Abstract
Human immunodeficiency virus (HIV) infection is associated with dysplastic changes in oral human papilloma virus (HPV) lesions, suggesting changes in keratinocytes. In the present study, we seek to identify proteomic changes in oral HPV lesions between HIV(+) and HIV(-) patients. While fresh tissues represent the most desirable samples for proteomic investigations, they are often difficult to obtain in large numbers under clinical settings. We therefore have developed a new method to identify protein changes in formalin fixed and paraffin-embedded (FFPE) oral HPV lesions utilizing iTRAQ™ technology in conjunction with Liquid Tissue® sample preparation method. Using this method, we identified nine proteins that were differentially expressed in oral HPV lesions as a result of HIV infection. The quantitative proteomic method presented here will be valuable for others who plan to analyze FFPE tissues.
Introduction
HPV is a DNA virus that infects the keratinocytes of stratified squamous epithelium, resulting in the development of cutaneous or mucosal warts (papillomas). Several oral papilloma variants are recognized, including conventional squamous papilloma, verruca vulgaris, and focal epithelial hyperplasia. The clinical and histological diversity of these lesions may be correlated to the viral subtypes present. The growth potential of papillomas in immunocompetent individuals is limited, and it is unusual to find more than one lesion at the time of examination. However, those infected with HIV can exhibit numerous lesions throughout the oral mucosa [1]. Atypical HPV subtypes have been reported in HIV patients [2, 3], and papillomas in these individuals may also show dysplastic epithelial changes [4]. The aggressive potential of HPV-induced lesions in the immunosuppressed suggests underlying genomic and proteomic changes in keratinocytes in the settings of HIV infection. In the present study, we seek to identify these proteomic changes in oral HPV lesions as a function of patient HIV-diagnostic-state HIV(+) or HIV(-).
Performing proteomic studies on tissues is important in order to dissect the specificity of protein involvement to the histological manifestation of diseases. While fresh and/or frozen tissues appear to represent the most desirable samples for proteomic investigations, they are often difficult to obtain in large numbers under clinical settings. Formalin fixed and paraffin embedded (FFPE) tissues have been used for decades to evaluate tissue histology and diagnose disease. Existing extensive archives of FFPE papilloma tissues at New Jersey Diagnostic Services represent a very accessible resource for this investigation. However, it is known that formaldehyde fixation process could cause intermolecular and intramolecular crosslinking of proteins and thus the crosslinked proteins would not be amenable to extraction and solubilization for biochemical analysis [5]. Recently, a new methodology, Liquid Tissue™, for extraction and procurement of soluble peptides directly from FFPE tissues has been developed by Expression Pathology (Gaithersburg, MD). Proteins are extracted and digested by trypsin from deparaffinized sections of FFPE tissues and the resulting peptides can be extracted and further analyzed by mass spectrometry (MS). Remarkable improvements in MS instrumentation and the rapid growth of genomic databases have made it possible to quantify large numbers of proteins from very complex samples. This can be achieved through multiple approaches including protein labeling with isotopically distinct “tags” and subsequent protein quantification by comparing the MS ion intensities [6, 7].
Various studies have attempted to perform protein quantification of FFPE tissues. For example, in a strategy known as AQUA (absolute quantification of protein) [8], using a synthetic tryptic peptide of a Prostate Specific Antigen, quantification has been performed on proteins extracted from FFPE prostate cancer tissue arrays [9]. However, this approach inherently focuses on the determination of quantities of only one or very few particular known proteins of interest. This is not a nonbiased discovery method, but may be very useful for biomarker validation of a large number of sample repeats, addressing the statistical limitation of the shotgun proteomics methods. Recently, using a spectral counting method, Patel et al. [10] have compared the protein abundance in FFPE tissues in normal and tumor head and neck squamous epithelium. Although very attractive, spectrum counting approach is contentious in the sense that for a reliable quantification, many spectra need to be observed for a given protein [11] and may not be sensitive enough for low abundant protein quantification. A primary amine group based isotope labeling technique, termed isobaric tags for relative and absolute quantification (iTRAQ™), has been developed [12]. It allows the comparison of up to four samples in a single experiment. In MS mode, differentially labeled versions of a peptide are indistinguishable. However, in tandem MS mode (MS/MS) peptides are isolated and fragmented and peptide quantification is achieved by comparing the intensities of the four iTRAQ™ reporter ions (mass/charge, m/z of 114, 115, 116 and 117). This permits the simultaneous determination of both identification and relative abundance of protein in up to four samples. This method can quantify more proteins than AQUA method, and is more sensitive than the spectral counting method for low abundant protein analysis.
