Abstract
Joubert syndrome (JBTS) is an autosomal recessive disorder characterized by cerebellum and brainstem malformations. Individuals with JBTS have abnormal breathing and eye movements, ataxia, hypotonia, and cognitive difficulty, and they display mirror movements. Mutations in the Abelson-helper integration site-1 gene (AHI1) cause JBTS in humans, suggesting that AHI1 is required for hindbrain development; however AHI1 may also be required for neuronal function. Support for this idea comes from studies demonstrating that the AHI1 locus is associated with schizophrenia. To gain further insight into the function of AHI1 in both the developing and mature CNS, we determined the spatial and temporal expression patterns of the gene products of AHI1 orthologs throughout development, in human, mouse, and zebrafish. Murine Ahi1 was distributed throughout the cytoplasm, dendrites, and axons of neurons, but was absent in glial cells. Ahi1 expression in the mouse brain was observed as early as embryonic day 10.5 and persisted into adulthood, with peak expression during the first post-natal week. Murine Ahi1 was observed in neurons of the hindbrain, midbrain, and ventral forebrain. Generally, the AHI1/Ahi1/ahi1 orthologs had a conserved distribution pattern in human, mouse, and zebrafish, but mouse Ahi1 was not present in the developing and mature cerebellum. Ahi1 was also observed consistently in the stigmoid body, a poorly characterized cytoplasmic organelle found in neurons. Overall, these results suggest roles for AHI1 in neurodevelopmental processes that underlie most of the neuroanatomical defects in JBTS, and perhaps in neuronal functions that contribute to schizophrenia.
Keywords: mouse, human, zebrafish, hindbrain
Introduction
Joubert syndrome (JBTS) is a rare autosomal recessive neurodevelopmental disorder characterized by multiple behavioral and neuroanatomical abnormalities. Individuals with classic JBTS have ataxia, hyperpnea/apnea, abnormal eye movements, hypotonia (weakness), developmental delay, and varying degrees of cognitive difficulties (Cantani et al., 1990; Chance et al., 1999; Joubert et al., 1968; Joubert et al., 1969; Kendall et al., 1990; Maria et al., 1999; Parisi et al., 2007; Saraiva and Baraitser, 1992; van Dorp et al., 1991). Classic JBTS is characterized neuroradiographically by cerebellar vermal aplasia/hypoplasia and by the molar tooth sign (an abnormal configuration of the superior cerebellar peduncles) (Friede and Boltshauser, 1978; Joubert et al., 1968; Joubert et al., 1969; Kendall et al., 1990; Maria et al., 1999; Niesen, 2002; Parisi et al., 2007; Quisling et al., 1999; van Dorp et al., 1991; Yachnis and Rorke, 1999). Neuropathological analyses of brains from individuals with JBTS catalogued multiple abnormalities in axonal tracts that cross the posterior midline of the brain (e.g. the superior cerebellar peduncles and the corticospinal tract). However, not all midline crossing tracts are abnormal (e.g. the corpus callosum and optic chiasm properly decussate) (Friede and Boltshauser, 1978; Joubert et al., 1968; Joubert et al., 1969; Kendall et al., 1990; Maria et al., 1999; Niesen, 2002; Quisling et al., 1999; van Dorp et al., 1991; Yachnis and Rorke, 1999). Other major neuroanatomical abnormalities in JBTS, particularly in the hindbrain, include dysplasia/hypoplasia of the inferior olives, accessory olives, the solitary nuclei and tracts, and the nucleus and spinal tracts of cranial nerve V (Friede and Boltshauser, 1978; Maria et al., 1999; Quisling et al., 1999; Yachnis and Rorke, 1999). Several non-brain features are associated with JBTS, such as retinal dysplasia and pathology of the kidney and liver (Gleeson et al., 2004; Liao et al., 2005; Parisi et al., 2007; Utsch et al., 2006; Valente et al., 2006). The differences in clinical and neuropathological characteristics observed in the various subclasses of JBTS suggest that JBTS is a heterogeneous family of disorders (Gleeson et al., 2004; Parisi et al., 2007).
One of the first genes implicated in JBTS was the Abelson-helper integration site-1 gene, AHI1 (Dixon-Salazar et al., 2004; Ferland et al., 2004) in the JBTS3 locus on chromosome 6q23 (Lagier-Tourenne et al., 2004). Individuals with AHI1 mutations have the classic brain malformations of cerebellar vermis aplasia, the molar tooth sign, and limited involvement of the cerebral cortex. However, kidney disease in patients with AHI1 mutations is relatively rare (Parisi et al., 2006; Utsch et al., 2006; Valente et al., 2006). The human AHI1 gene encodes a cytoplasmic multi-domain protein containing an N-terminal coiled-coil domain, multiple WD40 repeats, and a C-terminal SH3 domain (Jiang et al., 2002). The combination of protein-binding domain motifs indicates that AHI1 is likely a signaling or scaffolding protein involved in protein-protein interactions (Ferland et al., 2004; Jiang et al., 2002).
Prior expression studies suggest that AHI1 is involved most specifically in the development of the hindbrain (Dixon-Salazar et al., 2004; Ferland et al., 2004). Northern blot analysis demonstrates that AHI1 is expressed in human fetal brain and kidney (Dixon-Salazar et al., 2004; Ferland et al., 2004). In whole mouse brain, Ahi1 mRNA is found as early as embryonic day 10.5 with Ahi1 expression peaking during the first week of life and then continuing at lower levels into adulthood (Ferland et al., 2004). In situ hybridization studies confirmed this temporal pattern of expression of Ahi1 mRNA (Ferland et al., 2004). Moreover, Ahi1 mRNA is expressed throughout the brain, with the highest expression in the deep cerebellar nuclei, brainstem, tectum, hypothalamus, and amygdala, but very weak expression in the cerebellum and cerebral cortex (Ferland et al., 2004). Given these temporal and spatial expression patterns of murine Ahi1 mRNA, these data suggest that Ahi1 is involved in the development of the hindbrain and ventral forebrain, and possibly the cerebellum. Our present study has sought to determine the expression profiles of the gene product, AHI1/Ahi1/ahi1 in human, mouse, and zebrafish, respectively.
Ahi1 mRNA expression in the mouse brain persists into adulthood, suggesting that Ahi1 has functions beyond its role in the development of the central nervous system (Ferland et al., 2004). Association studies, using single nucleotide polymorphism (SNP) genotyping, found a high association between SNPs in human AHI1 and individuals with schizophrenia (Amann-Zalcenstein et al., 2006; Ingason et al., 2007). These studies suggest that AHI1 has additional roles in brain development, and possibly in the functioning of the mature nervous system, separate from the events during development in which mutations to AHI1 lead to the malformations of the posterior hindbrain associated with JBTS.
No formal examination of the temporal and spatial patterns of protein expression of the AHI1 orthologs has been conducted previously. Comparisons of the gene products of the AHI1 orthologs (primate, rodent, and zebrafish) indicate that, relative to the human and zebrafish protein, the rodent Ahi1 protein lacks an N-terminal, 140 amino acid putative coiled-coil domain (5′-RACE analysis, database ESTs, known mRNAs for Ahi1, all fail to identify this domain in mouse/rat)(Jiang et al., 2002). These differences suggest that AHI1 may have been under evolutionary selection. To address this idea, comparisons of the AHI1 gene sequences of various human and non-human primates were made and demonstrate that the primate AHI1 gene appears to be under positive selection along the human lineage (Ferland et al., 2004). This signifies that an understanding of species differences among the AHI1 orthologs will aid in the elucidation of the function of this gene in brain development. To explore species differences in the AHI1 family of genes, we sought to determine the localization pattern of AHI1 orthologs in multiple species. An understanding of the differences in AHI1-isoform expression at the neuroanatomical, cellular, and biochemical levels will not only aid in elucidating the role of the protein in brain development, but will also help in determining how mutations in AHI1 give rise to JBTS, and possibly schizophrenia, in humans.
MATERIAL AND METHODS
Production and characterization of antibodies against human AHI1 and mouse Ahi1
Rabbit polyclonal antibodies specific for human AHI1 or mouse Ahi1 were raised against various peptides derived from the human or mouse protein sequence, respectively. Specifically, peptides generated for the human AHI1 protein (Ab-813; PAPQKQSINKNKSQ, amino acids 1138-1151) and to four distance separate amino acid locations in the mouse Ahi1 protein (N-terminus: Ab-82/83, PTADDSDDSREKTGIE, amino acids 21-36; C-terminus: Ab-809, RSPPLTPKEKTKPE, amino acids 973-986; C-terminus: Ab-84/85, SEKGKDQNVEDRGHK, amino acids 1014-1028; C-terminus: Ab-05/06, KKSEPVVRKVTLIE, amino acids 1034-1047). The synthesis and conjugation of the peptide and production of rabbit polyclonal antibodies were performed by either Covance (Denver, PA) or Sigma-Genosys (The Woodlands, TX). All antibodies were affinity purified using the corresponding peptide and the SulfoLink Kit (Pierce, Rockford, IL) according to the manufacturer’s instructions.
The specificity of each antibody was determined using standard Western blotting and immunostaining techniques. Full-length human AHI1 and mouse Ahi1 sequences were cloned in-frame into an EGFP expression plasmid containing a CMV promoter (Human: N-terminal EGFP-AHI1 and C-terminal EGFP-AHI1; Mouse: N-terminal EGFP-Ahi1 and C-terminal EGFP-Ahi1). HEK293 cells (ATCC, Manassas, VA) were transiently transfected with each of these constructs using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). After EGFP-expressing cells were observed (approximately 70% of the cells at confluence in each culture dish expressed EGFP), they were lysed with boiling 2X sample buffer (62.5 mM Tris (pH 6.8), 2% SDS, 20% glycerol, 0.01% bromophenol blue, 5% β-mercaptoethanol (BME)), yielding protein lysates for separation on SDS/8% PAGE gels with electroblotting onto PVDF transfer membrane (Millipore, Billerica, MA). Blots were probed with each AHI1/Ahi1 antibodies (rabbit IgG) in combination with EGFP antibodies in order to determine localization of both AHI1 and EGFP; co-immunoreactivity indicates the specificity of each of the AHI1/Ahi1 antibodies (EGFP antibody: immunogen: full-length Aequorea victoria green fluorescent protein; mouse IgG; 1:2000; Cat. # 632381; Lot # 8010026; Clontech, Mountain View, CA); this antibody recognizes native and denatured GFP and EGFP as well as N- and C-terminal fusion proteins; addition of this antibody to non-EGFP containing cultured neuronal cells or brain tissue results in no specific signal (data not shown; see manufacturer’s technical data sheet). The N-terminal EGFP-human-AHI1 and C-terminal EGFP-human-AHI1 lysates were probed with Ab-813; and the N-terminal EGFP-mouse-Ahi1 and C-terminal EGFP-mouse-Ahi1 lysates were probed with Ab-82/83, Ab-809, Ab-84/85, and Ab-05/06 at varying concentrations. The primary antibodies were detected with goat anti-mouse IgG-Cy3 (for EGFP) and goat anti-rabbit IgG-Cy5 (for AHI1) fluorescent secondary antibodies (1:500; GE Healthcare, Piscataway, NJ). The signals were analyzed with a Typhoon Trio+ scanner and ImageQuant analysis software (GE Healthcare). Western blot analysis of these lysates demonstrated that the human AHI1 antibody recognized a band of ∼190 kDa corresponding to the size of the EGFP-human-AHI1 fusion protein. Similar results were obtained for the mouse Ahi1 antibodies and the EGFP-mouse-Ahi1 fusion protein (∼160 kDa). The EGFP antibody detected an overlapping molecular mass signal with the AHI1/Ahi1 antibody signals (data not shown). In addition, incubation of each of the AHI1/Ahi1 antibodies with a 5X excess of the corresponding peptide was able to completely abolish the signal corresponding to the AHI1/Ahi1 bands (data not shown).
In addition, HEK293 cells that expressed EGFP-tagged human AHI1 or mouse Ahi1 were also immunostained with the corresponding species AHI1/Ahi1 antibodies in conjunction with an Alexa-546 goat anti-rabbit IgG secondary antibody (Invitrogen), to determine whether AHI1/Ahi1 and EGFP co-localized in these cells. Over-expression of human AHI1-EGFP and mouse Ahi1-EGFP by transient transfections resulted in cytoplasmic expression of the EGFP-tagged AHI1/Ahi1 proteins. In these transfected cell lines, immunostaining with the corresponding human AHI1 antibody or mouse Ahi1 antibodies demonstrated overlapping localization of EGFP and human AHI1 and mouse Ahi1, respectively (data not shown).
