Abstract
Amelogenin is the predominant matrix protein in developing dental enamel. Making extensive use of residue-specific 15N-labeled amino acids samples, the majority of the main and side chain resonances for murine amelogenin were assigned in 2% aqueous acetic acid at pH 3.0.
Keywords: Enamel, Amelogenin, Biomineralization, Unfolded protein, Polyproline type II structure
Biological context
The hardest tissue in the human body is the dental enamel on the outer layer of the tooth (Ten Cate 1994). Enamel’s strength and exceptional functional properties are due to a combination of high mineral content and structural organization. Ninety-five percent of mature enamel consists of long and narrow crystals of carbonated hydroxyapatite packed into parallel arrays, called enamel rods, that are intricately interwoven together (Margolis et al. 2006). While there is little, if any, matrix protein remaining in mature enamel, the nucleation, growth, and organization of enamel formation (amelogenesis) are extracellular processes orchestrated by the enamel matrix proteins, predominantly made up of amelogenin (Termine et al. 1980). Specific mutations to amelogenin and amelogenin deficient mice both result in dramatic enamel phenotypes similar to amelogenesis imperfecta in humans (Margolis et al. 2006), observations that strongly suggest amelogenin plays an essential role in amelogenesis. Despite much effort, the precise mechanisms by which amelogenin regulates biomineralization to form mature enamel is not well-understood.
Mouse amelogenin is a 180-residue protein (Snead et al. 1986) that has been studied extensively by circular dichroism (CD), Infrared Spectroscopy and small angle X-ray scattering (SAXS) (Margolis et al. 2006 and references within). The general consensus is that in monomeric form, amelogenin adopts an extended β-strand/β-spiral/polyproline type II “structure” (Margolis et al. 2006). This “structure” appears to be very sensitive to pH, temperature, ionic strength, and metal ions. To date there has been no corroboration of the CD and SAXS defined structures for amelogenin with NMR spectroscopy largely because the highly repetitive nature of the core HQP-region made backbone assignment a daunting task. By preparing nine residue-specific, 15N-labeled amino acid samples, collecting data at high spectrometer frequencies (1H resonance frequencies between 750 and 900 MHz), and making extensive use of the HNN experiment (Planchal et al. 2001), we have successfully assigned the majority of the backbone and side chain 1H, 13C and 15N resonances for murine amelogenin.
Materials and methods
Cloning, expression, and purification
Recombinant mouse amelogenin containing a 12-residue, N-terminal histidine tag (MRGSHHHHHHGS-) to assist protein purification (rp(H)M180) was expressed and purified from Escherichia coli BL21(DE3) cells using methods previously described (Simmer et al. 1994) and exchanged into the final NMR buffer (2% CD3CO2D, 5% D2O/95% H2O, pH 3.0). To assist the assignments, nine residue-specific, 15N-labeled amino acid (L, V, H, R, K, M, F, Y, and E/Q) samples were prepared by growing the cells in “Redfield-medium” (Griffey et al. 1985; Buchko et al. 2005), a medium where 19 out of the 20 common amino acids are included. To prevent 15N-isotopic scrambling of the target, residue-specific, 15N-labeled amino acid due to endogenous amino acid biosynthesis (Waugh 1996), only ~50% of the unlabeled amino acid of interest was initially added to the medium. At an OD600 of ~0.8 the remaining 25% of the targeted amino acid containing 15N at the amide position (CIL, Andover, MA) was added and protein expression induced at 298 K (Buchko et al. 2005, 2007). The logic in keeping the target amino acid at 75% of the level called for in the “Redfield-medium” recipe (Griffey et al. 1985) is that it minimizes the likelihood that the cells will use the 15N-labeled amino acid as a precursor for other amino acid residues in endogenous amino acid biosynthetic pathways (Waugh 1996). Approximately 25 min after induction the cells were harvested and processed.
Nuclear magnetic resonance spectroscopy
The NMR data on the double-labeled samples (~2 mM) were collected at 298 K using Varian Inova-600, and -750, -800, and -900 spectrometers equipped with triple resonance probes, cyroprobes (Inova-600 and -800), and pulse field gradients. Pulse sequences for the two- and three-dimensional experiments were from BioPack. Twodimensional 1H–15N HSQC spectra of the residue-specific labeled samples (~0.5 mM) were collected in 2–12 h depending on the sample concentration, degree of labeling, and the resolution required. Carbon chemical shifts were obtained from three-dimensional HNCACB, CCC-TOCSY, HNCO, and HNCACO data collected on the double-labeled sample at 1H resonance frequencies of 750, 800, and 900 MHz. Proton chemical shifts were extracted primarily from a 3D 15N-edited TOCSY experiment. Due to the unstructured nature of M180, the HNN experiment proved especially useful for making sequence specific amide assignments (Planchal et al. 2001). All NMR data was processed using Felix 2007 (Felix NMR, Inc., San Diego, CA) software and the 1H, 13C, and 15N chemical shifts were referenced to DSS (DSS = 0 ppm) using indirect methods. The assigned 1H, 13C, and 15N chemical shifts have been deposited in the BioMagResBank database (www.bmrb.wisc.edu) under the accession number BMRB-15662.