As a novel approach for quantitative proteomic studies of FFPE tissues, we have been able to conduct iTRAQ™ analysis of peptides extracted with Liquid Tissue® technology. Successful application of this approach has enabled us to identify novel proteomic changes in oral HPV lesions in HIV patients.
Materials and Methodology
Cell Procurement
Following approval by the IRB, cases of oral papillomas were selected from the records of New Jersey Diagnostic Services. The patient's charts were reviewed to determine HIV status. For fixation, after collection from patients, samples were immersed immediately in freshly prepared solution of 10% neutral buffered formalin and fixed for 24 hours at room temperature. After fixation, the specimens were dehydrated through a graded alcohol series before embedding in paraffin wax which, following slide preparation and diagnosis, were archived at room temperature in New Jersey Diagnostic Services. For current studies, we used the samples which were archived for between two months to seven years. Total of ten representative cases were selected, creating one HIV(+) group containing 5 cases, and one HIV(-) group containing 5 cases. Ten micron thick sections of oral papillomas were cut from two HIV(+) cases and two HIV(-) cases using sterile microtome blades and placed on untreated microscope slides. The slides were deparaffinized in xylene, rehydrated to 70% ethanol, and placed in distilled water for 1 minute. Sections were dipped in hematoxylin for 5 minutes to allow subsequent visualization of the epithelium, and washed in double-distilled water for 5 minutes. Sixteen mm2 of epithelium (approximately 60,000 cells) were needle dissected for preparation of protein lysate.
Protein Extraction and Digestion
Protein lysates were prepared according to the manufacturer's recommendations (Expression Pathology Inc, Gaithersburg, MD). Procured cells were suspended in 20 μl of proprietary Liquid Tissue® buffer (US Patent application WO/2004/080579) and heated at 95°C for 90 minutes. After cooling for 2 minutes on ice, 1 μl of trypsin reagent was added and incubated at 37°C for one hour with vigorous shaking for one minute at 20 minute intervals. Samples were further incubated at 37°C for 18 hours followed by heating at 95°C for 5 minutes. In preparation for labeling, the samples were harvested via centrifugation at 10,000 g, dried completely using a Speed-Vac and re-suspended in 100 μl of 0.5% trifluoroacetic acid (TFA) in 5% acetonitrile, and were desalted via PepClean C-18 spin columns (Pierce Biotechnology, Rockford, IL) and dried for iTRAQ™ processing.
iTRAQ™ Sample Preparation and Tandem Mass Spectrometry (MS/MS)
Each peptide sample was processed according to manufacturer's protocol with minor modifications (Applied Biosystems Inc., ABI, Framingham, MA, USA). iTRAQ™ reagents were dissolved in 280 μl of ethanol. The peptides were labeled with the iTRAQ™ reagents as follows: two groups of HPV(+)/HIV(-) samples with iTRAQ™ tags 114 and 115 and two groups of HPV(+)/HIV(+) samples with iTRAQ™ tags 116 and 117. A mixture containing small aliquots from each labeled sample was analyzed by MS/MS to determine a proper mixing ratio to correct for unevenness [13] in peptide yield from Liquid Tissues procedures. Labeled peptides were then mixed in 1:1:1:1 ratio and analyzed as described previously [14]. Briefly, peptide mixture was dried completely using a Speed-Vac and re-suspended in 0.5 ml of cation exchange mobile phase A (10 mM KH2PO4 and 20% acetonitrile (ACN) (pH 3.0)). The peptides were first fractionated on a BioCAD™ Perfusion Chromatography System (ABI) equipped with a polysulfoethyl A column (4.6 mm × 200 mm, 5 μm, 300 Å, Poly LC Inc., Columbia, MD, USA) plus an upstream guard column (4 mm × 10 mm). The column was first washed