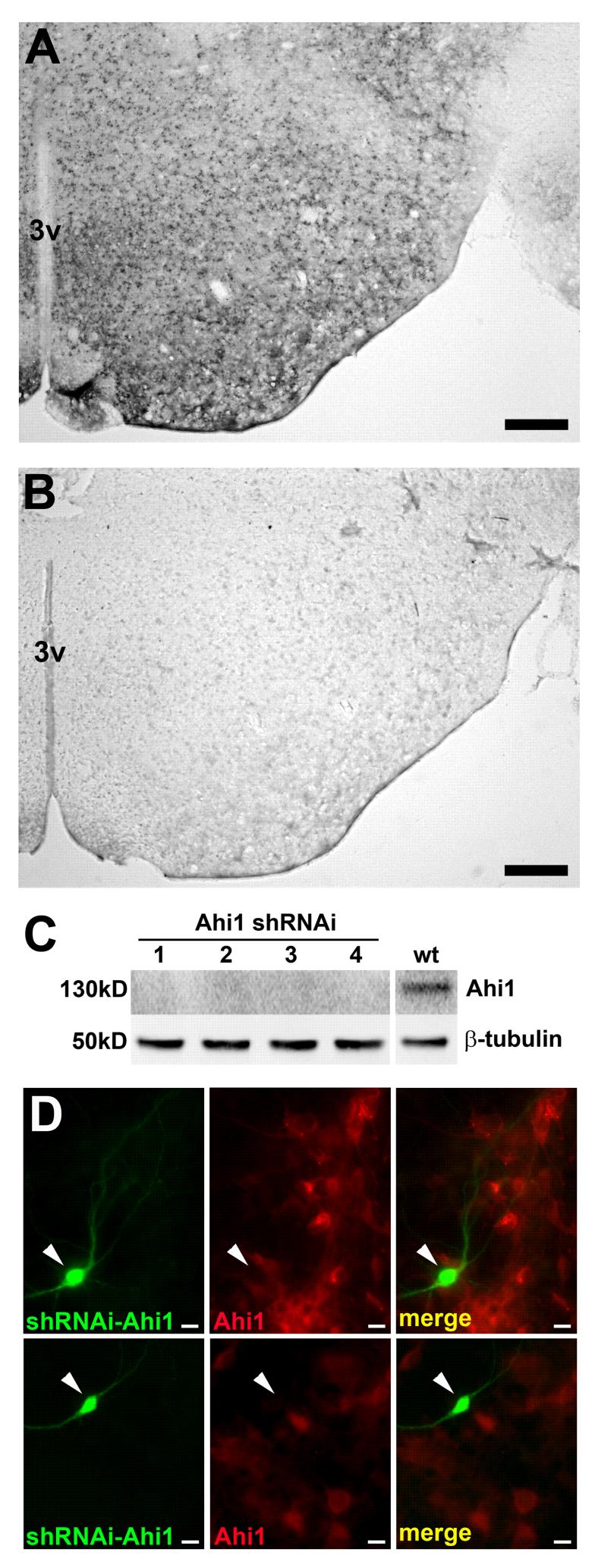
To further confirm the specificity of the mouse Ahi1 antibodies, we performed peptide blocking experiments in which the mouse Ahi1 antibodies were pre-incubated with a 5X excess of each corresponding peptide that was used to raise the antibodies. The antibody-peptide mixture was then added to mouse brain sections using standard immunohistochemistry (as described below). Excess peptide was able to completely abolish the immunostaining observed with the mouse Ahi1 antibodies alone (without peptide) (Fig. 1a, b).
Figure 1.
Characterization of the specificity of Ahi1 antibodies. A, Immunohistochemical detection of Ahi1 shows diffuse cytoplasmic staining in the mature murine hypothalamus. Bar indicates 200 μm. B, Lack of detection of Ahi1 immunoreactivity in the presence of a 5X excess of the peptide, used to generate the antibody; this result demonstrates the specificity of the Ahi1 antibody. Scale bar = 200 μm. 3V: third ventricle C, Knockdown of Ahi1 by shRNAi in IMCD3 cells results in an almost complete absence of the 130 kDa Ahi1 band as compared to non-transfected cells (WT), in four independent shRNAi experiments; this result further demonstrates the specificity of the Ahi1 antibody. D, Knockdown of Ahi1 by siRNA in cultured hypothalamic neurons also shows the specificity of the Ahi1 antibody. Left panels show hypothalamic neurons that have been transfected with, and express, EGFP and siRNAs against Ahi1 (green cells). Middle panels ilustrate the typical distribution of Ahi1 immunoreactivity in these cells (red cells). Right panels are a merge of the previous panels indicating a lack of Ahi1 expression in cells that have had Ahi1 knockdown. Note the lack of Ahi1 (in red) in the cells transfected with RNAis against Ahi1 (green). Bar indicates 10 μm. Magenta-green converted images are available as supplemental figures (Supplementary Figure 1).
An additional confirmation method used to determine the specificity of each mouse Ahi1 antibody was Western blot analyses of IMCD3 cells (a mouse inner medullary collecting duct cell line; ATCC, Manassas, VA). IMCD3 cells were grown according to the manufacturer’s instructions. IMCD3 cells were transfected with a pSUPER.gfp+neo RNAi construct (Oligoengine, Seattle, WA) containing a shRNAi sequence directed against mouse Ahi1 or a scrambled sequence. After 3-5 days in culture, the IMCD3 cells were lysed and processed for mouse Ahi1 Western immunoblotting as described above. Wild-type IMCD3 cells or cells transfected with scrambled sequence expressed full-length Ahi1 with a molecular mass of 130 kDa as determined by Western blotting with Ahi1 antibodies (Fig. 1c). One additional ∼150 kDa protein band was also observed by Western blotting in wild-type IMCD3 cells and cells transfected with scrambled sequence (data not shown). No bands were found with a molecular mass below 130 kDa. IMCD3 cells transfected with shRNAi constructs against mouse Ahi1 demonstrated no detectable levels of the 130 kDa, full-length mouse Ahi1 protein (Fig. 1c). However, the additional ∼150 kDa protein band still remained in Ahi1-knockdown cells (data not shown). Given this result, we performed immunocytochemistry for Ahi1 in Ahi1-knockdown IMCD3 cells. Significant decreases in Ahi1 immunoreactivity were observed in these cells (data not shown). This suggests that this ∼150 kDa band observed by Western blotting was not observed in non-denatured, immunocytochemical experiments.
The final confirmation method used to determine the specificity of each mouse Ahi1 antibody was immunostaining of primary mouse neurons that had been transfected with mouse Ahi1 siRNAs (details below). Primary neuronal cultures from mouse hypothalamus were grown using modified neuronal culturing techniques (Banker and Goslin, 2002). Briefly, hypothalami from embryonic day 17.5 (E17.5) mice were dissected and placed into ice-cold Ca++/Mg++ free PBS. The tissue was minced into smaller pieces and added to a trypsin/EDTA solution for 30 minutes at 37°C. Following trypsin digestion, the tissue was rinsed with a trypsin inhibitor and placed in Neurobasal medium supplemented with B27, glucose, sodium pyruvate, glutamine, and gentamicin (all at manufacturer’s suggested concentrations (Invitrogen (GIBCO), Carlsbad, CA)). The tissue was triturated multiple times, to liberate individual cells from the tissue. Viable cells were counted on a hemocytometer, using the trypan blue exclusion assay, and were then plated onto poly-d-lysine treated glass coverslips at density of 2000-5000 cells/mm2. Following incubation for 1-3 days, hypothalamic cell cultures were transfected, using Lipofectamine 2000 (Invitrogen), with both an ON-TARGETplus SMARTpool of siRNAs directed against mouse Ahi1 (Dharmacon, Chicago, IL) and an EGFP-expressing plasmid, all according to the manufacturers’ suggested protocols. Neuronal cultures were allowed to grow for an additional 3-5 days and were then fixed in cold 4% paraformaldehyde (PFA) and processed for mouse Ahi1 immunostaining, as described below. The transfected cells were visualized on a Zeiss AxioImager-A1 microscope with AxioVision Rel. 4.5 software (Carl Zeiss Microimaging, Thornwood, NY), with EGFP used as the indicator of any cell that was co-transfected with mouse Ahi1 siRNAs.
Primary cultures of hypothalamic neurons were immunostained with each mouse Ahi1 antibody individually; they demonstrated diffuse cytoplasmic, dendritic, and axonal localization (Fig. 1d). However, knockdown of mouse Ahi1 expression in hypothalamic cultures by siRNA resulted in greatly reduced expression or absence of Ahi1 staining only with mouse Ahi1 antibodies, Ab-84/85 and Ab-05/06 (data for Ab-05/06 shown in Fig. 1d). Antibody Ab-82/83 continued to show immunostaining in cultured Ahi1-knockdown neurons, although at a reduced level, suggesting that this antibody was not entirely specific for mouse Ahi1 in non-denatured protein preparations (this antibody is specific for mouse Ahi1 protein by Western blotting). Lastly, Ab-809 did not show any detectable immunostaining of cultured neurons suggesting the epitope for this antibody is hidden in the folded protein, under non-denaturing conditions (since this antibody is specific by Western blotting). Given that all of the mouse Ahi1 antibodies are specific by Western blotting, but only mouse Ab-84/85 and mouse Ab-05/06 are specific in immunohistochemical assays, we present all of following mouse experiments using Ab-05/06; however, Ab-84/85 gave similar results by immunohistochemistry and Western blotting. For experiments using human AHI1 protein, we utilized human AHI1 antibody, Ab-813.
Animals and histological procedures
All mice used to characterize the normal expression pattern of Ahi1 were wild-type Swiss Webster mice obtained from Taconic (Germantown, NY). The mice were sacrificed at various developmental ages: embryonic day (E) 10.5, E12.5, E14.5, E16.5, E18.5, postnatal day (P) 0.5, P1.5, P3.5, P7.5, P10.5, P14.5, P21.5, and adult.
To obtain tissue for immunohistochemistry (IHC), we administered an overdose of either sodium pentobarbital or avertin to mice, which were then perfused transcardially with 0.1M phosphate-buffered saline (PBS; pH 7.4). This was followed by cold 4% PFA (made in PBS). Brains were dissected, post-fixed in 4% PFA, and stored at 4°C for up to 7 days. Following post-fixation, brains were cryoprotected in a 30% sucrose solution made in PBS.
Brains were sectioned on a Microm cryostat (Richard-Allan Scientific, Kalamazoo, MI), in either the sagittal or coronal plane, at a thickness of 25-40 μm (depending on the developmental stage). All sections were mounted on Superfrost Plus microscope slides (Fisher Scientific, Pittsburg, PA) and allowed to air dry for 30 minutes before being stored at -80°C.
All mouse procedures were performed under approval from the Institutional Animal Care and Use Committees of both Rensselaer Polytechnic Institute and the Wadsworth Center (NY State Department of Health), in accordance with The National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Zebrafish husbandry
AB and TL Zebrafish were maintained under a 10 hour dark to 14 hour light cycle at 28.5°C using standard laboratory practices (Nüsslein-Volhard and Dahm, 2002). Zebrafish embryos were obtained by natural mating and were staged by days post-fertilization (dpf) and according to standard morphological criteria (Kimmel et al., 1995). Embryos were raised at 28.5°C until sacrificed and fixed at the desired stage. 1-phenyl-2-thiourea (0.003%) was used to suppress pigmentation of embryos older than 24 hours post-fertilization (hpf). All fish procedures were performed under approval from the Institutional Animal Care and Use Committee of Rensselaer Polytechnic Institute.
Western blotting
Tissue was obtained for Western blotting from additional mice at the ages described previously. Mice were administered an overdose of either sodium pentobarbital or avertin, and their brains and peripheral organs were harvested and immediately frozen in liquid nitrogen. For some animals, brains were dissected into sub-regions (cerebral cortex, hippocampus, striatum, thalamus, hypothalamus, brainstem, and cerebellum) and frozen in liquid nitrogen. All samples were stored at -80°C until processed into lysates.
Whole mouse brain (or sub-regions) or peripheral tissues were homogenized in RIPA buffer (50 mM Tris (pH 8), 150 mM NaCl, 1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride and 1X protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN)) with a plastic mortar and pestle. Homogenized lysates were incubated on ice for 30 minutes and centrifuged at 10,000 x g for 30 minutes to obtain the supernatant. The protein concentration was determined for each supernatant sample using the Advanced Protein Assay reagent (Cytoskeleton Inc., Denver, CO) according to the manufacturer’s protocol. Protein lysates (5 μg of protein), isolated from brains of variously aged mice, from various regions of mouse brain, or from various peripheral organs, were resolved by SDS/8% PAGE gel electrophoresis and electroblotted onto PVDF transfer membrane. The membrane was blocked with 3% non-fat dried milk in TBS-TX (100 mM Tris (pH 7.4), 150 mM NaCl, 0.01% Triton X-100) for 1 hour at room temperature and then probed with the various mouse Ahi1 antibodies (at 1:1000 to 1:4000 dilution in blocking solution) and with anti-βIII tubulin antibodies (chicken IgY; 1:4000; Chemicon, Temecula, CA) as a loading control, at room temperature overnight.
For human brain tissue Western blotting, a custom human normal brain tissue blot was obtained (ProSci, Poway, CA) containing the following tissues: adult human amygdala, cerebellar peduncles, cerebellar hemispheres, cerebellar vermis, cerebral cortex, and medulla oblongata; and fetal human cerebellum and brainstem. Briefly, 15 μg of each protein lysate were separated on a 4-20% gradient SDS-PAGE gel and transferred onto nitrocellulose membranes. The membrane was blocked with 3% non-fat dried milk in TBS-TX for 1 hour at room temperature and then probed with our human AHI1 antibody (Ab-813; 1:100) and with anti-βIII tubulin antibodies (chicken IgY; 1:10000; Chemicon) as a loading control, at room temperature overnight.
Primary antibodies were detected with the SuperSignal West Femto Maximum Sensitivity Substrate Chemiluminescence Kit (Pierce, Rockford, IL). The signal was analyzed with a Typhoon Trio+ scanner and ImageQuant analysis software (GE Healthcare).
Immunostaining
All tissue sections were allowed to dry at room temperature before being permeabilized for 10 minutes in 0.04% Triton X-100 in PBS (pH 7.4; PBS-TX). Endogenous peroxidases were removed via incubation in 0.03% hydrogen peroxide/methanol for 30 minutes. This hydrogen peroxide step was excluded if antibody detection was accomplished through fluorescently tagged antibodies. Sections were then washed in PBS-TX and blocked for 1 hour in 10% normal goat serum (NGS), followed by an overnight incubation with the primary antibody. For the majority of the immunohistochemistry, we utilized the mouse Ahi1 antibody designated Ab-06 (as described above) at a concentration of 1:1000 in 1% NGS/PBS-TX. However, the other Ahi1 antibodies, Ab-05 and Ab-84/85, gave similar results. Tissue sections were again washed in PBS-TX, followed by primary antibody detection using the Vector Elite ABC kit and a 3,3′-diaminobenzidine (DAB) kit (Vector Labs, Burlingame, CA) according to the manufacturer’s instructions. The slides were allowed to dry overnight before a coverslip was applied with Cytoseal-60 (Richard-Allan Scientific, Kalamazoo, MI); slides were visualized with a Zeiss AxioImager-Z1 microscope and imaged with an AxioCam MRc camera with AxioVision Rel. 4.5 software and MosaiX (Carl Zeiss Microimaging). All images were processed using Adobe Photoshop CS2 (version 9.0.2; Adobe Systems Inc., San Jose, CA). Contrast and brightness of images were adjusted through linear level adjustments, as needed, to optimize the clarity of the images presented.
