Abstract
Chromosomal aneuploidy is the major reason why couples opt for prenatal diagnosis. Current methods for definitive diagnosis rely on invasive procedures, such as chorionic villus sampling and amniocentesis, and are associated with a risk of fetal miscarriage. Fetal DNA has been found in maternal plasma but exists as a minor fraction among a high background of maternal DNA. Hence, quantitative perturbations caused by an aneuploid chromosome in the fetal genome to the overall representation of sequences from that chromosome in maternal plasma would be small. Even with highly precise single molecule counting methods such as digital PCR, a large number of DNA molecules and hence maternal plasma volume would need to be analyzed to achieve the necessary analytical precision. Here we reasoned that instead of using approaches that target specific gene loci, the use of a locus-independent method would greatly increase the number of target molecules from the aneuploid chromosome that could be analyzed within the same fixed volume of plasma. Hence, we used massively parallel genomic sequencing to quantify maternal plasma DNA sequences for the noninvasive prenatal detection of fetal trisomy 21. Twenty-eight first and second trimester maternal plasma samples were tested. All 14 trisomy 21 fetuses and 14 euploid fetuses were correctly identified. Massively parallel plasma DNA sequencing represents a new approach that is potentially applicable to all pregnancies for the noninvasive prenatal diagnosis of fetal chromosomal aneuploidies.
Keywords: Down syndrome, Solexa sequencing, trisomy 21
The testing of fetal chromosomal aneuploidies is the predominant reason why many pregnant women opt for prenatal diagnosis. Conventional methods for definitive prenatal diagnosis of these disorders involve the invasive sampling of fetal materials through amniocentesis and chorionic villus sampling, with a risk for the fetus (1). Many workers tried to develop noninvasive approaches. Methods based on ultrasound scanning and maternal serum biochemical markers (2) have proved to be useful screening tests. However, they detect epiphenomena instead of the core pathology of chromosomal abnormalities. They have limitations such as a narrow gestational window of applicability and the need to combine multiple markers, even over different time points, to arrive at a clinically useful sensitivity and specificity profile.
For the direct detection of fetal chromosomal and genetic abnormalities from maternal blood, early work focused on the relatively difficult isolation of the rare fetal nucleated cells from maternal blood (3–5). The discovery of cell-free fetal nucleic acids in maternal plasma in 1997 opened up new possibilities (6, 7). However, the fact that fetal DNA represents only a minor fraction of total DNA in maternal plasma (8), with the majority being contributed by the pregnant woman herself, has offered considerable challenge. Recently, a number of approaches have been developed. One strategy targets a fetal-specific subset of nucleic acids in maternal plasma, e.g., placental mRNA (9–11) and DNA molecules bearing a placental-specific DNA methylation signature (12–14). The fetal chromosomal dosage is then assessed by allelic ratio analysis of SNPs within the targeted molecules. These strategies are called the RNA–SNP allelic ratio approach (11) and the epigenetic allelic ratio approach (14). These allelic ratio-based methods can be used only for fetuses heterozygous for the analyzed SNPs. Thus, multiple markers are needed to enhance the population coverage of the methods.
To develop a polymorphism-independent method for the detection of fetal chromosomal aneuploidies from maternal plasma, our group has recently outlined the principles for the measurement of relative chromosome dosage (RCD) using digital PCR (15). Digital RCD aims to measure the total (maternal plus fetal) amount of a specific locus on a potentially aneuploid chromosome in maternal plasma, e.g., chromosome 21 (chr21) in trisomy 21 (T21), and compares it to that on a reference chromosome. Hence, fetal T21 is diagnosed by detecting the small increment in the total amount of the chr21 gene locus contributed by the trisomic chr21 in the fetus as compared with a gene locus on a reference chromosome. The proportional increment in chr21 sequences is expectedly small because fetal DNA contributes only a minor fraction of DNA in maternal plasma (8). To reliably detect the small increase, a large absolute number of chr21 and reference chromosome sequences of the loci targeted by the digital PCR assays need to be analyzed and quantified with high precision. The number of molecules required for RCD increases by four times, for every twofold reduction in the fractional concentration of circulating fetal DNA. Thus, for cases in which the fractional concentration for circulating fetal DNA is low, e.g., during early gestation, relatively large volumes of maternal plasma may be needed. One way is to perform multiplex analysis of multiple genetic loci. However, the optimization of highly multiplexed digital PCR might be challenging. If fluorescence reporters are used, one would also quickly run out of reporters for distinguishing the products from the various loci.
To overcome the above limitations, we propose to use a method independent of any particular gene locus to quantify the amount of chr21 sequences in maternal plasma. When a locus-independent method is used, potentially every DNA fragment originating from the aneuploid chromosome could contribute to the measurement of the amount of that chromosome. Therefore, for any fixed volume of maternal plasma, the number of quantifiable sequences would be much greater than the number of DNA molecules that could serve as templates for detection by gene locus-specific assays. Hence, precise detection of the over- or underrepresentation of sequences from an aneuploid chromosome could be more readily achieved. We previously (15) proposed that the recently available massively parallel genomic sequencing (MPGS) platforms (16, 17) might be adaptable as an approach to quantify DNA sequences for the noninvasive prenatal diagnosis of fetal chromosomal aneuploidy. In this study, we demonstrate the use of the “Solexa” sequencing technique (Illumina) (18) for this purpose.