isocratically with mobile phase A for 10 min at 1.0 ml/min to remove unbound materials. Bound peptides were then eluted with a 40 min linear gradient from 0 to 50% Mobile phase B (600 mM KCl, 10 mM KH2PO4 and 20% ACN (pH 3.0), followed by a 10 min linear gradient from 50 to 100% Mobile phase B. Two minute fractions were collected and desalted via PepClean™ C18 spin columns (Pierce, Rockford, IL, USA). Peptides in each ion exchange fraction were further fractionated on an Ultimate™ Chromatography System equipped with a Probot matrix-assisted laser desorption ionization (MALDI) spotting device (Dionex, Sunnyvale, CA, USA). Peptides were captured onto a reversed phase 0.3 mm × 5 mm trapping column and resolved on a 75 μm × 150 mm capillary PepMap column (3 μm, 100 Å, C18, Dionex) with a 70 min gradient of solvent A (5% ACN, 0.1% trifluoroacetic acid, TFA) and solvent B (95% ACN, 0.1% TFA): 0–4 min, from 5 to 8% B, at 34 min, to 18% B, at 57 min, to 35% B and at 64 min to 95% B. The HPLC eluent was mixed in a 1:3 ratio with matrix (7 mg/ml α-cyano-4-hydroxycinnamic acid in 60% ACN, 5 mM ammonium monobasic phosphate and the internal calibrants (50 fmol/μl each of GFP and ACTH, 18–39)) through a 30 nl mixing tee, and spotted onto MALDI plates in a 18 × 18 spot array format. The peptides were analyzed on a 4700 Proteomics Analyzer tandem mass spectrometer (ABI) in a data-dependent fashion. MS spectra (m/z 800–3600) were acquired in positive ion mode with internal mass calibration. Ten most intense ions per spot were selected for subsequent MS/MS analysis in 1 keV mode. Each spectrum was averaged over 4000 laser shots.
Bioinformatics
Peptide identification and quantification were analyzed as reported earlier with minor modifications [15]. Each protein was identified with at least two peptides with confidence interval (C.I.) values of no less than 98%. The relative expression of a peptide was calculated by dividing the normalized iTRAQ™ reporter peak areas of m/z 115, 116 or 117 by that of m/z 114. Protein expression ratios were computed as the average of all corresponding peptides [15]. P-values were generated via Student's t-test. This is a good choice of statistic, given the small sample size. To search for formaldehyde-modified peptides, the MS/MS spectra were re-searched again with formaldehyde modified (+12 or +30 amu) arginine, lysine, tryptophan set as additional variable modifications and trypsin cleavage with two possible miss-cleavages. The peptide was considered to have formaldehyde modification, if its spectrum had a C.I. value above 98 % and the identification score was higher than the score matched to peptides without formaldehyde modifications. The spectra of all formaldehyde modified peptides were manually validated.
Immunohistochemistry
Slides from the diseased group along with corresponding negatives controls were heated and de-paraffinized from xylene to water and then pretreated with Target Retrieval Solution (Dako, Carpinteria, CA). Endogenous peroxidases were quenched by immersing sections in 3% hydrogen peroxide and Tris Buffered Saline (TBS) for 5 minutes, followed by placement in a TBS bath for 5 minutes. Sections were incubated for 30 minutes with 100 μl of monoclonal CK17 antibody (ab2502, Abcam, Inc., Cambridge, MA) at a 1:100 dilution. The primary antibody was replaced with TBS for the negative controls. Following 10 minute incubation with a rabbit anti-mouse secondary antibody and streptavidin, DAB substrate-chromogen was placed on the sections for 3 minutes, and the slides were then rinsed in water. After hematoxylin counterstaining, the sections were dipped in ammonia and then rinsed with water, dehydrated in alcohol, and cover slipped. Each slide with its negative control was evaluated by two investigators (JR and VF).