For fluorescence immunohistochemistry, we used the following antibodies: NeuN (immunogen: purified cell nuclei from mouse brain; clone: A60, mouse IgG; Cat. # MAB377; Lot # 060109159; Chemicon, Temecula, CA); this antibody recognizes the neuron-specific protein NeuN present in most neurons of the vertebrate CNS (Bulloch et al., 2008) and shows immunoreactivity in morphologically distinct neurons in culture (data not shown) and in brain tissue (Ferland et al., 2003); glial fibrillary acidic protein (GFAP)(immunogen: purified bovine GFAP; chicken IgY; Cat. # AB5541; Lot # 0605030036; Chemicon); this antibody recognizes GFAP in most astrocytes of the vertebrate CNS, reacts with both native and recombinant protein, and it shows immunoreactivity in morphologically distinct glia in culture (data not shown; see manufacturer’s technical data sheet); LR11 (immunogen: human LR11 a.a. 1220-1337; clone: 48, mouse IgG; Cat. # 611860; Lot # 74851; BD Transduction Laboratories, San Jose, CA); this antibody recognizes the lipoprotein receptor homologue that is expressed at high levels in the brain (Posse De Chaves et al., 2000) and it recognizes a 250 kDa band corresponding to the predicted mass of LR11. Increased dilutions of the primary antibody result in decreasing immunoreactivity (see manufacturer’s technical data sheet). LR11 antibodies also detect morphologically distinct organelles known as stigmoid bodies (Gutekunst et al., 2003), therefore making LR11 an ideal marker for the stigmoid body; dynein (immunogen: bovine brain cytoplasmic dynein (the antigenic site has been localized to the first 60 a.a. of the N-terminus of dynein); clone: 74.1, mouse IgG; Cat. # MAB1618; Lot # 0512018659; Chemicon); this antibody recognizes the 74 kDa intermediate chain subunit of cytoplasmic dynein (see manufacturer’s technical data sheet). Dynein antibodies demonstrate immunoreactivity to neuronal growth cones (Grabham et al., 2007). The final working concentrations for the primary antibodies were as follows: Ahi1 (1:1000), NeuN (1:1000), GFAP (1:200), LR11 (1:1000), and dynein (1:100). Omission of all primary antibodies, used in the present studies, resulted in the absence of any immunoreactivity (data not shown).
Tissue sections or cells on slides were washed extensively in 0.1M PBS (pH 7.4). Then, they were washed in PBS-TX followed by blocking in 10% NGS made in PBS-TX, for at least 1 hour. Primary antibodies were diluted to their final working concentrations in PBS-TX containing 1% NGS. The slides were incubated overnight at 4°C. The following day, all slides were washed in PBS-TX and then were incubated for at least 2 hours with the appropriate fluorophore-labeled secondary antibodies: Alexa Fluor 488 goat anti-mouse IgG, Alexa Fluor 546 goat anti-rabbit IgG, and Alexa Fluor 488 goat anti-chicken IgY (all from Invitrogen (Molecular Probes), Carlsbad, CA). Each fluorophore-labeled secondary antibody was diluted 1:500 in PBS-TX containing 1% NGS. Following the 2 hour incubation with the fluorophore-labeled secondary antibodies, all sections were rinsed in PBS and mounted on slides, and a coverslip was applied with Fluoromount-G (Southern Biotechnology, Birmingham, AL). All images were visualized with a Zeiss AxioImager-Z1 microscope and imaged with an AxioCam MRm camera and AxioVision Rel. 4.5 software. All images were processed using Adobe Photoshop CS2. Contrast and brightness of images were adjusted through linear level adjustments, as needed, to optimize the intensity range of the images.
In situ hybridization probes for zebrafish ahi1
Total RNA was isolated from zebrafish embryos at various stages using TRIzol reagent according to the manufacturer’s instructions. PCR and genomic analysis indicated that ahi1 is a single gene in zebrafish. Gene-specific PCR primers were designed according to Ensembl (Ensembl release 47, Assembly Zv7) for the zebrafish ahi1 transcript (Ensembl ID: ENSDARG00000044056). Zebrafish ahi1 cDNA was obtained by reverse transcription, 3′-RACE, and 5′-RACE, following manufacturer’s instructions (GeneRace Kit, Invitrogen, Carlsbad, CA). PCR reactions were carried out using primers with T3/T7 polymerase binding sites, and with zebrafish ahi1 cDNA as template. Digoxigenin-11-uridine-5′-triphosphate (DIG)/NTP mix (10 mM ATP, 10 mM GTP, 10 mM CTP, 6.5 mM UTP, 3.5 mM DIG-11-UTP in Tris-neutralized solution, pH 7.5)(Roche, Indianapolis, IN) was used instead of dNTP. PCR products with the correct sizes were transcribed with T3/T7 polymerase following the manufacturer’s instructions (MEGAscript T3 Kit, MEGAscript T7 Kit, Roche). Primer pairs used to generate the five riboprobes used in this study are available upon request. The probes were, in order from the 5′- to the 3′-end of the zebrafish cDNA: probe 1: nt 15-468; probe 2: nt 15-589; probe 3: nt 15-661; probe 4: nt 389-661; probe 5: nt 15-389. Staining patterns for the individual probes were indistinguishable.
Whole-mount in situ hybridization
Zebrafish embryos from 48 hpf to 5 dpf were sacrificed and fixed overnight in 4% PFA/PBT (0.1% Tween-20 in PBS) at 4°C, and were then transferred to methanol for at least 16 hours at -20°C. Embryos were washed in 75%-, 50%-, 25%-methanol/PBT gradients, followed by washes in PBT. Embryos at 2 dpf, 3 dpf, 4 dpf, 5 dpf were treated with 10 μg/ml proteinase K for 20 , 22 , 24 , 26 minutes, respectively, followed by a 30 minute incubation in 4% PFA/PBT and washes in PBT at room temperature. Embryos were prehybridized in HYB buffer (50% formamide, 5X SSC, 500 μg/ml torula RNA, 50 μg/ml heparin, 0.1% Tween-20, 9 mM citric acid at pH 6.0-6.5) for 2 hours at 68°C and then hybridized with ∼150 ng of probe in HYB buffer overnight at 68°C.
On the next day, embryos were washed sequentially with 75% HYB/2X SSC, 50% HYB/2X SSC, 25% HYB/2X SSC, 2X SSC, 10 minutes for each step, followed by two 30 minute washes with 0.2X SSC at 68°C. Embryos were then equilibrated to room temperature and washed sequentially with 75% 0.2X SSC/PBT, 50% 0.2X SSC/PBT, 25% 0.2X SSC/PBT, and then PBT, 5 minutes for each wash. Embryos were incubated in 0.1 M glycine (pH 2.2) + 0.1% Tween-20 at room temperature, to remove endogenous alkaline phosphatases; this was followed by washes in PBT. Blocking was performed in PBT with 2 mg/ml BSA and 5% normal calf serum for 2 hours at room temperature. After blocking, embryos were incubated in PBT/BSA with anti-DIG antibody (Anti-digoxigenin, Fab fragments; Roche) at 1:2000 overnight at 4°C.
Embryos were washed in PBT/BSA and then further washed in NTMT buffer (0.1 M Tris-Cl (pH 9.5), 0.1 M NaCl, 0.05 M MgCl2, 0.1% Tween-20) at room temperature. Staining was then carried out in the dark at room temperature with 225 μg/ml NBT (4-Nitro blue tetrazolium chloride; Roche) and 175 μg/ml BCIP (5-bromo-4-chloro-3-indolyl-phosphate; Roche) in NTMT, until staining became visible. Embryos were imaged on a Zeiss AxioImager-Z1 microscope with a Zeiss AxioCam MRc camera. Figures were prepared using AxioVision 4.5 software and Adobe Illustrator; image manipulations were limited to linear level adjustments, rotations and scaling.
Electron microscopy
The hypothalamus was dissected from E18.5 mouse brains and then fixed in either of two electron microscopy-compatible fixatives. The hypothalamus was processed either for immuno-electron microscopy or for preservation of fine structure.
For immuno-electron microscopy, the tissue was fixed with cold 4% PFA/0.1% glutaraldehyde in PBS (pH 7.4) for 2 hours, washed twice in water, dehydrated in a graded ethanol series, and embedded in LR White. Semi-thin (0.08-0.20 μm) sections were cut using a Diatome diamond knife on a Reichert Ultracut E ultramicrotome and were placed on carbon-coated, Formvar-coated hexagonal nickel grids. The sections were blocked for 2 hours in a blocking solution containing 40 μg/ml goat sera, 20 μg/ml BSA in TBS buffer (containing 1 μg/ml BSA, 0.05% Tween-20, 0.5 M NaCl, 20 mM NaN3, pH 7.4), exposed to the anti-Ahi1 rabbit polyclonal antibody (Ab-05) in TBS buffer (4.0 μg/ml, pH 7.4). Following an overnight incubation in the Ahi1 antibody, the sections were washed four times in TBS, and then labeled with a 1:100 dilution of 10-nm gold particles conjugated to goat anti-rabbit IgG antibodies (Ted Pella Inc., Redding, CA) in TBS buffer with 10 μg/ml goat sera for 2 hours. Sections then were washed five times in TBS, and the antibodies were covalently linked with 1% glutaraldehyde in water. The samples were stained with uranyl acetate for 15 minutes, followed by three water washes. Substitution of rabbit IgG for the primary antibody provided a negative control. Sections were analyzed at 80 kV with a Zeiss 910 transmission electron microscope (Carl Zeiss, Oberkochen, Germany).
For fine structure studies, the tissue was fixed in cold 6.5% glutaraldehyde in PBS (pH 7.4) for 2 hours, washed two times, 10 minutes per wash in PBS, and rinsed in 0.1 M sodium cacodylate buffer (pH 7.4) twice, 10 minutes per rinse. Samples were post-fixed in 1% osmium tetraoxide in cacodylate buffer for 1 hour, washed in water overnight, dehydrated in a graded acetone series, and embedded in Epon 812/Araldite. Semi-thin (0.08-0.20 μm) sections were stained with uranyl acetate and Reynold’s lead. Sections were viewed on a Zeiss 910 transmission electron microscope at 80 kV.
Results
Expression of Ahi1 in various mouse organs
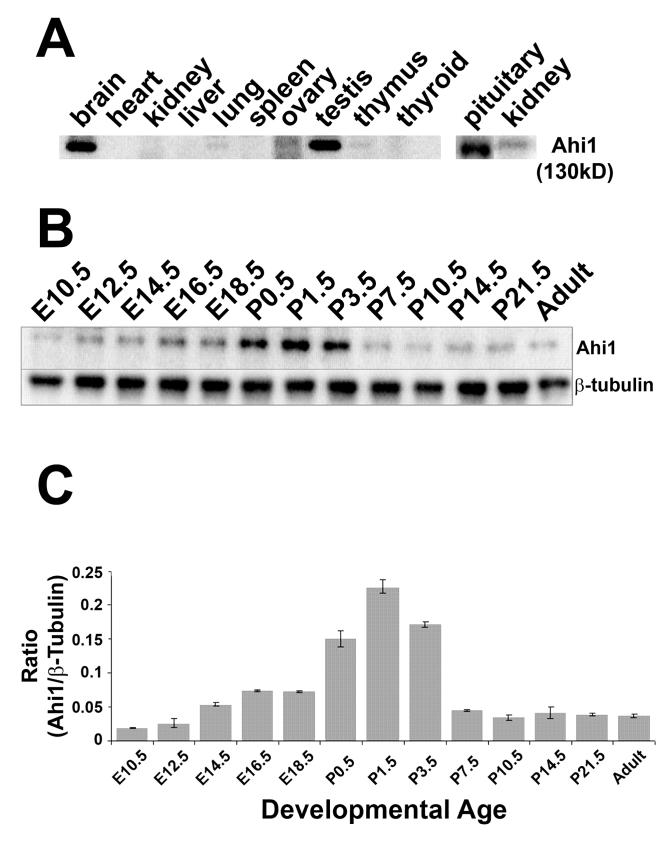
Following confirmation of the specificity of our Ahi1 antibodies (Fig. 1), we performed Western blot analysis of organs in the mouse and revealed widespread Ahi1 expression in a number of organ systems (Fig. 2A). Ahi1 expression was highest in the brain and testes. Expression occurred in the pituitary (also see Fig. 3F) and at lower levels in the thymus, lung, and kidney (Fig. 2A). Ahi1 was expressed in the pancreas at levels so low, it could only be detected by Ahi1 immunoprecipitation (data not shown). No obvious expression was found in heart, liver, spleen, and thyroid.
Figure 2.