Results and discussion
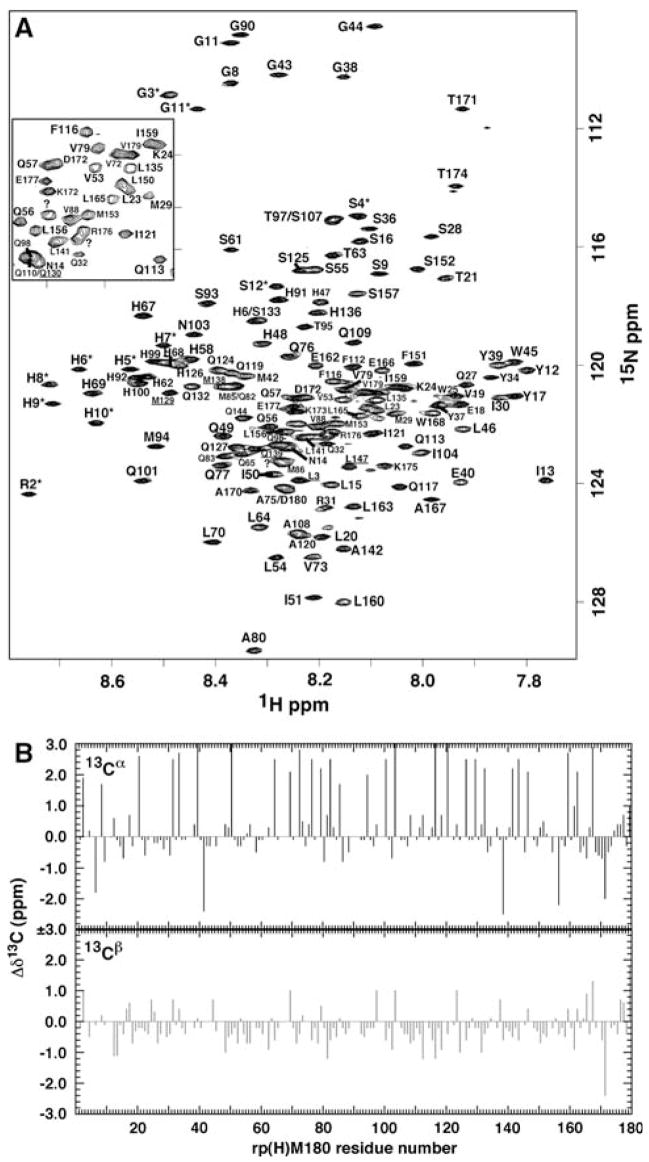
The 1H-15N HSQC spectrum for rp(H)M180 is shown in Fig. 1A. While the resolution in the spectrum was assisted by collecting the data at a 1H resonance frequency of 900 MHz, the most striking feature is that the majority of amide cross peaks in the 191-residue protein are well resolved. Indeed, using a strategy similar to the one employed to assign the backbone for the natively disordered protein HMGA1 (Buchko et al. 2007), it was possible to assign 143 out of the 146 expected amide cross peaks in rp(H)M180 (191 − (44 prolines + M1*)) as illustrated in Fig. 1A. Of these 143 amide cross peaks, six could not be unambiguously assigned due to degeneracy in the sequence, -PLP-, and -PMQP-. Note that while most of the amide cross peaks are resolved in the 1H–15N HSQC spectrum, an observation that suggests rp(H)M180 adopts some type of unique “structure” in solution, the chemical shift dispersion in the proton dimension is relatively narrow, at ~1.0 ppm, perhaps suggesting that any unique structural feature is common to the majority of the protein.
Fig. 1.
(A) The 1H–15N HSQC spectrum for double-labeled rp(H)M180 collected at a proton resonance frequency of 900 MHz, 298 K, in 2% aqueous (95% H2O/5% D2O) CD3CO2D, pH 3.0. The residues for the N-terminal affinity tag, MRGSHHHHHHGS-, are number 1–12 with an asterisk and the amelogenin residues are number sequentially starting from P2 through to D180. The underlined assignments for L3 and L15, M129 and M138, and Q130 and Q139 are ambiguous and may be reversed. The amide cross peak for Y26 is not shown (7.60 and 121.0 ppm). The three unassigned cross peaks are indicated with a “?” and correspond to residues Q114, Q154, and E178. Inset is an expansion of a congested region of the spectrum. (B) Analysis of the observed 13Cα and 13Cβ chemical shift deviations from random coil values for rp(H)M180 where Δδ13C = δRandom Coil − δObserved. Solid = 13Cα, Open = 13Cβ. Random coils values are from CNS (cns_solve_1.1)
A rapid insight into the nature of any unique, structural feature common to the majority of rp(H)M180 may be obtained through an analysis of the 13Cα and 13Cβ chemical shifts. Relative to a random coil, well-defined changes in the carbon chemical shifts are associated with α-helical (negative Δδ13Cα, positive Δδ13Cβ) and β-sheet (positive Δδ13Cα, negative Δδ13Cβ) secondary structure. As shown in Fig. 1B, the majority of the 13Cα and 13Cβ chemical shifts for rp(H)M180 do not significantly deviate from random coil values. However, there does appear to be a propensity towards positive Δδ13Cα values and negative Δδ13Cβ values, suggesting a predisposition towards β-strand character. This observation corroborates with CD and SAXS studies that suggest monomeric amelogenin adopts an extended β-strand/β-spiral/polyprolineII type structure in solution (Margolis et al. 2006). Efforts are in progress to address the extent of such an extended structure via the analysis of 3JHNα coupling constants and NOE data (Parrot et al. 2002).