Results
Peptides can be Identified from FFPE Papilloma
To develop a MS based peptide-labeling method for quantitative proteomic analysis of formalin fixed tissues, we first extracted peptides from FFPE oral HPV lesions from HIV(+) and HIV(-) patients and subsequently labeled them with iTRAQ™ reagents (Supplement Figure 1). The iTRAQ™ labeled peptides were identified and quantified by MS/MS. Under very stringent criteria (C.I. ≥ 98%), this analysis led to the identification of 632 unique peptides which rendered 114 proteins identified by at least two peptides (Supplement Table 1). When the MS/MS spectra were searched against a decoyed database with all Swiss-Prot protein sequences reversed [15, 16], we could identify only two proteins which corresponded to a false discovery rate of 3.5%. Various populations of unmodified, formalin (or other)-modified and formalin cross-linked peptides were also identified from these tissues. Unfortunately, the effects of formalin fixation and long term storage on samples are not very well understood yet. However, our results showed that 2.8% of the identified peptides likely contained formaldehyde modification; we have therefore focused the quantitative bioinformatics analysis of the resulting MS data solely based on the unmodified peptides.
The molecular functions and cellular localization of all proteins identified in FFPE oral HPV lesions were classified using Gene Ontology (GO) classification system (http://www.geneontology.org) (Supplement Figure 2). These proteins were associated with every cellular compartment (Supplement Figure 2A) with nearly half from the cytoplasm. These proteins found here were compared with all proteins across the entire human proteome and no significant bias, in terms of the enrichment of proteins from specific cellular compartment was observed (data not shown). Furthermore, proteins identified in this analysis represented a diverse array of molecular functions including signal transduction, cell adhesion and metabolism (Supplement Figure 2B). Interestingly, many of the proteins (e.g. vimentin, cytokeratin 19 and small proline-rich proteins) have previously been implicated in HPV infection in keratinocytes [17].
Differentially Expressed Proteins in Oral HPV Lesions from HIV(+) and HIV(-) Patients
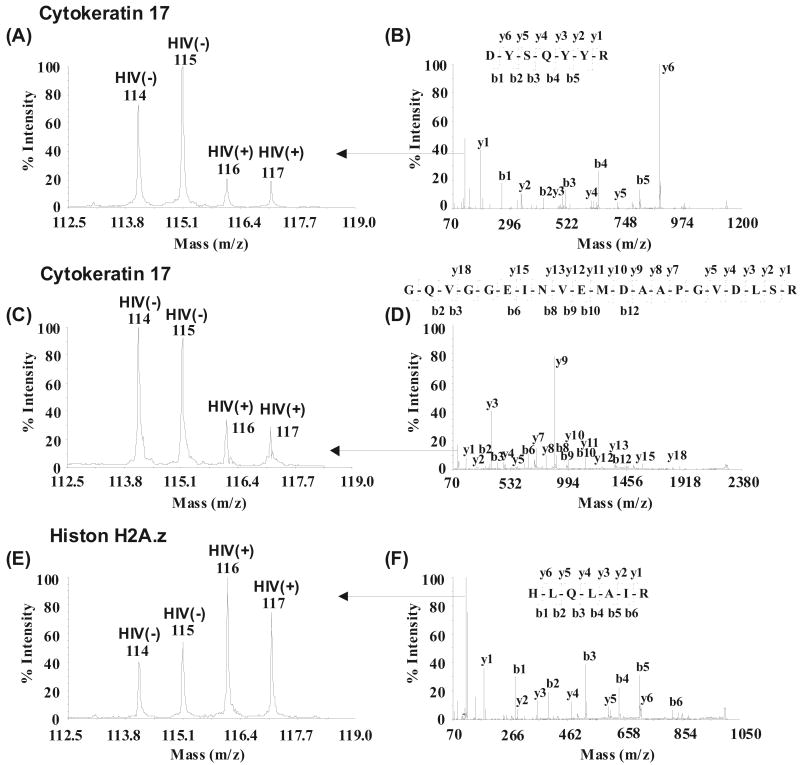
After confirming papillomal protein identities, we further analyzed their relative expression between HIV(+) and HIV(-) patients. Protein expression changes (in iTRAQ™ ratios) were considered significant if they were different by at least 20% (p≤0.05) [18]. Differentially expressed proteins that correlate with the HIV infection status in HPV oral lesions are shown in Table 1 which includes upregulation of CD44 antigen precursor, desmuslin, glutathione S-transferase P, histone H2A.z, histone H3.3, histone H4, lamin A/C and down regulation of involucrin and cytokeratin 17 (CK17). Unique peptides identified within these proteins are listed in Supplement Table 2. Fig. (1) provides examples of MS/MS spectra for the identification and quantification of CK17 and histone H2A.z. CK17 was downregulated (Fig. 1A-D) while histone H2A.z was upregulated (Fig. 1E and F) in the HIV(+) samples. Two peptides corresponding to CK17 (Fig. 1A and C) are shown to indicate quantification reproducibiligy in iTRAQ™ analysis.