Western blot analyses of Ahi1 expression in mouse tissues. A, Ahi1 is expressed in various organs of the mouse. The highest levels of expression occur in brain and testes. High Ahi1 expression is also observed in the pituitary gland (note that this blot was from another experiment and expression levels cannot be directly compared) with lower Ahi1 levels in the thymus, lung, and kidney (higher concentrations of protein are needed to reveal Ahi1 expression in the kidney (compare left kidney blot to right). B, Representative Western blot of the temporal expression pattern of Ahi1, in whole mouse brain, indicates that the highest levels of expression occur during the first 4-days following birth. β-tubulin expression serves as a loading control. C, Quantification of Ahi1 levels in whole mouse brains across developmental ages. Ahi1 and β-tubulin levels are quantified and standardized according to β-tubulin expression, averaged across 3 experiments, and presented as the ratio of Ahi1/ β-tubulin. E: embryonic day; P: post-natal day
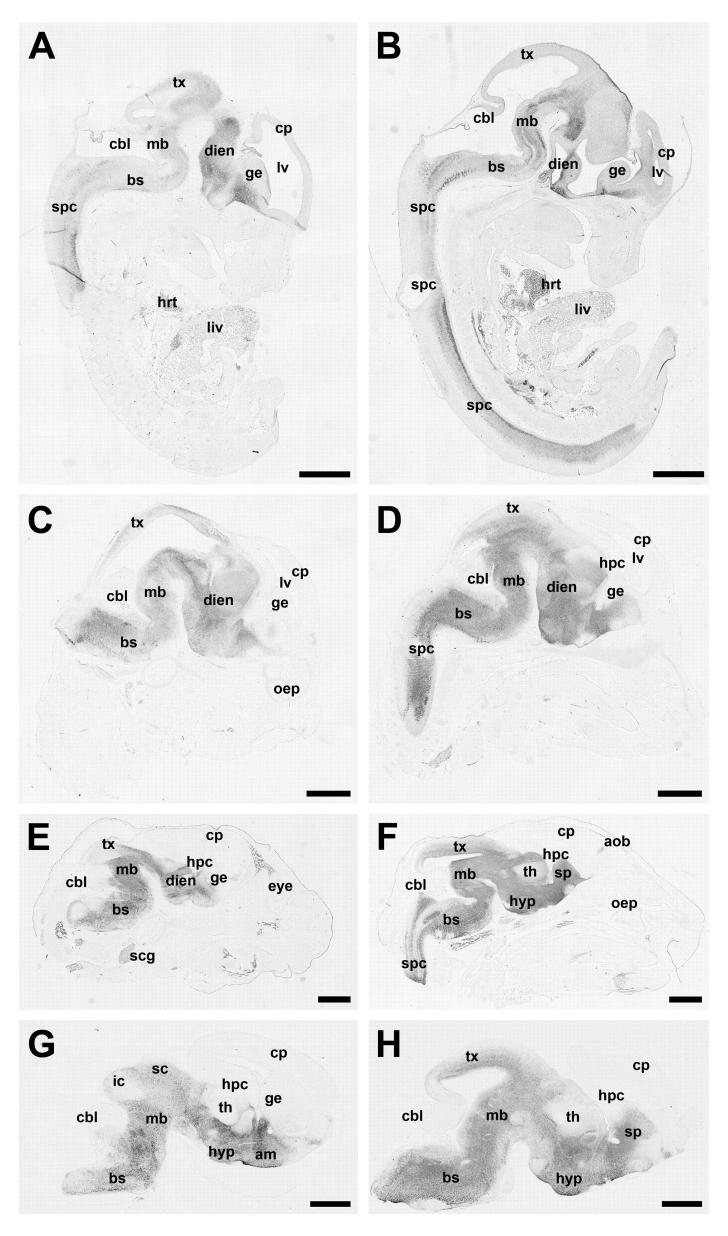
Figure 3.
Representative photomicrographs demonstrate the expression pattern of Ahi1 in the developing mouse at embryonic day (E) 12.5 (A,B), E14.5 (C,D), E16.5 (E,F) and E18.5 (G,H) in lateral sections through the brain (A,C,E,G) and in midsagittal sections through the brain (B,D,F,H). Ahi1 expression is most pronounced in the ventral aspects of the developing brain (developing amygdala, hypothalamus, midbrain, brainstem, and spinal cord) with no detectable immunoreactivity in the developing cortical plate or cerebellum. Bar indicates 1 mm. am: amygdala; aob: accessory olfactory bulb; bs: brainstem; cbl: cerebellum; cp: cortical plate; dien: diencephalon; ge: ganglionic eminences; hpc: hippocampus; hyp: hypothalamus; ic: inferior colliculus; lv: lateral ventricle; mb: midbrain; oep: olfactory epithelium; sc: superior colliculus; spc: spinal cord; scg: superior cervical ganglion; sp: septal nucleus; th: thalamus; tx: tectum
Temporal expression of Ahi1 in the mouse brain
Western blot analysis of Ahi1 in whole organs clearly demonstrated high levels of Ahi1 in the brain. Therefore, we further examined the patterns of spatial and temporal expression of Ahi1 in the mouse brain. Western blot analysis of whole mouse brain indicated that Ahi1 was expressed at all ages examined (E10.5-adult). At E10.5, there was relatively weak expression of Ahi1; expression gradually increased until birth (Fig. 2B,C). From P0.5 to P3.5, Ahi1 expression was maximal, but then from P3.5 to adult, decreased to levels comparable to the level seen in embryogenesis (Fig. 2B,C). These results suggest that Ahi1 levels are regulated during brain development, but the continued expression in the mature brain may indicate an additional role of Ahi1 beyond that in brain development.
Spatial expression of Ahi1 in the mouse brain
Analysis of Ahi1 immunolocalization in the mouse brain indicated that the Ahi1 protein had a very restricted expression pattern, principally in the ventral regions of the developing and mature forebrain, midbrain, and hindbrain (Figs. 3&4). Ahi1 expression was found in the cytoplasm of neurons and in neuronal processes.
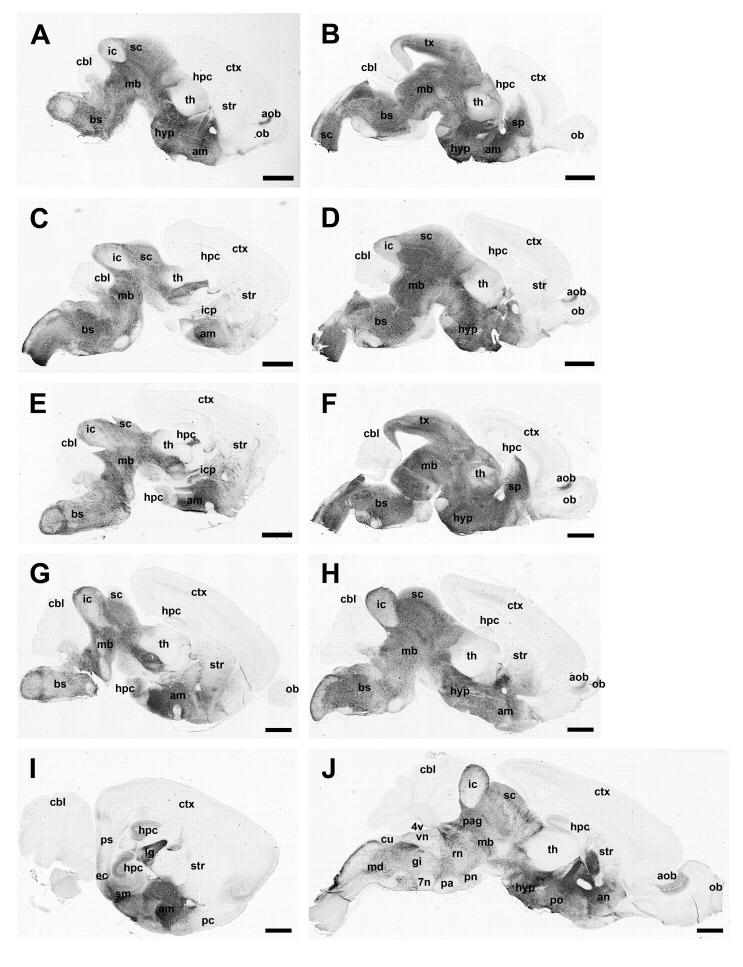
Figure 4.
Representative photomicrographs demonstrate the expression pattern of Ahi1 in the developing and mature mouse brain at post-natal day (P) 0.5 (A,B), P1.5 (C,D), P3.5 (E,F), P7.5 (G,H), P21.5 (I,J) in lateral sections through the brain (A,C,E,G,I) and in midsagittal sections through the brain (B,D,F,H,J). Ahi1 expression is most pronounced in the accessory olfactory bulb, amygdala, hypothalamus, ventral hippocampus (with weak expression in the dorsal hippocampus), midbrain (various nuclei of the superior and inferior colliculus), and brainstem, with no detectable immunoreactivity in the cerebellum and most of the cerebral cortex (exception entorhinal cortex). Bar indicates 1 mm. am: amygdala; an: nucleus accumbens; aob: accessory olfactory bulb; bs: brainstem; cbl: cerebellum; ctx: cerebral cortex; cu: cuneate nucleus; ec: entorhinal cortex; gi: gigantocellular reticular nucleus; hpc: hippocampus; hyp: hypothalamus; ic: inferior colliculus; icp: internal capsule; lg: lateral geniculate nucleus; mb: midbrain; md: medullary reticular nucleus; ob: olfactory bulb; pag: periaqueductal gray; pc: piriform cortex; pn: pontine nucleus; po: preoptic area; pa: periolivary area; ps: parasubiculum; rn: reticular nucleus (pontine); sc: superior colliculus; sm: subiculum; sp: septal nucleus; str: striatum; th: thalamus; vn: vestibular nucleus, 7n: cranial nerve seven; 4v: fourth ventricle
The olfactory bulb showed little Ahi1 immunoreactivity, except in the accessory olfactory bulb (Figs. 3F&4A,D,F,H,J). Accessory olfactory bulb immunoreactivity initially appeared in a single group of cells early in development. This expression gradually expanded, around P10.5, to two parallel groups of cells, the mitral and granular layers of the accessory olfactory bulb, a pattern that continued through adulthood.
Cerebral cortical expression of Ahi1 was almost completely absent, at each developmental age examined (Figs. 3&4). The exceptions were the medial entorhinal cortex, perirhinal cortex, and piriform cortex, all of which showed low levels of Ahi1 immunoreactivity. The ventrocaudal region of the cortex also had very low levels of Ahi1 immunoreactivity, beginning around E16.5.
The hippocampus had a restricted pattern of Ahi1 immunoreactivity, with expression of Ahi1 occurring specifically in the dentate gyrus (Figs. 3&4). Ahi1 expression in the dentate gyrus was higher in the ventral hippocampus than in the dorsal hippocampus. During early developmental ages, dorsal hippocampal expression of Ahi1 was entirely absent (data not shown). Expression of Ahi1 in the ventral hippocampal dentate gyrus became evident around E14.5 and continued to increase into adulthood (Fig. 4E,G,I). The distribution became more comparable between dorsal and ventral hippocampus, by P21.5, but it was skewed toward the ventral hippocampus at all time points (Fig. 4I,J). The CA fields showed low levels of Ahi1 immunoreactivity, but only in the ventral hippocampus. Moderate Ahi1 expression was observed in the fimbria and subiculum (Fig. 4I).
The amygdala and hypothalamus were the two brain structures with the highest levels of Ahi1 immunoreactivity (Figs. 3&4). In both amygdala and hypothalamus, Ahi1 expression was observed as early as E10.5; expression rose to its highest levels in the period from P0.5 to P7.5. Significant reductions at P10.5 were noted in both amygdala and hypothalamus (data not shown). However, whereas levels of Ahi1 immunoreactivity in the hypothalamus remained steady into adulthood, levels of Ahi1 expression in the amygdala tapered off at P14.5 and remained constant into adulthood (data not shown). The hypothalamus had high, fairly uniform Ahi1 immunoreactivity in the majority of sub-nuclei (see Table 1) with the highest levels of Ahi1 immunoreactivity in the ventromedial, dorsomedial, arcuate hypothalamic nuclei, dorsal nucleus, and the septohypothalamic nucleus. In the amygdala, Ahi1 was most preferentially observed in the central nucleus of the amygdala, the medial amygdaloid nucleus, the amygdalostriatal transition area, and the intercalated nuclei (see Table 1 for other regions of Ahi1 expression in amygdala).
Table 1.
Detailed expression patterns of Ahi1 in the Adult Mouse Forebrain
| Anatomical Structure | Expression Levels |
Presence in Stigmoid Bodies |
|---|---|---|
| Olfactory bulb | ||
| Accessory olfactory bulb | ++ | - |
| Main olfactory bulb | - | - |
| Cerebral cortex | ||
| Entorhinal cortex | + | - |
| Perirhinal cortex | + | - |
| Piriform cortex | + | - |
| Other cortical regions | - | - |
| Hippocampus | ||
| Dentate gyrus | ||
| dorsal | + | - |
| ventral | +++ | - |
| Hilus | - | - |
| CA1 region | ||
| dorsal | - | - |
| ventral | + | - |
| CA3 region | ||
| dorsal | - | - |
| ventral | + | - |
| Subiculum | ++ | + |
| Fimbria | ++ | + |
| Amygdala | ||
| Basolateral nucleus | - | - |
| Basomedial nucleus | ++ | + |
| Central nucleus | +++ | + |
| Medial nucleus | +++ | + |
| Lateral nucleus | + | - |
| Cortical nucleus | ||
| Anterior | ++ | + |
| Posteromedial, ventral | + | + |
| Posterior | ++ | - |
| Intercalated cells | +++ | - |
| Amygdalopiriform transition area | + | - |
| Amygdalostriatal transition area | +++ | + |
| Amygdalohippocampal area | + | + |
| Interstitial nucleus of the posterior limb of the anterior commissure |
+ | + |
| Septal and basal magnocellular nuclei | ||
| Bed nucleus of stria terminalis | +++ | + |
| Nucleus accumbens | + | + |
| Substantia innominata | + | + |
| Septal lateral nucleus | +++ | + |
| Septal medial nucleus | +++ | + |
| Striatopallidal system | ||
| Caudate putamen | + | - |
| Ventral pallidum | ++ | + |
| Thalamus | ||
| Anterior area | ++ | + |
| Dorsal area | +++ | + |
| Lateral area | + | + |
| Posterior area | ++ | + |
| Parafascicular nucleus | +++ | + |
| Paraventricular nucleus | +++ | + |
| Reticular nucleus | + | + |
| Ethmoid nucleus | ++ | + |
| Subthalamic nucleus | + | - |
| Zona incerta | ++ | + |
| Posterior intralaminar nucleus | ++ | + |
| Posterior triangular nucleus | ++ | + |
| Stria medullaris nucleus | +++ | + |
| Stria terminalis | ++ | + |
| Dorsal lateral geniculate nucleus, lateral aspects |
+++ | + |
| Ventrolateral geniculate nucleus, lateral aspects |
+++ | + |
| Medial geniculate nucleus | ||
| dorsal | +++ | + |
| ventral | +++ | + |
| Lateral habenular nucleus | + | + |
| Hypothalamus | ||
| Preoptic nucleus | ++ | + |
| Paraventricular nucleus | ++ | + |
| Dorsomedial nucleus | +++ | + |
| Ventromedial nucleus | +++ | + |
| Arcuate nucleus | +++ | + |
| Premammillary nucleus, ventral | +++ | + |
| Lateroanterior nucleus | ++ | + |
| Dorsal nucleus | +++ | + |
| Stigmoid nucleus | ++ | + |
| Septohypothalamic nucleus | +++ | + |
| Striohypothalamic nucleus | ++ | + |
| Tuberal nucleus | + | + |
| Terete nucleus | + | + |
| Parasubthalamic nucleus | ++ | + |
| Supraoptic nucleus | +++ | + |
| Medial forebrain bundle | + | + |
+, present (for expression level, low); ++, moderate expression; +++, high expression; -, not present.
