon et al., 2008). Therefore, this phylogenetic conservation probably reflects an important role for NMD in the degradation of unspliced precursors of suboptimally spliced transcripts. This function is important for cell survival as the deletion of the NMD component Upf1p is synthetic lethal with the inactivation of the splicing/nuclear retention factor BBP/ScSF1 (Rutz and Seraphin, 2000). This suggests that NMD might be required to prevent the cellular toxicity generated by high amounts of unspliced cytoplasmic transcripts.
Ribosomal protein genes are highly and tightly regulated at many levels in yeast, including splicing (Warner, 1999; Zhao et al., 2003). The large number of RPGs identified here as NMD targets suggests that NMD might be involved in preventing the accumulation of unspliced RPG transcripts generated during regulated splicing throughout various physiological conditions. To illustrate this point, we found that NMD limits the levels of unspliced RPS29B precursors generated by splicing inhibition during amino acid starvation (Pleiss et al., 2007). Thus NMD plays a role in limiting accumulation of unspliced precursors during a known physiological stress, and it could play a similar role under other conditions that have yet to be determined, such as other stresses, or life stages that result in modulations of splicing efficiency.
Experimental Procedures
Strains and media
The list of yeast strains used in this study is provided in the Supplemental materials. Strains used for tiling array profiling were grown in synthetic complete medium. All other strains were grown in YPD. Cells were harvested during log phase (O.D.600 0.4 to 0.8). Amino acid starvation experiments were performed as described (Pleiss et al., 2007a). Strains were constructed via one-step PCR product-based gene disruption (Longtine et al., 1998) or by delitto perfetto (Storici et al., 2001).
Microarray Analysis
Four biological replicates of wild-type, upf1Δ and xrn1Δ from the BY4741 background were grown independently and analyzed by Affymetrix yeast tiling microarray analysis. Probes preparation, hybridization and tiling array scanning were performed according to the manufacturer’s instructions. Methods used for bioinformatics analysis of the tiling arrays are described in Supplemental materials. Microarray data are accessible in the GEO database (Accession Number GSE11621).
Northern blots and Fluorescent In Situ Hybridization
Northern blots were hybridized with radioactive probes in Church buffer (250mM sodium phosphate, pH 7.2; 1% bovine serum albumin; 7% sodium dodecyl sulfate, and 1mM EDTA) at 67–70°C overnight, washed twice at room temperature with 2X SSPE with 0.1% SDS for 20 minutes, then at room temperature in 0.1XSSPE, 0.1% SDS. A last wash with pre-warmed 0.1X SSPE buffer with 0.1%SDS was performed at 67°C for 5 minutes. All probes were antisense riboprobes with the exception of the G3PDH and SCR1 probes, generated by random priming from PCR products. Riboprobes were synthesized using Ambion T3 MaxiScript kits (Supplemental Materials) from PCR templates in which a T3 RNA polymerase promoter had been incorporated through the reverse primer. Blots were imaged using a Molecular Dynamics Phosphorimager Scanner or a compatible Bio-Rad scanner system. FISH with the RPS10B intronic probe was performed as described in Supplemental Materials. FISH with the U14 probe was performed as described (Henras et al., 2004).
Supplementary Material
Acknowledgements
We thank the UCLA Microarray Core Facility for help with microarray hybridization and scanning, Lauren May for constructing the strain carrying a double mutation in Rrp6 and the 5’ splice site of RPS10B, J. Gober for use of his fluorescence microscope, J. Pleiss and C. Guthrie for communication of results prior to publication and D.L. Black, C. Coffinier and D. Egecioglu for critical reading of the manuscript. Supported by NIH grant GM61518 to G.C. The authors declare that they do not have a conflict of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3'-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- Arciga-Reyes L, Wootton L, Kieffer M, Davies B. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 2006;47:480–489. doi: 10.1111/j.1365-313X.2006.02802.x. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–2853. doi: 10.1016/j.febslet.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- Burckin T, Nagel R, Mandel-Gutfreund Y, Shiue L, Clark TA, Chong JL, Chang TH, Squazzo S, Hartzog G, Ares M., Jr Exploring functional relationships between components of the gene expression machinery. Nat Struct Mol Biol. 2005;12:175–182. doi: 10.1038/nsmb891. [DOI] [PubMed] [Google Scholar]
- Chanfreau G, Legrain P, Dujon B, Jacquier A. Interaction between the first and last nucleotides of pre-mRNA introns is a determinant of 3' splice site selection in S. cerevisiae. Nucleic Acids Res. 1994;22:1981–1987. doi: 10.1093/nar/22.11.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF. The Nonsense-Mediated Decay RNA Surveillance Pathway. Annu Rev Biochem. 2007 doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Clark TA, Sugnet CW, Ares M., Jr Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr Opin Cell Biol. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Danin-Kreiselman M, Lee CY, Chanfreau G. RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol Cell. 2003;11:1279–1289. doi: 10.1016/s1097-2765(03)00137-0. [DOI] [PubMed] [Google Scholar]