Table 1. Differentially Expressed Proteins in Oral HPV Lesions from HIV(+) and HIV(-) Patients.
| Swiss-Prot Accession no. | Protein | No. of Unique Peptides | Average Protein Ratio (HIV+)/HIV(-) | p-value |
|---|---|---|---|---|
| O15061 | Desmuslin | 2 | 2.84 | <0.03 |
| P17317 | Histone H2A.z | 2 | 1.92 | <0.00 |
| P84243 | Histone H3.3 | 4 | 1.68 | <0.05 |
| P02545 | Lamin A/C | 5 | 1.42 | <0.05 |
| P62805 | Histone H4 | 4 | 1.33 | <0.05 |
| P09211 | Glutathione S-transferase P | 2 | 1.31 | <0.02 |
| P16070 | CD44 antigen precursor | 2 | 1.27 | <0.03 |
| P07476 | Involucrin | 10 | 0.72 | <0.04 |
| Q04695 | Cytokeratin 17 | 12 | 0.27 | <0.02 |
Fig. (1).
Examples of quantification and identification of proteins. Cytokeratin 17 (A-D) was downregulated and histone H2A.z (E and F) was upregulated in HPV(+) HIV(+) samples. Two peptides corresponding to cytokeratin 17 (A and C) are shown to indicate similar trends in peptide quantification. Peptide sequences were deduced from the MS/MS spectra, (B, D and F) based on the observation of continuous series of either N-terminal (b series) or C-terminal (y series) ions. Quantification of peptides was based on the relative iTRAQ reporter peak areas (A, C and E).
Corroboration of iTRAQ™ Quantification Results by Immunohistochemistry (IHC)
In present analysis, CK17 was found to be down regulated in oral HPV lesions in HIV(+) patients. To evaluate the iTRAQ™ quantification results, we compared CK17 expression in additional FFPE oral HPV lesions from HIV(+) and HIV(-) patients by IHC. Fig. (2) visually demonstrates the general trend of CK17 downregulation in representative papilloma sections from HIV(+) (Fig. 2B) patients as compared to HIV(-) (Fig. 2A) patients, overall corroborating quantitative proteome analysis of FFPE tissues using iTRAQ™ technology.
Fig. (2).
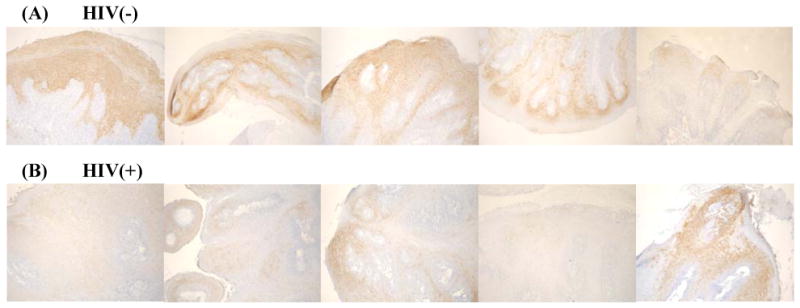
Immunoperoxidase staining for cytokeratin 17. Variable staining of suprabasal keratinocytes is shown in papillomas removed from the oral mucosa. The overall observation is that of decreased staining intensity of oral papilloma keratinocytes in the HIV(+) patients (B) as compared to papillomas found in the HIV(-) patients (A) (100X), corroborating the iTRAQ quantification trend.