Other areas of the forebrain had varying levels of Ahi1 immunoreactivity (Figs. 3&4; Table 1). Thalamic expression of Ahi1 was observed mainly around the exterior borders of the thalamus, including the lateral and medial geniculate nuclei (Fig. 3&4; Table 1). The supraoptic nucleus had very high levels of Ahi1 immunoreactivity, while only moderate expression was noted in the median preoptic nucleus. The septal nucleus also demonstrated high Ahi1 immunoreactivity, particularly in the lateral nucleus. Moderate immunoreactivity was observed in the ventral pallidum, but low Ahi1 levels were seen in the caudate-putamen. The stria terminalis had moderate Ahi1 immunoreactivity, but the bed nucleus of the stria terminalis showed significantly higher levels. The premammillary nucleus had relatively high levels, particularly in ventral regions, but the remaining areas of the mammillary nuclei were devoid of immunoreactivity. Lastly, the medial forebrain bundle and the nucleus accumbens (core and shell) both had low levels of Ahi1, similar to the levels observed in the substantia innominata and the lateral habenural nucleus.
Ahi1 immunoreactivity was seen in numerous areas of the midbrain, including the tectum and a number of tegmental nuclei (Figs. 3&4; Table 2). High expression of Ahi1 was found in the inferior colliculus, particularly in the exterior and dorsal cortex of this structure. The central nucleus almost completely lacked expression. Ahi1 immunoreactivity tended to be higher in the ventral aspects of the inferior colliculus than in the dorsal inferior colliculus. Ahi1 levels in the superior colliculus varied depending on the layer and the age of the mouse. The superficial gray layer had its highest Ahi1 levels at E14.5 and at P14.5, while Ahi1 immunoreactivity in the intermediate white layer was highest at E18.5 (Figs. 3&4). The optic nerve layer of the superior colliculus had one of the highest levels of Ahi1 immunoreactivity of any region in the adult brain, although the deep gray layer, the intermediate gray layer, and the superficial gray layer all showed only moderate levels of Ahi1. High Ahi1 immunoreactivity was also observed in the caudal central gray and central gray pons. The intercollicular nucleus as well as a number of tegmental nuclei had moderate levels of Ahi1 expression. Other areas of the midbrain that had low levels of Ahi1 immunoreactivity included the rhabdoid nucleus and the median raphe nucleus. Lastly, the deep mesencephalic nucleus, the retrorubral field, and the locus coeruleus all had relatively low levels of Ahi1 immunoreactivity.
Table 2.
Detailed expression patterns of Ahi1 in the Adult Mouse Midbrain and Hindbrain
| Anatomical Structure | Expression Levels |
Presence in Stigmoid Bodies |
|---|---|---|
| Midbrain | ||
| Pretectal nucleus | + | + |
| Precommissural nucleus | ++ | + |
| Intercollicular nucleus | ++ | + |
| Inferior Colliculus | ||
| Brachium nucleus | + | + |
| Central nucleus | + | - |
| Dorsal cortex | +++ | + |
| External cortex | +++ | + |
| Superior Colliculus | ||
| Optic nerve layer | +++ | + |
| Deep Gray layer | ++ | + |
| Intermediate gray/white layers | ++ | - |
| Superficial gray layer | ++ | - |
| Periaqueductal gray, dorsomedial and lateral |
++ | + |
| Central Gray | +++ | + |
| Cuneiform nucleus | ++ | + |
| Retrorubral field | + | + |
| Deep mesencephalic nucleus | + | + |
| Dorsal tegmental nucleus (central/pericentral) |
++ | + |
| Microcellular tegmental nucleus | ++ | + |
| Pedunculopontine tegmental nucleus | + | + |
| Ventral tegmental area | + | + |
| Rhabdoid tegmental nucleus | + | + |
| Retroparafascicular nucleus | ++ | + |
| Cerebellum | ||
| Hemispheres | - | - |
| Vermis | - | - |
| Dentate nucleus | + | - |
| Interposed nucleus | + | + |
| Fastigial nucleus | + | - |
| Superior cerebellar peduncle | +++ | + |
| Pons and medulla oblongata | ||
| Locus coeruleus | + | + |
| Median raphe nucleus | + | + |
| Reticular nucleus | ++ | + |
| Solitary (tract) nucleus | ++ | + |
| Inferior olive | + | + |
| Lateral lemniscus | + | + |
| Parabrachial nucleus | +++ | + |
| External cuneate nucleus | + | + |
| Paralemniscal nucleus | ++ | + |
| Spinal trigeminal nucleus | +++ | + |
| Dorsomedial spinal 5 nucleus | + | + |
+, present (for expression level, low); ++, moderate expression; +++, high expression; -, not present.
Surprisingly, the majority of the developing and mature cerebellum, including the cerebellar vermis, was devoid of Ahi1 immunoreactivity; however, Ahi1 expression was seen in the deep cerebellar nuclei, particularly the dentate nucleus (Figs. 3, 4, and 5A; Table 2). As a whole, the developing brainstem showed a more widespread expression of Ahi1 that became restricted by adulthood (Figs. 3&4). The decrease in Ahi1 immunoreactivity became obvious at P14.5; from this age onward Ahi1 levels lessened.
Figure 5.
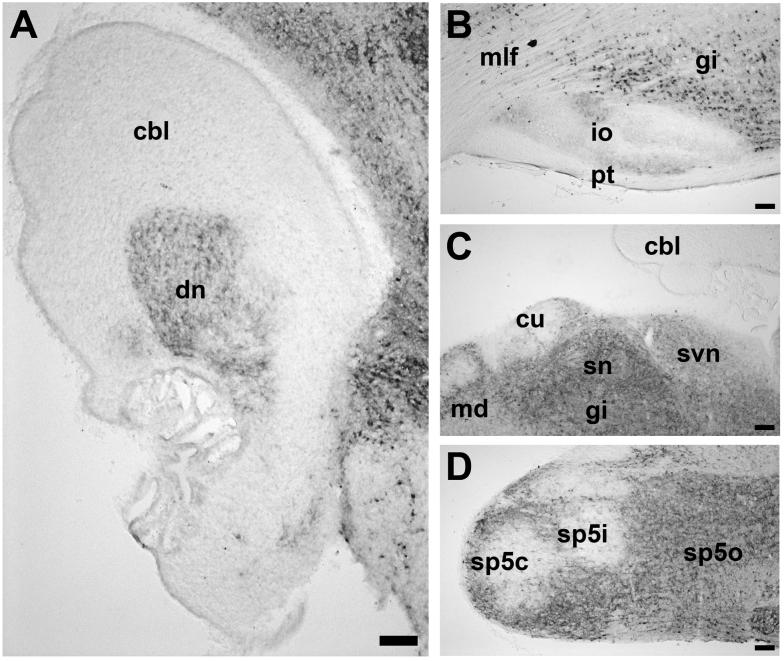
High magnification photomicrographs demonstrate the expression of Ahi1 in sagittal sections through the dentate nucleus of the cerebellum (A), the inferior olive (B), the solitary nucleus (C), and the spinal 5 nucleus (D) from P0.5 brains (A,C,D) and P21.5 brain (B). Bar indicates 100 μm. cbl: cerebellum; cu: cuneate nucleus; dn: dentate nucleus of the cerebellum; io: inferior olive; gi: gigantocellular reticular nucleus; md: medullary reticular nucleus; mlf: medial longitudinal fasciculus; pt: pyramidal tract; sn: solitary nucleus; svn: spinal vestibular nucleus; sp5c: spinal 5 nucleus, caudal; sp5i: spinal 5 nucleus, interpolar; sp5o: spinal 5 nucleus, oral.
Ahi1 immunoreactivity in the spinal trigeminal nucleus was restricted to the outer margins, forming a distinct band around the nucleus and leaving the center virtually devoid of immunoreactivity (Fig. 5D). The reticular nucleus, including the ventrolateral reticular nucleus and the gigantocellular reticular nucleus, had moderate Ahi1 immunoreactivity (Fig. 5B,C). The solitary nucleus had moderate levels of Ahi1 immunoreactivity (Fig. 5C); these levels were consistent throughout development and into adulthood. Ahi1 immunoreactivity was also noted in the inferior olives (Fig. 5B). Moderate Ahi1 expression was also observed in the interior lateral parabrachial nucleus and the external cuneate nucleus. Low Ahi1 immunoreactivity was found in the lateral lemniscus nuclei and the dorsomedial spinal 5 nucleus.
Among the cranial nerve nuclei, the motor trigeminal nucleus, the facial nucleus, and the hypoglossal nucleus all displayed moderate Ahi1 levels beginning around P3.5, but immunoreactivity had disappeared by adulthood. The motor trigeminal and facial nuclei had their highest Ahi1 expression around P7.5, while the expression in the hypoglossal nucleus peaked around P10.5.
AHI1 expression in the developing and mature human brain
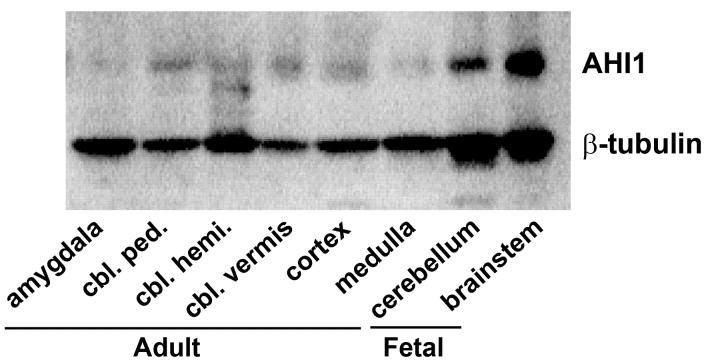
The lack of Ahi1 expression that we found in the cerebellum of the mouse was striking, given that the cerebellum is one of the neuroanatomical structures malformed in humans with JBTS. As mentioned previously, the gene and protein structures of human AHI1 are distinct from those for rodent Ahi1, particularly with respect to the N-terminus of the gene product, a region containing a putative coiled-coil domain. Therefore, we determined the expression profile of AHI1 in the developing and mature human brain. AHI1 was expressed in human fetal cerebellum and brainstem (Fig. 6). Similarly, AHI1 expression was observed in the adult cerebellar hemispheres, cerebellar peduncles, cerebellar vermis, and medulla. The adult cerebral cortex and amygdala also had AHI1 expression (Fig. 6). The highest expression of AHI1 in adult tissue (normalized to β-tubulin levels) was in the mature cerebellar vermis and cerebellar peduncles, followed by cerebellum and cerebral cortex (data not shown). Amygdala and medulla had modest levels of AHI1 in our analysis.
Figure 6.
AHI1 expression in human central nervous system. A, AHI1 expression in the adult and fetal human brain demonstrates that AHI1 is present in the developing cerebellum and brainstem, and is also present in the adult brain.
ahi1 mRNA in situ hybridization in the developing zebrafish
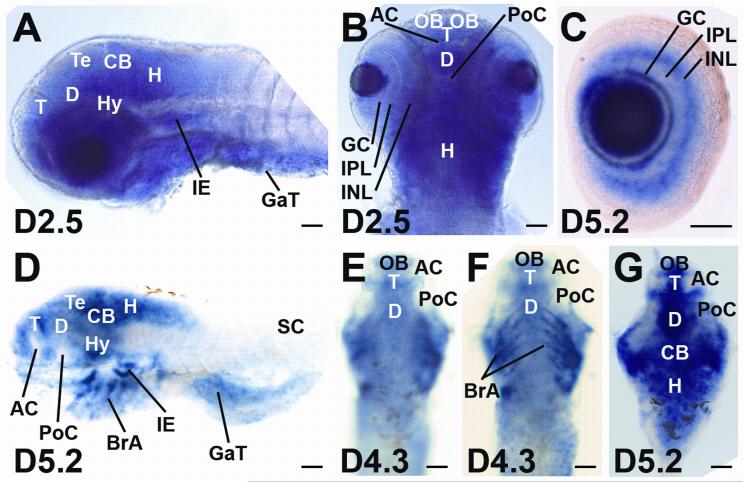
AHI1 orthologs are found in all vertebrates. Previous work demonstrates that zebrafish ahi1 is expressed in adult zebrafish gonads, specifically in the primary spermatocytes and oocytes (Zhou and Song, 2006). To determine whether expression in the cerebellum is primate-specific, we examined the pattern of ahi1 mRNA distribution in the zebrafish. Whole-mount in situ hybridization for ahi1 mRNA in late embryo and larval-stage zebrafish (2.5 dpf, 3 dpf, 3.2 dpf, 4.3 dpf, 4.5 dpf, and 5.2 dpf) demonstrated expression throughout the brain and retina, but expression was absent in the spinal cord (Fig. 7A-G). All antisense probes gave the same expression pattern. In brain, ahi1 expression was detected in the olfactory bulb, telencephalon, diencephalon, tectum, and cerebellum, while no staining was seen in the anterior or postoptic commissure (Fig. 7A,B,D,E,G). In the retina, ahi1 expression was observed in the ganglion cell layer and the inner nuclear layer, but not in the inner plexiform layer (Fig. 7C). Expression of ahi1 was also seen in the posterior brachial arches, the gastrointestinal tract, and the inner ear (Fig. 7A,D,F). Staining of the brachial arches occurred in cells surrounding the cartilage cells, but apparently not in the cells of the cartilage itself, suggesting that these ahi1-expressing cells are derived from the neural crest (Fig. 7D,F).