- Dong S, Li C, Zenklusen D, Singer RH, Jacobson A, He F. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol Cell. 2007;25:559–573. doi: 10.1016/j.molcel.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 2004;116:63–73. doi: 10.1016/s0092-8674(03)01026-2. [DOI] [PubMed] [Google Scholar]
- Goguel V, Wang Y, Rosbash M. Short artificial hairpins sequester splicing signals and inhibit yeast pre-mRNA splicing. Mol Cell Biol. 1993;13:6841–6848. doi: 10.1128/mcb.13.11.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Lewis BP, Hillman RT, Blanchette M, Lareau LF, Garnett AT, Rio DC, Brenner SE. Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics. 2003;19 Suppl 1:i118–i121. doi: 10.1093/bioinformatics/btg1015. [DOI] [PubMed] [Google Scholar]
- Guan Q, Zheng W, Tang S, Liu X, Zinkel RA, Tsui KW, Yandell BS, Culbertson MR. Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast. PLoS Genet. 2006;2:e203. doi: 10.1371/journal.pgen.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Jacobson A. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol Cell Biol. 2001;21:1515–1530. doi: 10.1128/MCB.21.5.1515-1530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5' to 3' mRNA decay pathways in yeast. Mol Cell. 2003;12:1439–1452. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1 - mutant. Proc Natl Acad Sci U S A. 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Bertrand E, Chanfreau G. A cotranscriptional model for 3'-end processing of the Saccharomyces cerevisiae pre-ribosomal RNA precursor. RNA. 2004;10:1572–1585. doi: 10.1261/rna.7750804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren PJ, Parker R. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol Cell. 2003;12:1453–1465. doi: 10.1016/s1097-2765(03)00488-x. [DOI] [PubMed] [Google Scholar]
- Hillman RT, Green RE, Brenner SE. An unappreciated role for RNA surveillance. Genome Biol. 2004;5:R8. doi: 10.1186/gb-2004-5-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Watanabe Y. UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J. 2005;43:530–540. doi: 10.1111/j.1365-313X.2005.02473.x. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Bouhouche K, Gout JF, Aury JM, Noel B, Saudemont B, Nowacki M, Serrano V, Porcel BM, Segurens B, et al. Translational control of intron splicing in eukaryotes. Nature. 2008;451:359–362. doi: 10.1038/nature06495. [DOI] [PubMed] [Google Scholar]
- Johansson MJ, He F, Spatrick P, Li C, Jacobson A. Association of yeast Upf1p with direct substrates of the NMD pathway. Proc Natl Acad Sci USA. 2007;104:20872–20877. doi: 10.1073/pnas.0709257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneau K, Palm C, Miranda M, Davis RW. High-density yeast-tiling array reveals previously undiscovered introns and extensive regulation of meiotic splicing. Proc Natl Acad Sci USA. 2007;104:1522–1527. doi: 10.1073/pnas.0610354104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Leeds P, Wood JM, Lee BS, Culbertson MR. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Lelivelt MJ, Culbertson MR. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol. 1999;19:6710–6719. doi: 10.1128/mcb.19.10.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Paulovich AG, Woolford JL., Jr Feedback inhibition of the yeast ribosomal protein gene CRY2 is mediated by the nucleotide sequence and secondary structure of CRY2 pre-mRNA. Mol Cell Biol. 1995;15:6454–6464. doi: 10.1128/mcb.15.11.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- Mitrovich QM, Anderson P. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev. 2000;14:2173–2184. doi: 10.1101/gad.819900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovich QM, Anderson P. mRNA surveillance of expressed pseudogenes in C. elegans. Curr Biol. 2005;15:963–967. doi: 10.1016/j.cub.2005.04.055. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Schwartzfarb EM, Silver PA, Yu MC. Differential recruitment of the splicing machinery during transcription predicts genome-wide patterns of mRNA splicing. Mol Cell. 2006;24:903–915. doi: 10.1016/j.molcel.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol Cell. 2007;27:928–937. doi: 10.1016/j.molcel.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker PJ, Guthrie C. Autoregulation of the mRNA export factor Yra1p requires inefficient splicing of its pre-mRNA. RNA. 2006;12:994–1006. doi: 10.1261/rna.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz B, Seraphin B. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J. 2000;19:1873–1886. doi: 10.1093/emboj/19.8.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer FW, Jr, Spingola M. A subset of Mer1p-dependent introns requires Bud13p for splicing activation and nuclear retention. RNA. 2006;12:1361–1372. doi: 10.1261/rna.2276806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F, Lewis LK, Resnick MA. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 2001;19:773–776. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
- Tardiff DF, Lacadie SA, Rosbash M. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol Cell. 2006;24:917–929. doi: 10.1016/j.molcel.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan U, Parker R, Tamm J, Iimura Y, Rossi J, Abelson J, Guthrie C. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986;5:1683–1695. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Hesselberth JR, Fields S. Genome-wide identification of spliced introns using a tiling microarray. Genome Res. 2007;17:503–509. doi: 10.1101/gr.6049107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sohn JH, Warner JR. Autoregulation in the biosynthesis of ribosomes. Mol Cell Biol. 2003;23:699–707. doi: 10.1128/MCB.23.2.699-707.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.