Discussion
Until recently, IHC has been the primary technique for quantitative analysis of protein expression in formalin fixed tissues. However this technique lacks the throughput for unbiased discovery of unforeseen proteomic changes. This study demonstrates a quantitative proteomic method for analyzing FFPE tissues by combining iTRAQ™ and Liquid Tissue® technologies. With this approach, we have successfully compared the expression of 114 proteins from oral HPV lesions between HIV(+) and HIV(-) patients. To the best of our knowledge, no other study has reported quantitative analysis of more than 100 proteins from FFPE tissues, considering the paucity of materials available. This study was performed on cells that were needle dissected directly from standard formalin fixed tissue sections, yet use of laser-based tissue microdissection to focus on specific cell populations would provide even greater specificity of data. The impact of any new technology is based upon the new insights it can provide into the biology of the system under investigation. In this case, a variety of differentially expressed proteins were identified in oral papillomas from HIV(+) samples as compared to HIV(-) samples. These included the upregulation of desmuslin, lamin A/C, histones H2A.z, H3.3, H4, glutathione S-transferase P, CD44 and the down regulation of involucrin and CK17. CK17 is a member of the cytokeratin family of intermediate filaments and cytoskeletal proteins which are found in all cells, but are particularly prevalent in epithelial cells. They provide structural stability to the cell, as well as assistance with intercellular communication and attachment, the trafficking of membrane proteins, and the regulation of apoptosis [19, 20]. CK17 is normally expressed within certain epidermal appendages, such as the hair follicle and nail bed, the epidermal and mucosal stratum basale, and hyperproliferating squamous epithelia [20-23]. Histologic examination of oral papillomas has revealed a substantial proliferation of keratinocytes resulting in clinically apparent warty mucosal growths. The higher degree of expression of CK17 that we observed in the papilloma keratinocytes of the HIV(-) population may reflect a greater degree of differentiation of these cells when compared to the HIV(+) patients. These differences point to an underlying modification in cell proliferation and maturation due to the influence of HIV on the patient's immune system, allowing a less restrictive environment for the replication of HPV. It is conceivable that such a situation leads to an increased emphasis on viral replication and assembly at the expense of CK17 production. Modulation of CK17 and above mentioned proteins in HIV(+) samples may represent an underlying mechanism(s) for interaction of HIV with HPV. To explore these possibilities, further studies may be carried out.
The ability to quantify the proteome of diseased oral tissues will be of valuable assistance to our understanding of oral diseases that will also assist with both diagnosis and patient management. Many oral conditions, such as autoimmune vesiculobullous and preneoplastic diseases present with similar, if not identical, clinical and histologic manifestations. This often creates considerable difficulty in arriving at a definitive diagnosis and treatment plan, which is of concern with differing management regimens and prognoses.
Historically, immunohistochemistry has been developed as a qualitative tool complementing the morphologic interpretation and is difficult to be implemented as a quantitative technique. Attempts to quantify using immunohistochemical staining intensity have typically relied on determining the overall staining intensity across the breadth of the tissue in question, and attributing a grade of low, moderate, intense, or negative by practitioners. Given the immense variability in procedural and interpretive protocols across the labs, staining intensity may not be simply used as a sole criterion for quantification. Although, we have found that IHC staining with CK17 antibody on a representative set of samples corroborated our iTRAQ quantification results, we wish to further pursue to validate these iTRAQ results with a larger set of samples. By detecting protein expression differences in lesional tissue, it may be possible to reliably discriminate between similar diseases and manage patients appropriately. In addition, the presence of certain proteins may yield clues to subsequent disease behavior, allowing health care workers to target those individuals who are more likely to experience disease progression or relapse.
Conclusion
FFPE tissues provide a convenient medium to analyze a wide variety of pathological materials. The ability to perform large scale quantitative proteomic analysis of formalin fixed tissue would lead to greater understanding of the biological processes of disease and lead to improvements in diagnosis and treatment. Results reported here demonstrate that quantification of proteins using iTRAQ™ reagents can be applied to conduct large scale quantitative proteomics analysis of the vast archives of formalin fixed tissues.
Supplementary Material
Acknowledgments
This project is supported in part by an NIH grant, NS046593 to HL and a grant # 27-06 from the UMDNJ Foundation. We thank Ms. Dana Settembre for her valuable assistance.