Figure 7.
ahi1 expression in zebrafish central nervous system. A-G, Whole-mount in situ hybridization was performed using D2.5 (A, B) D4.3 (E, F) and D5.2 (C, D, G) zebrafish embryos with ahi1 antisense riboprobe 1 (A, B) and riboprobe 2 (C-G). Eyes of the embryos were dissected in D-F and only the brain is removed and shown in G. Bar indicates 50 μm. AC: anterior commissure; BrA: brachial arches; CB: cerebellum; D: diencephalon; GaT: gastrointestinal tract; GC: granule cell layer; H: hindbrain: Hy: hypothalamus; IE: inner ear; INL: inner nuclear layer; IPL: inner plexiform layer; M: midbrain; PoC: postoptic commissure; OB: olfactory bulb; SC: spinal cord; T: telencephalon; Te: tectum
Cellular localization of Ahi1: Neuronal cell bodies and processes
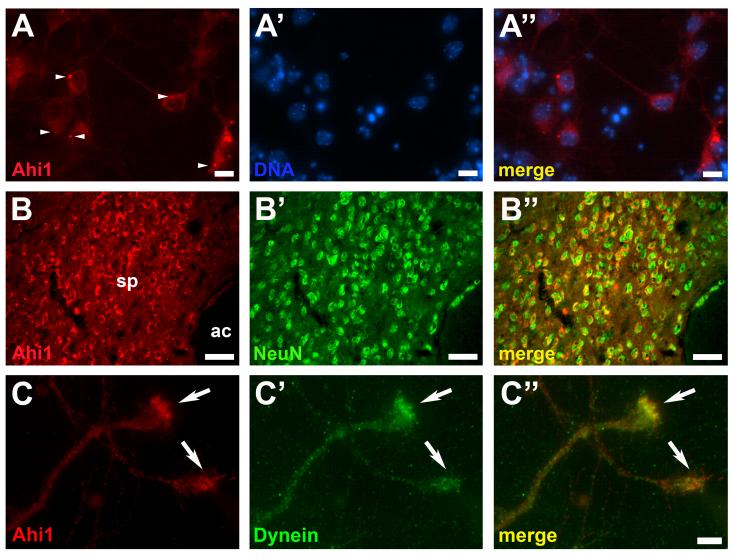
To assess the cellular localization of mouse Ahi1, we performed immunocytochemical staining in neuronal cultures and murine brain tissue sections. Ahi1 was localized in primary hypothalamic neuronal cultures (E17.5) principally in the cytoplasm and the processes of neurons (as determined by morphology and use of the nuclear neuronal marker, NeuN [data not shown]), with little nuclear expression (Fig. 8A). Ahi1 expression in the mouse brain was limited to post-mitotic, differentiated neurons, as indicated by the labeling of Ahi1 and NeuN in the same cells (Fig. 8B). To determine whether Ahi1 is associated with glial cells, we labeled mouse brain sections with the glial marker, GFAP. No match in labeling patterns was observed between Ahi1 and GFAP (data not shown) indicating that Ahi1 was not present in glial cells. Ahi1 was also consistently found in axons with concentrated immunoreactivity in the axonal growth cone and in co-localization with a growth cone marker, dynein (Fig. 8C)(Grabham et al., 2007).
Figure 8.
Representative fluorescent photomicrographs demonstrate the expression of Ahi1 (red) in cultured primary hypothalamic neurons (A), in the septal nucleus of mouse brain (B), and in cultured primary hypothalamic neuronal axon growth cones (C). Ahi1 (red) is localized to the cytoplasm of neurons as well as to neuronal processes (A) with low expression in the nucleus (A’,A”). Arrowheads indicate Ahi1-positive stigmoid bodies. Bar indicates 10 μm. Ahi1 primarily is expressed in neurons given the co-immunoreactivity of Ahi1 (red) (B) with NeuN (green), a marker of differentiated neurons (B’,B”). Bar indicates 50μm. ac: anterior commissure; sp: septal nucleus. Ahi1 (red) was observed in axons and axonal growth cones (C) with co-localization with a growth cone marker, dynein (C’,C”). Arrows indicate Ahi1-positive growth cones. Bar indicates 5 μm. Magenta-green converted images are available as supplemental figures (Supplementary Figure 2).
Subcellular localization of Ahi1: Stigmoid bodies
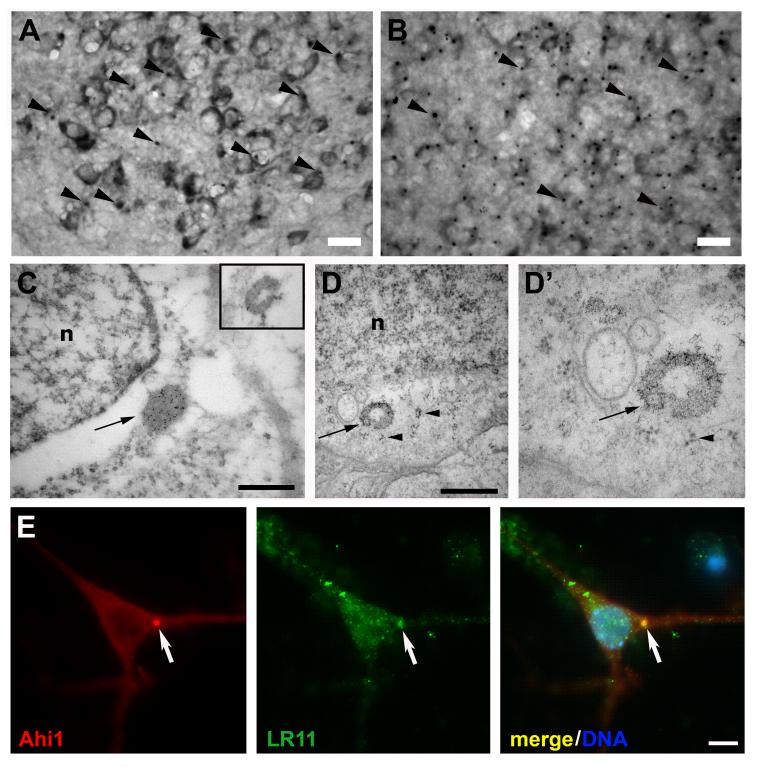
To determine the subcellular distribution of Ahi1 within neurons, we carried out Ahi1 immunostaining and electron microscopy. Our initial staining for Ahi1 consistently showed high expression levels of Ahi1 in distinct spherical punctate foci in the cytoplasm of neurons, usually in close proximity to the nucleus, in many brain regions (Figs. 8A and 9A,B). These foci were typically on the order of 1-3 μm in diameter. We eliminated the possibility that these foci represent centrosomes or basal bodies, on the basis that they were never found to contain centrosomal or basal body markers such as pericentrin and γ-tubulin (data not shown). Immuno-electron microscopy was conducted to identify any structures within the cell that were associated with these foci. Immunolabeling of Ahi1 on sections through the mouse hypothalamus identified a structure (Fig. 9C) that corresponds spatially and morphologically to the foci observed with immunohistochemistry and light microscopy (Fig. 9A,B). Since immuno-electron microscopy requires a balance between cell preservation and antigenicity, we also performed electron microscopy on tissue fixed for ultrastructural analysis, in the absence of antibody. The candidate structure, which was usually found proximal to the nucleus and close to polyribosomes (Fig. 9C,D,D’), was an apparently proteinaceous mass consisting in part of the Ahi1 protein (Fig. 9C). This structure seemed to correspond to previously reported non-membrane bound cytoplasmic organelles called stigmoid bodies, also known as nematosomes, nucleolus-like bodies, or nucleolus-like inclusions (Gutekunst et al., 1998; Li et al., 1998; Shinoda et al., 1992). One of the proteins that has been reported to uniquely localize to the stigmoid body was LR11, a lipoprotein receptor homologue that contains 11 low-density lipoprotein receptor repeats, 5 low-density lipoprotein receptor YWTD repeats, a large fibronectin-type III hexarepeat domain, and a domain similar to a yeast receptor involved in vacuolar protein sorting (Gutekunst et al., 2003). To further confirm the identity of this structure as a stigmoid body, we performed multiple marker immunocytochemistry for both Ahi1 and LR11. Ahi1 and LR11 both co-localized to the same cytoplasmic structure, the stigmoid body, in cultured hypothalamic cells (Fig. 9E), confirming that this structure is a stigmoid body. Moreover, the structural characteristics of this organelle in our electron micrographs (Fig. 9D,D’) were consistent with previous descriptions of stigmoid bodies (Gutekunst et al., 1998; Li et al., 1998; Shinoda et al., 1992).
Figure 9.
Localization of Ahi1 to a non-membrane limited organelle called a stigmoid body. Immunohistochemical localization of Ahi1 to stigmoid bodies (black arrowheads) in the (A) amygdala and the (B) hypothalamus of the mouse brain. Bar indicates 20 μm. C, EM immunolocalization of Ahi1 shows that Ahi1 antibodies (gold particles) localize to an organelle, often found near the nucleus (n), called the stigmoid body (see black arrow). C, inset, The stigmoid body (gold-labeled) is ring-shaped in cross-section. D, D’, In well-preserved material, fixed for fine structure analysis, the ring-shaped stigmoid body (arrow) is confirmed not to be membrane bound, but has a chromatin-like appearance, whose components were packed together with a certain periodicity, with nearby polyribosomes (arrowheads). n, nucleus. Bars in C, D indicate 0.5 μm. LR11 also demonstrated co-localization to stigmoid bodies (Gutekunst et al., 2003) and therefore we performed co-labeling experiments for both LR11 and Ahi1. E, LR11 immunostaining (green) had identical labeling of stigmoid bodies as Ahi1 (red) demonstrating that Ahi1 is localized to stigmoid bodies (white arrows point to LR11/Ahi1 localization to stigmoid bodies). Bar in E indicates 5 μm. Magenta-green converted images are available as supplemental figures (Supplementary Figure 3).
Ahi1 localization to stigmoid bodies in the mouse brain
Ahi1 is associated with stigmoid bodies in some, but not all, neurons (and in some, but not all, brain regions) (Tables 1&2). Most neurons had only one Ahi1-immunoreactive stigmoid body, but there were occasional neurons that had 2-3 stigmoid bodies. No obvious temporal (developmental) Ahi1 expression variations were noted for the stigmoid bodies, with the exception that Ahi1-immunoreactive stigmoid bodies were only present in post-mitotic, differentiated neurons. There were a few areas that lacked Ahi1-immunoreactive stigmoid bodies entirely, despite having Ahi1 expression (see Tables 1&2) while a number of structures were remarkable in their abundance of Ahi1-immunoreactive stigmoid bodies (see Tables 1&2). There did not appear to be any correlation between the levels of Ahi1 immunoreactivity and the presence of stigmoid bodies. Since Ahi1-immunoreactive stigmoid bodies were not observed in every neuron that had Ahi1 expression, we need to determine whether non-immunoreactive stigmoid bodies are nevertheless present in these cells, albeit not detected by the present methods.
Discussion
Expression analysis of Ahi1 indicates that the preponderance of the protein is found in the developing and mature ventral forebrain (particularly in the amygdala and hypothalamus), midbrain, and hindbrain. However, cerebellar expression of Ahi1 was absent in the mouse, but expression was present in human (AHI1) and zebrafish (ahi1); this difference in pattern suggests differential regulation of the AHI1 gene orthologs among species. Mouse Ahi1 was expressed predominantly in the cytoplasm and processes of neurons, as well as in a novel organelle called a stigmoid body. Moreover, the distinct expression patterns for Ahi1 suggest a role for Ahi1 in the signaling pathways of the ventral forebrain and hindbrain, possibly explaining the association of AHI1 with schizophrenia in humans (Amann-Zalcenstein et al., 2006; Ingason et al., 2007). Our human AHI1 and mouse Ahi1 expression results are a first step in deciphering the role of AHI1 in the clinical and neuroanatomical abnormalities observed in humans with JBTS, while uncovering unexpected areas of expression in the brain.
The developmental expression of Ahi1 in regions of the brain affected in individuals with JBTS suggests that AHI1 is critically involved in aspects of the development of these brain structures. The spatial expression of Ahi1 in brain regions, such as the inferior olives, solitary nuclei, and nucleus of cranial nerve V, structures known to be malformed in individuals with JBTS (i.e. (Yachnis and Rorke, 1999)), further demonstrates the importance of Ahi1 for the development of these structures. Mouse Ahi1 and human AHI1 expression was also observed in the deep cerebellar nuclei and superior cerebellar peduncles. Deep cerebellar nuclei localization of AHI1 is noteworthy given that individuals with JBTS have the presence of the molar tooth sign. The molar tooth sign is the hallmark radiographic sign for the diagnosis of JBTS, and is formed by an abnormal configuration of the tracts of the superior cerebellar peduncles projecting from the dentate nucleus of the cerebellum (Maria et al., 1997). The presence of AHI1, in the dentate nuclei of the cerebellum, suggests a developmental role for AHI1 in the formation of this hindbrain tract.
Previous mRNA expression analyses for Ahi1 in mouse (Ferland et al., 2004) and our current Ahi1 protein expression data demonstrated consistent temporal and spatial expression patterns. The two regions that did not share this pattern were regions of the cerebral cortex (not including the entorhinal, perirhinal, and piriform cortices) and the cerebellum. Whereas Ahi1 mRNA expression was observed in the mouse cerebral cortex and cerebellum, albeit at a weak level, Ahi1 protein was absent in these structures. There are three possible explanations for these discrepancies. First, the lack of an immunoreactive signal in the cerebral cortex and cerebellum could be the result of failure to detect very low levels of the protein. Second, the failure to detect protein in these regions may result from an absence of the expression of the particular Ahi1 epitope(s) found in the cerebral cortex and cerebellum, despite the presence of Ahi1 protein isoforms in these areas. Finally, the differences between Ahi1 mRNA and Ahi1 protein may reflect the inherent background signals that can be observed with in situ hybridization preparations, especially considering that the Ahi1 mRNA expression levels were very weak in mouse cerebral cortex and developing cerebellum (no Ahi1 mRNA was observed in more mature mouse cerebellum)(Ferland et al., 2004). Overall, we favor the third explanation since we have used multiple antibodies raised in different animals with 2 unique peptide antigens directed at alternative regions of Ahi1. Antibodies against these 2 peptides gave consistent expression profiles in mouse brain suggesting that failure to observe Ahi1 protein with these antibodies is unlikely due to the absence of both epitopes. These antibodies all have been tested using immunohistochemistry and Western blot analysis, and we have observed no Ahi1 protein in cortical or cerebellar tissue. Although we cannot completely rule out cerebral cortical (except for the areas mentioned previously) or cerebellar expression in mouse at the present time, we do not believe full-length Ahi1 protein to be expressed in these tissues.