In summary, we have shown that it is possible to assign the fingerprint 1H–15N HSQC spectrum of a ~20 kDa “unstructured” protein using a combination of residue specific labels, novel pulse programs, and high-field NMR spectrometers. More importantly, the NMR resonance assignments reported here for amelogenin will allow us to better define how “unstructured” amelogenin is before amelogenesis begins. Indeed, these assignments may allow us to closely follow the initial steps of biomineralization.
Acknowledgments
This work was supported by NIH-NIDCR Grant DE-015347. The research was performed at the Pacific Northwest National Laboratory (PNNL), a facility operated by Battelle for the U.S. Department of Energy. Part of the research was performed at the W.R. Wiley Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by U.S. Department of Energy’s Office of Biological and Environmental Research (BER) program.
References
- Buchko GW, McAteer K, Wallace SS, Kennedy MA. Solutionstate NMR investigation of DNA binding interactions in Escherichia coli formamidopyrimidine-DNA glycosylase (Fpg): a dynamic description of the DNA/protein interface. DNA Repair. 2005;4:327–339. doi: 10.1016/j.dnarep.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Buchko GW, Ni S, Lourette NM, Reeve R, Kennedy MA. NMR resonance assignments of the human high mobility group protein HMGA1. J Biomol NMR. 2007;38:185. doi: 10.1007/s10858-006-9116-8. [DOI] [PubMed] [Google Scholar]
- Griffey RH, Redfield AG, Loomis RE, Dahlquist FW. Nuclear magnetic resonance observations and dynamics of specific amide protons in T4 lysozyme. Biochemistry. 1985;24:817–822. doi: 10.1021/bi00325a001. [DOI] [PubMed] [Google Scholar]
- Margolis HC, Baniash E, Fowler CE. Role of macromolecular assembly of enamel matrix proteins in enamel formation. J Dent Res. 2006;85:775–793. doi: 10.1177/154405910608500902. [DOI] [PubMed] [Google Scholar]
- Parrot I, Huang PC, Khosla C. Circular dichroism and nuclear magnetic resonance analysis of immunogenic gluten peptides and their analogs. J Biol Chem. 2002;277:45572–45578. doi: 10.1074/jbc.M207606200. [DOI] [PubMed] [Google Scholar]
- Planchal SC, Bhavesh NS, Hosur RV. Improved 3D triple resonance HNN and HN(C)N, for HN and 15N sequential correlations in (13C, 15N) labeled proteins: application to unfolded proteins. J Biomol NMR. 2001;20:135–147. doi: 10.1023/a:1011239023422. [DOI] [PubMed] [Google Scholar]
- Simmer JP, Lau EC, Hu CC, Bringas P, Santos V, Aoba T, Lacey M, Nelson D, Zeichner-David M, Snead ML, Slavkin HC, Fincham AG. Isolation and characterization of a mouse amelogenin expressed in Escherichia coli. Calcif Tissue Int. 1994;54:312–319. doi: 10.1007/BF00295956. [DOI] [PubMed] [Google Scholar]
- Snead ML, Lau EC, Zeichner-David M, Fincham AG, Woo SL, Slavkin HC. DNA sequence for cloned cDNA for murine amelogenin reveals the amino acid sequence for enamel-specific proteins. Biochem Biophys Res Commun. 1986;129:812–818. doi: 10.1016/0006-291x(85)91964-3. [DOI] [PubMed] [Google Scholar]
- Ten Cate RA. Oral histology: development, structure, and function. Mosby-Year Book; St. Louis: 1994. [Google Scholar]
- Termine JD, Belcourt AB, Christner PJ, Conn KM, Nylen MU. Properties of dissociatively extracted fetal tooth matrix proteins. J Biol Chem. 1980;255:9760–9768. [PubMed] [Google Scholar]
- Waugh DS. Genetic tools for selective labeling of proteins with α-15N amino acids. J Biomol NMR. 1996;8:184–192. doi: 10.1007/BF00211164. [DOI] [PubMed] [Google Scholar]