Abbreviations
- HPV
Human papilloma virus
- HIV
Human immunodeficiency virus
- FFPE
Formalin fixed and paraffin-embedded
- iTRAQ™
Isobaric tags for relative and absolute quantitation
References
- 1.Rinaggio J, Glick M, Lambert WC. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006;101:328–32. doi: 10.1016/j.tripleo.2005.02.066. [DOI] [PubMed] [Google Scholar]
- 2.Greenspan D, de Villiers EM, Greenspan JS, de Souza YG, zur Hausen H. J Oral Pathol. 1988;17:482–8. doi: 10.1111/j.1600-0714.1988.tb01321.x. [DOI] [PubMed] [Google Scholar]
- 3.Volter C, He Y, Delius H, Roy-Burman A, Greenspan JS, Greenspan D, de Villiers EM. Int J Cancer. 1996;66:453–6. doi: 10.1002/(SICI)1097-0215(19960516)66:4<453::AID-IJC7>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 4.Regezi JA, Dekker NP, Ramos DM, Li X, Macabeo-Ong M, Jordan RC. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2002;94:724–31. doi: 10.1067/moe.2002.129760. [DOI] [PubMed] [Google Scholar]
- 5.Ahram M, Flaig MJ, Gillespie JW, Duray PH, Linehan WM, Ornstein DK, Niu S, Zhao Y, Petricoin EF, Emmert-Buck MR. Proteomics. 2003;3:413–21. doi: 10.1002/pmic.200390056. [DOI] [PubMed] [Google Scholar]
- 6.Julka S, Regnier FE. Brief Funct Genomic Proteom. 2005;4:158–77. doi: 10.1093/bfgp/4.2.158. [DOI] [PubMed] [Google Scholar]
- 7.Yan W, Chen SS. Brief Funct Genom Proteom. 2005;4:27–38. doi: 10.1093/bfgp/4.1.27. [DOI] [PubMed] [Google Scholar]
- 8.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Proc Natl Acad Sci. 2003;100:6940–5. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang SI, Thumar J, Lundgren DH, Rezaul K, Mayya V, Wu L, Eng J, Wright ME, Han DK. Oncogene. 2007;26:65–76. doi: 10.1038/sj.onc.1209755. [DOI] [PubMed] [Google Scholar]
- 10.Patel V, Hood BL, Molinolo AA, Lee NH, Conrads TP, Braisted JC, Krizman DB, Veenstra TD, Gutkind JS. Clin Cancer Res. 2008;14:1002–14. doi: 10.1158/1078-0432.CCR-07-1497. [DOI] [PubMed] [Google Scholar]
- 11.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Mol Cell Proteom. 2005;4:1487–502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Mol Cell Proteom. 2004;3:1154–69. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Boehm AM, Putz S, Altenhofer D, Sickmann A, Falk M. BMC Bioinformatics. 2007;8:214. doi: 10.1186/1471-2105-8-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu T, D'Mello V, Deng L, Hu J, Ricardo M, Pan S, Lu X, Wadsworth S, Siekierka J, Birge R, Li H. J Neurosci Methods. 2006;158:22–9. doi: 10.1016/j.jneumeth.2006.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu J, Qian J, Borisov O, Pan S, Li Y, Liu T, Deng L, Wannemacher K, Kurnellas M, Patterson C, Elkabes S, Li H. Proteomics. 2006;6:4321–34. doi: 10.1002/pmic.200600026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 17.Duffy CL, Phillips SL, Klingelhutz AJ. Virology. 2003;314:196–205. doi: 10.1016/s0042-6822(03)00390-8. [DOI] [PubMed] [Google Scholar]
- 18.Karp NA, Lilley KS. Proteomics. 2007;7:42–50. doi: 10.1002/pmic.200700683. [DOI] [PubMed] [Google Scholar]
- 19.Coulombe PA, Tong X, Mazzalupo S, Wang Z, Wong P. Eur J Cell Biol. 2004;83:735–46. doi: 10.1078/0171-9335-00443. [DOI] [PubMed] [Google Scholar]
- 20.Kirfel J, Magin TM, Reichelt J. Cell Mol Life Sci. 2003;60:56–71. doi: 10.1007/s000180300004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu PG, Weiss LM. Histopathology. 2002;40:403–39. doi: 10.1046/j.1365-2559.2002.01387.x. [DOI] [PubMed] [Google Scholar]
- 22.Lane EB, McLean WH. J Pathol. 2004;204:355–66. doi: 10.1002/path.1643. [DOI] [PubMed] [Google Scholar]
- 23.van der Velden LA, Schaafsma HE, Manni JJ, Ruiter DJ, Ramaekers CS, Kuijpers W. Eur Arch Otorhinolaryngol. 1997;254:376–83. doi: 10.1007/BF01642554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.