Although our studies did not reveal any significant mouse Ahi1 immunoreactivity in the mouse cerebral cortex, except for low immunoreactivity in the entorhinal, perirhinal, and piriform cortices, human AHI1 expression was observed in human cerebral cortical protein lysates, by Western blotting. This suggests that AHI1 is expressed in the cerebral cortex of the developing and adult human brain. Unfortunately, our human AHI1 antibodies did not recognize AHI1 in fixed human brain sections, barring us from examining the spatial expression pattern of AHI1 in human brain tissue.
Individuals with JBTS and AHI1 mutations have agenesis of the cerebellar vermis, suggesting an involvement of AHI1 in cerebellar vermis development (Dixon-Salazar et al., 2004; Ferland et al., 2004). Surprisingly, mouse Ahi1 protein was not found to localize to the developing cerebellar hemispheres or vermis in mouse, but it was observed in the developing and mature human and zebrafish cerebellum. These results suggest significant species differences in AHI1 ortholog expression in the cerebellum. The differences in AHI1 ortholog expression likely result from divergence in the genomic structure of AHI1 between human/zebrafish and mouse, since the mouse Ahi1 gene is lacking part of the 5′-end of the Ahi1 gene sequence (which in part encodes a putative coiled-coil domain) (Ferland et al., 2004; Jiang et al., 2002). One possible explanation for the lack of Ahi1 expression in the mouse cerebellum is that the coiled-coil domain of human/zebrafish AHI1, or more likely the intronic regions adjacent to the region coding for the coiled-coil domain, contains regulatory elements responsible for human/zebrafish AHI1 expression in the developing and mature cerebellum. This could explain the lack of mouse Ahi1 immunoreactivity in the mouse cerebellum, but presence of it in the human and zebrafish cerebellum. A second explanation could be differences in the promoter region of AHI1 among human, zebrafish, and mouse that possibly resulted when mouse lost the 5′-end of the Ahi1 gene. This loss of a 5′-promoter region in mouse Ahi1, but not in human AHI1 or zebrafish ahi1 could have resulted in differential regulation of AHI1 expression among these species. Future studies are necessary to determine the exact causes of the species differences in expression of AHI1 in the developing and mature cerebellum.
Given our current understanding of the neuropathological and behavioral abnormalities in JBTS, unexpected regions of Ahi1 expression were identified in the mouse that should initiate further studies addressing whether these areas are also affected in individuals with JBTS and AHI1 mutations. Many brain areas that had high Ahi1 immunoreactivity are regions involved in either the relaying/processing of sensory information or are responsible for maintaining homeostasis. Among such regions were the amygdala, stria terminalis, ventral amygdalofugal pathway, and hypothalamus, suggesting a potential role of the AHI1 orthologs in the development of these structures. In addition, Ahi1 expression continued in the adult brain in these regions, suggesting a potential role for Ahi1 in signal transduction events involved in sensory and homeostatic processing.
The hypothalamus had the highest levels of mouse Ahi1 among all regions of the brain that we analyzed. One of the main effectors of hypothalamic function, the pituitary, also had extensive Ahi1 immunoreactivity. This suggests a potential role of Ahi1 in aspects of the hypothalamic-pituitary-adrenal axis and in hormone release from the pituitary. The hypothalamus is a very complex region of the brain that receives inputs from the brainstem, olfactory bulb, amygdala, septum, and cerebral cortex (Canteras and Swanson, 1992; Cliffer et al., 1991; Risold et al., 1997). The majority of these regions showed Ahi1 expression in our mouse experiments. Since the hypothalamus is considered the control center for most autonomic regulation, Ahi1 may play a role in modulating appetite, feeding, homeostasis, sexual activity, and hormonal regulation, and it may modulate development of the hypothalamus and pituitary. Whether individuals with JBTS have corresponding abnormalities in hypothalamic or pituitary function remains unknown.
In the mouse hippocampus, we observed an interesting developmental and dorsal/ventral expression pattern of Ahi1, mainly in the granule cells of the dentate gyrus. Early stage Ahi1 immunoreactivity was observed at a low level and only in the ventral hippocampus. As development continued, hippocampal Ahi1 expression became more pronounced in the ventral hippocampus, with still only a low level of Ahi1 immunoreactivity in the dorsal hippocampus. Dorsal/ventral hippocampal differences have been noted in rodents for such processes as adult hippocampal neurogenesis and plasticity (Burnham, 1975; Cameron et al., 1995; Ferland et al., 2002; Gould et al., 1998; Izaki et al., 2003; Maruki et al., 2001; Racine et al., 1977). Moreover, the entorhinal cortex, which projects to the hippocampus, was one of the few regions of the cerebral cortex that we found having Ahi1 immunoreactivity. Output regions of the hippocampus, such as the subiculum and fimbria, also showed Ahi1 immunoreactivity. Our data suggest that Ahi1 is present in many of the structures involved in the beginning and ending of the classic hippocampal tri-synaptic loop; only the hippocampal CA fields showed relatively little, to no, expression of Ahi1. The significance of this differential expression pattern for mouse Ahi1 remains to be determined, not only for the development of the hippocampus and the formation of efferent/afferent pathways, but also with respect to the functional role of this differential expression in the mature nervous system.
Mouse Ahi1 immunoreactivity and human AHI1 expression were observed in the developing and mature brainstem, suggesting the importance of AHI1 in the development of this structure. Moreover, absence of expression of AHI1 in the brainstem of individuals with JBTS may account for the brainstem abnormalities associated with JBTS, such as abnormal breathing and eye movements. The majority of the nuclei in the murine brainstem demonstrated widespread Ahi1 immunoreactivity until P21.5; then, the expression became restricted to areas such as the external cuneate nucleus, the reticular nucleus, the spinal trigeminal nucleus, and the caudal nucleus. The human counterparts of these nuclei, unlike the inferior olives, the solitary nuclei, and the nucleus of cranial nerve V, have not been described as being malformed, in individuals with JBTS. Whether individuals with JBTS have deficits in fine touch, in pain or temperature perception, or in regulating head movement, as would be expected from disruption of such pathways (Rub et al., 2003), has yet to be determined.
The stigmoid body was originally described in the 1960s (Toro and Rohlich, 1966) and has been reported to contain both RNA and protein, but no evidence of any DNA content (Ockleford et al., 1987). It has been given multiple names over the years, such as a nematosome, a nucleolus-like body, and a nucleolus-like inclusion. Stigmoid bodies are typically found near the nucleus and the perinuclear Golgi apparatus in differentiated, post-mitotic neurons, as well as in the cells of the placenta (Jones and Ockleford, 1985). Not every neuron in the brain has a stigmoid body that can be recognized by Ahi1 immunoreactivity, as evidenced by the results of our expression studies. Why neurons that show Ahi1 immunoreactivity do not have Ahi1-positive stigmoid bodies remains unknown. The presence of stigmoid bodies that do not show Ahi1 immunoreactivity (Gutekunst et al., 2003; Li et al., 1998; Muneoka and Takigawa, 2003; Shinoda et al., 1992) suggests that Ahi1 is not a structural component of stigmoid bodies; but instead, it may associate with the stigmoid body, so as to mediate a cell-signaling or post-translational processing function.
Other proteins have been found to label the stigmoid body (HAP1, sortilin, 5-HT7 receptor, and hPAX-P2) (Gutekunst et al., 2003; Li et al., 1998; Muneoka and Takigawa, 2003; Shinoda et al., 1992). Why such diverse proteins are partitioned to the stigmoid body is unknown, but it may be that these proteins participate in similar biological signaling pathways, either directly or indirectly. Various hypotheses concerning the exact function of these organelles have suggested a role as: a storage center for RNA, an inactive microtubule organizing center, an organelle involved in vesicular trafficking, and either a direct inhibitor of mitosis or a marker of a post-mitotic event (Ockleford et al., 1987). While the role of the stigmoid body remains elusive, the partitioning of Ahi1 to the stigmoid body and the association of Hap1 and Ahi1 with vesicular fractions (Gutekunst et al., 1998)(unpublished observations), suggest a possible link between these processes.
The association of particular SNPs in AHI1 with schizophrenia (Amann-Zalcenstein et al., 2006; Ingason et al., 2007) suggests either a direct or indirect link of genes involved in this disorder with AHI1. However, the observation of human AHI1 (and mouse Ahi1) expression both during development and in the mature brain, particularly in regions thought to be implicated in schizophrenia (cerebral cortex, hippocampus, and the mesolimbic pathway)(Hirayasu, 2007; White et al., 2008), suggests an important role of AHI1 in neuronal function. Moreover, structures such as the medial geniculate nucleus, the dorsal cochlear nucleus, and the auditory cortex have been implicated in schizophrenia (Roger and Arnault, 1989; Winer, 1985). Unlike the cells of the auditory cortex, which do not appear to express Ahi1, the cells of the inferior colliculus and dorsal cochlear nucleus do express Ahi1. Ahi1 immunoreactivity in these subcortical/brainstem structures suggest a potential role of Ahi1 in the subcortical processing of auditory information. The effects of Ahi1 in the auditory pathway may have direct relevance to the auditory processing deficits that can occur in individuals with schizophrenia (Shergill et al., 2000).
Given the relative paucity of neuropathological cases for JBTS that have been published to date (Friede and Boltshauser, 1978; Maria et al., 1999; Quisling et al., 1999; Yachnis and Rorke, 1999), our current study entailing localizations in human and zebrafish tissue, in addition to the mouse experiments, reveals important information on expression changes in the brain, through development and into adulthood. As a whole, our study demonstrated Ahi1 expression in many brain regions that have been observed to be malformed in individuals with JBTS. We also found expression in other brain regions that are not known to be malformed in JBTS. It is possible that subtle defects in these brain regions or in the signaling of cells in these areas, could lead to some of the behavioral impairments observed in the human disorder. These structures require detailed examination, to elucidate whether they are also involved in the JBTS phenotype.
Acknowledgements
This work was supported by NIMH grant 1K01MH71801 (to RJF). The authors wish to thank Dr. Adriana Verschoor (Wadsworth Center) for critical reading and input on our manuscript. The electron microscopy portion of this work was done at the Wadsworth Center’s Electron Microscopy Core Facility.
Support (grant information): This work was supported by NIMH grant 1K01MH71801 (to RJF).
LITERATURE CITED
- Amann-Zalcenstein D, Avidan N, Kanyas K, Ebstein RP, Kohn Y, Hamdan A, Ben-Asher E, Karni O, Mujaheed M, Segman RH, Maier W, Macciardi F, Beckmann JS, Lancet D, Lerer B. AHI1, a pivotal neurodevelopmental gene, and C6orf217 are associated with susceptibility to schizophrenia. Eur J Hum Genet. 2006;14(10):1111–1119. doi: 10.1038/sj.ejhg.5201675. [DOI] [PubMed] [Google Scholar]
- Banker G, Goslin K. In: Rat hippocampal neurons in low-density culture. Goslin K, Asmussen H, Banker G, editors. MIT Press; Cambridge: 2002. pp. 339–370. [Google Scholar]
- Bulloch K, Miller MM, Gal-Toth J, Milner TA, Gottfried-Blackmore A, Waters EM, Kaunzner UW, Liu K, Lindquist R, Nussenzweig MC, Steinman RM, McEwen BS. CD11c/EYFP transgene illuminates a discrete network of dendritic cells within the embryonic, neonatal, adult, and injured mouse brain. J Comp Neurol. 2008;508(5):687–710. doi: 10.1002/cne.21668. [DOI] [PubMed] [Google Scholar]
- Burnham WM. Primary and “transfer” seizure development in the kindled rat. Can J Neurol Sci. 1975;2(4):417–428. doi: 10.1017/s0317167100020564. [DOI] [PubMed] [Google Scholar]
- Cameron HA, McEwen BS, Gould E. Regulation of adult neurogenesis by excitatory input and NMDA receptor activation in the dentate gyrus. J Neurosci. 1995;15(6):4687–4692. doi: 10.1523/JNEUROSCI.15-06-04687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantani A, Lucenti P, Ronzani GA, Santoro C. Joubert syndrome. Review of the fifty-three cases so far published. Ann Genet. 1990;33(2):96–98. [PubMed] [Google Scholar]
- Canteras NS, Swanson LW. Projections of the ventral subiculum to the amygdala, septum, and hypothalamus: a PHAL anterograde tract-tracing study in the rat. J Comp Neurol. 1992;324(2):180–194. doi: 10.1002/cne.903240204. [DOI] [PubMed] [Google Scholar]
- Chance PF, Cavalier L, Satran D, Pellegrino JE, Koenig M, Dobyns WB. Clinical nosologic and genetic aspects of Joubert and related syndromes. J Child Neurol. 1999;14(10):660–666. doi: 10.1177/088307389901401007. [DOI] [PubMed] [Google Scholar]
- Cliffer KD, Burstein R, Giesler GJ., Jr. Distributions of spinothalamic, spinohypothalamic, and spinotelencephalic fibers revealed by anterograde transport of PHA-L in rats. J Neurosci. 1991;11(3):852–868. doi: 10.1523/JNEUROSCI.11-03-00852.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon-Salazar T, Silhavy JL, Marsh SE, Louie CM, Scott LC, Gururaj A, Al-Gazali L, Al-Tawari AA, Kayserili H, Sztriha L, Gleeson JG. Mutations in the AHI1 gene, encoding jouberin, cause Joubert syndrome with cortical polymicrogyria. Am J Hum Genet. 2004;75(6):979–987. doi: 10.1086/425985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferland RJ, Cherry TJ, Preware PO, Morrisey EE, Walsh CA. Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. J Comp Neurol. 2003;460(2):266–279. doi: 10.1002/cne.10654. [DOI] [PubMed] [Google Scholar]
- Ferland RJ, Eyaid W, Collura RV, Tully LD, Hill RS, Al-Nouri D, Al-Rumayyan A, Topcu M, Gascon G, Bodell A, Shugart YY, Ruvolo M, Walsh CA. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat Genet. 2004;36(9):1008–1013. doi: 10.1038/ng1419. [DOI] [PubMed] [Google Scholar]
- Ferland RJ, Gross RA, Applegate CD. Differences in hippocampal mitotic activity within the dorsal and ventral hippocampus following flurothyl seizures in mice. Neurosci Lett. 2002;332(2):131–135. doi: 10.1016/s0304-3940(02)00808-x. [DOI] [PubMed] [Google Scholar]
- Friede RL, Boltshauser E. Uncommon syndromes of cerebellar vermis aplasia. I: Joubert syndrome. Dev Med Child Neurol. 1978;20(6):758–763. doi: 10.1111/j.1469-8749.1978.tb15307.x. [DOI] [PubMed] [Google Scholar]
- Gleeson JG, Keeler LC, Parisi MA, Marsh SE, Chance PF, Glass IA, Graham JM, Jr, Maria BL, Barkovich AJ, Dobyns WB. Molar tooth sign of the midbrainhindbrain junction: occurrence in multiple distinct syndromes. Am J Med Genet A. 2004;125(2):125–134. doi: 10.1002/ajmg.a.20437. [DOI] [PubMed] [Google Scholar]
- Gould E, Tanapat P, McEwen BS, Flugge G, Fuchs E. Proliferation of granule cell precursors in the dentate gyrus of adult monkeys is diminished by stress. Proc Natl Acad Sci USA. 1998;95(6):3168–3171. doi: 10.1073/pnas.95.6.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabham PW, Seale GE, Bennecib M, Goldberg DJ, Vallee RB. Cytoplasmic dynein and LIS1 are required for microtubule advance during growth cone remodeling and fast axonal outgrowth. J Neurosci. 2007;27(21):5823–5834. doi: 10.1523/JNEUROSCI.1135-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutekunst CA, Li SH, Yi H, Ferrante RJ, Li XJ, Hersch SM. The cellular and subcellular localization of huntingtin-associated protein 1 (HAP1): comparison with huntingtin in rat and human. J Neurosci. 1998;18(19):7674–7686. doi: 10.1523/JNEUROSCI.18-19-07674.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutekunst CA, Torre ER, Sheng Z, Yi H, Coleman SH, Riedel IB, Bujo H. Stigmoid bodies contain type I receptor proteins SorLA/LR11 and sortilin: new perspectives on their function. J Histochem Cytochem. 2003;51(6):841–852. doi: 10.1177/002215540305100615. [DOI] [PubMed] [Google Scholar]
- Hirayasu Y. Brain imaging in schizophrenia. Neuropathology. 2007;27(6):601–603. doi: 10.1111/j.1440-1789.2007.00827.x. [DOI] [PubMed] [Google Scholar]
- Ingason A, Sigmundsson T, Steinberg S, Sigurdsson E, Haraldsson M, Magnusdottir BB, Frigge ML, Kong A, Gulcher J, Thorsteinsdottir U, Stefansson K, Petursson H, Stefansson H. Support for involvement of the AHI1 locus in schizophrenia. Eur J Hum Genet. 2007;15(9):988–991. doi: 10.1038/sj.ejhg.5201848. [DOI] [PubMed] [Google Scholar]
- Izaki Y, Takita M, Nomura M, Akema T. Differences between paired-pulse facilitation and long-term potentiation in the dorsal and ventral hippocampal CA1-prefrontal pathways of rats. Brain Res. 2003;992(1):142–145. doi: 10.1016/s0006-8993(03)03538-8. [DOI] [PubMed] [Google Scholar]
- Jiang X, Hanna Z, Kaouass M, Girard L, Jolicoeur P. Ahi-1, a novel gene encoding a modular protein with WD40-repeat and SH3 domains, is targeted by the Ahi-1 and Mis-2 provirus integrations. J Virol. 2002;76(18):9046–9059. doi: 10.1128/JVI.76.18.9046-9059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CJ, Ockleford CD. Nematosomes in the human placenta. Placenta. 1985;6(4):355–361. doi: 10.1016/s0143-4004(85)80045-x. [DOI] [PubMed] [Google Scholar]
- Joubert M, Eisenring JJ, Andermann F. Familial dysgenesis of the vermis: a syndrome of hyperventilation, abnormal eye movements and retardation. Neurology. 1968;18(3):302–303. [PubMed] [Google Scholar]
- Joubert M, Eisenring JJ, Robb JP, Andermann F. Familial agenesis of the cerebellar vermis. A syndrome of episodic hyperpnea, abnormal eye movements, ataxia, and retardation. Neurology. 1969;19(9):813–825. doi: 10.1212/wnl.19.9.813. [DOI] [PubMed] [Google Scholar]
- Kendall B, Kingsley D, Lambert SR, Taylor D, Finn P. Joubert syndrome: a clinico-radiological study. Neuroradiology. 1990;31(6):502–506. doi: 10.1007/BF00340131. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Boltshauser E, Breivik N, Gribaa M, Betard C, Barbot C, Koenig M. Homozygosity mapping of a third Joubert syndrome locus to 6q23. J Med Genet. 2004;41(4):273–277. doi: 10.1136/jmg.2003.014787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Gutekunst CA, Hersch SM, Li XJ. Association of HAP1 isoforms with a unique cytoplasmic structure. J Neurochem. 1998;71(5):2178–2185. doi: 10.1046/j.1471-4159.1998.71052178.x. [DOI] [PubMed] [Google Scholar]
- Liao M, Shen J, Zhang Y, Li SH, Li XJ, Li H. Immunohistochemical localization of huntingtin-associated protein 1 in endocrine system of the rat. J Histochem Cytochem. 2005;53(12):1517–1524. doi: 10.1369/jhc.5A6662.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maria BL, Hoang KB, Tusa RJ, Mancuso AA, Hamed LM, Quisling RG, Hove MT, Fennell EB, Booth-Jones M, Ringdahl DM, Yachnis AT, Creel G, Frerking B. “Joubert syndrome” revisited: key ocular motor signs with magnetic resonance imaging correlation. J Child Neurol. 1997;12(7):423–430. doi: 10.1177/088307389701200703. [DOI] [PubMed] [Google Scholar]
- Maria BL, Quisling RG, Rosainz LC, Yachnis AT, Gitten J, Dede D, Fennell E. Molar tooth sign in Joubert syndrome: clinical, radiologic, and pathologic significance. J Child Neurol. 1999;14(6):368–376. doi: 10.1177/088307389901400605. [DOI] [PubMed] [Google Scholar]
- Maruki K, Izaki Y, Nomura M, Yamauchi T. Differences in paired-pulse facilitation and long-term potentiation between dorsal and ventral CA1 regions in anesthetized rats. Hippocampus. 2001;11(6):655–661. doi: 10.1002/hipo.1080. [DOI] [PubMed] [Google Scholar]
- Muneoka KT, Takigawa M. 5-Hydroxytryptamine7 (5-HT7) receptor immunoreactivity-positive ‘stigmoid body’-like structure in developing rat brains. Int J Dev Neurosci. 2003;21(3):133–143. doi: 10.1016/s0736-5748(03)00029-7. [DOI] [PubMed] [Google Scholar]
- Niesen CE. Malformations of the posterior fossa: current perspectives. Semin Pediatr Neurol. 2002;9(4):320–334. doi: 10.1053/spen.2002.32508. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Dahm R. Zebrafish. Oxford University Press; New York: 2002. [Google Scholar]
- Ockleford CD, Nevard CH, Indans I, Jones CJ. Structure and function of the nematosome. J Cell Sci. 1987;87(1):27–44. doi: 10.1242/jcs.87.1.27. [DOI] [PubMed] [Google Scholar]
- Parisi MA, Doherty D, Chance PF, Glass IA. Joubert syndrome (and related disorders) (OMIM 213300) Eur J Hum Genet. 2007;15(5):511–521. doi: 10.1038/sj.ejhg.5201648. [DOI] [PubMed] [Google Scholar]
- Parisi MA, Doherty D, Eckert ML, Shaw DW, Ozyurek H, Aysun S, Giray O, Al Swaid A, Al Shahwan S, Dohayan N, Bakhsh E, Indridason OS, Dobyns WB, Bennett CL, Chance PF, Glass IA. AHI1 mutations cause both retinal dystrophy and renal cystic disease in Joubert syndrome. J Med Genet. 2006;43(4):334–339. doi: 10.1136/jmg.2005.036608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Chaves EI Posse, Vance DE, Campenot RB, Kiss RS, Vance JE. Uptake of lipoproteins for axonal growth of sympathetic neurons. J Biol Chem. 2000;275(26):19883–19890. doi: 10.1074/jbc.275.26.19883. [DOI] [PubMed] [Google Scholar]
- Quisling RG, Barkovich AJ, Maria BL. Magnetic resonance imaging features and classification of central nervous system malformations in Joubert syndrome. J Child Neurol. 1999;14(10):628–635. doi: 10.1177/088307389901401002. [DOI] [PubMed] [Google Scholar]
- Racine R, Rose PA, Burnham WM. Afterdischarge thresholds and kindling rates in dorsal and ventral hippocampus and dentate gyrus. Can J Neurol Sci. 1977;4(4):273–278. doi: 10.1017/s0317167100025117. [DOI] [PubMed] [Google Scholar]
- Risold PY, Thompson RH, Swanson LW. The structural organization of connections between hypothalamus and cerebral cortex. Brain Res Brain Res Rev. 1997;24(2-3):197–254. doi: 10.1016/s0165-0173(97)00007-6. [DOI] [PubMed] [Google Scholar]
- Roger M, Arnault P. Anatomical study of the connections of the primary auditory area in the rat. J Comp Neurol. 1989;287(3):339–356. doi: 10.1002/cne.902870306. [DOI] [PubMed] [Google Scholar]
- Rub U, Schultz C, Del Tredici K, Gierga K, Reifenberger G, de Vos RA, Seifried C, Braak H, Auburger G. Anatomically based guidelines for systematic investigation of the central somatosensory system and their application to a spinocerebellar ataxia type 2 (SCA2) patient. Neuropathol Appl Neurobiol. 2003;29(5):418–433. doi: 10.1046/j.1365-2990.2003.00504.x. [DOI] [PubMed] [Google Scholar]
- Saraiva JM, Baraitser M. Joubert syndrome: a review. Am J Med Genet. 1992;43(4):726–731. doi: 10.1002/ajmg.1320430415. [DOI] [PubMed] [Google Scholar]
- Shergill SS, Brammer MJ, Williams SC, Murray RM, McGuire PK. Mapping auditory hallucinations in schizophrenia using functional magnetic resonance imaging. Arch Gen Psychiatry. 2000;57(11):1033–1038. doi: 10.1001/archpsyc.57.11.1033. [DOI] [PubMed] [Google Scholar]
- Shinoda K, Mori S, Ohtsuki T, Osawa Y. An aromatase-associated cytoplasmic inclusion, the “stigmoid body,” in the rat brain: I. Distribution in the forebrain. J Comp Neurol. 1992;322(3):360–376. doi: 10.1002/cne.903220306. [DOI] [PubMed] [Google Scholar]
- Toro I, Jr., Rohlich P. A new cytoplasmic component in the trophoblast cells of the rat and mouse. Anat Rec. 1966;155(3):385–399. doi: 10.1002/ar.1091550310. [DOI] [PubMed] [Google Scholar]
- Utsch B, Sayer JA, Attanasio M, Pereira RR, Eccles M, Hennies HC, Otto EA, Hildebrandt F. Identification of the first AHI1 gene mutations in nephronophthisis-associated Joubert syndrome. Pediatr Nephrol. 2006;21(1):32–35. doi: 10.1007/s00467-005-2054-y. [DOI] [PubMed] [Google Scholar]
- Valente EM, Brancati F, Silhavy JL, Castori M, Marsh SE, Barrano G, Bertini E, Boltshauser E, Zaki MS, Abdel-Aleem A, Abdel-Salam GM, Bellacchio E, Battini R, Cruse RP, Dobyns WB, Krishnamoorthy KS, Lagier-Tourenne C, Magee A, Pascual-Castroviejo I, Salpietro CD, Sarco D, Dallapiccola B, Gleeson JG. AHI1 gene mutations cause specific forms of Joubert syndrome-related disorders. Ann Neurol. 2006;59(3):527–534. doi: 10.1002/ana.20749. [DOI] [PubMed] [Google Scholar]
- van Dorp DB, Palan A, Kwee ML, Barth PG, van der Harten JJ. Joubert syndrome: a clinical and pathological description of an affected male and a female fetus from the same sibship. Am J Med Genet. 1991;40(1):100–104. doi: 10.1002/ajmg.1320400121. [DOI] [PubMed] [Google Scholar]
- White T, Cullen K, Rohrer LM, Karatekin C, Luciana M, Schmidt M, Hongwanishkul D, Kumra S, Schulz S Charles, Lim KO. Limbic Structures and Networks in Children and Adolescents With Schizophrenia. Schizophr Bull. 2008;34(1):18–29. doi: 10.1093/schbul/sbm110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer JA. The medial geniculate body of the cat. Adv Anat Embryol Cell Biol. 1985;86:1–97. [PubMed] [Google Scholar]
- Yachnis AT, Rorke LB. Neuropathology of Joubert syndrome. J Child Neurol. 1999;14(10):655–659. doi: 10.1177/088307389901401006. [DOI] [PubMed] [Google Scholar]
- Zhou W, Song P. Molecular cloning of a novel gene ZAhi-1 and its expression analysis during zebrafish gametogenesis. Mol Biol Rep. 2006;33(2):111–116. doi: 10.1007/s11033-006-0005-8. [DOI] [PubMed] [Google Scholar]