Results
Procedural Framework.
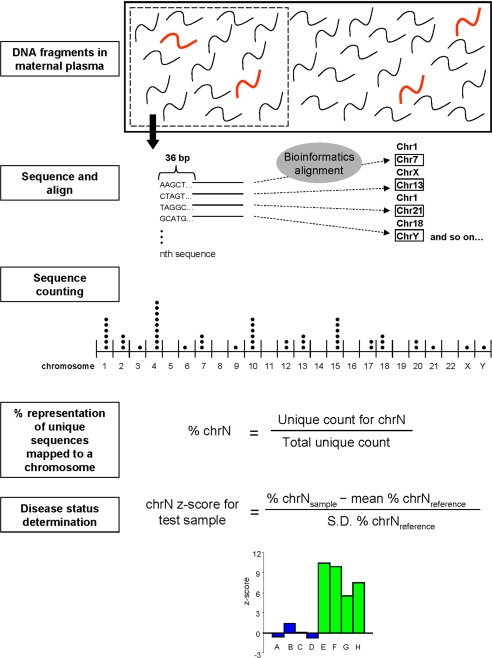
The procedural framework of using MPGS for noninvasive fetal chromosomal aneuploidy detection in maternal plasma is schematically illustrated in Fig. 1. In this study, we used the sequencing-by-synthesis Solexa method (18). As the maternal plasma DNA (maternal and fetal) molecules were already fragmented in nature (19), no further fragmentation was required. One end of the clonally expanded copies of each plasma DNA fragment was sequenced and processed by standard postsequencing bioinformatics alignment analysis for the Illumina Genome Analyzer, which uses the Efficient Large-Scale Alignment of Nucleotide Databases (ELAND) software. The purpose of the alignment was to simply determine the chromosomal origin of the sequenced plasma DNA fragments and details about their gene-specific location were not required. The number of sequence reads originating from any particular chromosome was then counted and tabulated for each human chromosome. In this study, we counted only sequences that could be mapped to just one location in the repeat-masked reference human genome with no mismatch, i.e., deemed as a “unique” sequence in the human genome. We termed these sequences as U0–1–0–0 on the basis of values in a number of fields in the data output files of the ELAND sequence alignment software (Illumina) (see Materials and Methods).
Fig. 1.
Schematic illustration of the procedural framework for using massively parallel genomic sequencing for the noninvasive prenatal detection of fetal chromosomal aneuploidy. Fetal DNA (thick red fragments) circulates in maternal plasma as a minor population among a high background of maternal DNA (black fragments). A sample containing a representative profile of DNA molecules in maternal plasma is obtained. In this study, one end of each plasma DNA molecule was sequenced for 36 bp using the Solexa sequencing-by-synthesis approach. The chromosomal origin of each 36-bp sequence was identified through mapping to the human reference genome by bioinformatics analysis. The number of unique (U0–1–0–0, see text) sequences mapped to each chromosome was counted and then expressed as a percentage of all unique sequences generated for the sample, termed % chrN for chromosome N. Z-scores for each chromosome and each test sample were calculated using the formula shown. The z-score of a potentially aneuploid chromosome is expected to be higher for pregnancies with an aneuploid fetus (cases E–H shown in green) than for those with a euploid fetus (cases A–D shown in blue).
We then determined the percentage contribution of unique sequences mapped to each chromosome by dividing the U0–1–0–0 count of a specific chromosome by the total number of U0–1–0–0 sequence reads generated in the sequencing run for the tested sample to generate a value termed % chrN, when the chromosome of interest is chrN. To determine if a tested maternal plasma sample belonged to a T21 pregnancy, we calculated the z-score of % chr21 of the tested sample. The z-score refers to the number of standard deviations from the mean of a reference data set. Hence, for a T21 fetus, a high z-score for % chr21 was expected when compared with the mean and standard deviation of % chr21 values obtained from maternal plasma of euploid pregnancies.
For this procedure to be effective for noninvasive prenatal fetal chromosomal aneuploidy detection, a number of assumptions need to be met. First, MPGS needs to be sensitive enough to capture and generate sequence reads for the small fraction of fetal DNA in maternal plasma alongside the background maternal DNA. Second, the pool of plasma DNA fragments captured for sequencing needs to be a representative sample of the total DNA pool with similar interchromosomal distribution to that in the original maternal plasma. Third, there should be no major bias in the ability to sequence DNA fragments originating from each chromosome. When these assumptions hold, then the % chrN values should be reflective of the genomic representation of the maternal and fetal DNA fragments in maternal plasma. Furthermore, if both the maternal and the fetal genomes are evenly represented in maternal plasma, the proportional contribution of plasma DNA sequences per chromosome should in turn bear correlation with the relative size of each chromosome in the human genome. If the % chrN values could be determined precisely enough by sequencing and counting a large enough pool of plasma DNA sequences, we hypothesize that we would be able to discriminate perturbations in the quantitative representation of sequences mapped to the aneuploid chromosomes in a maternal plasma sample from a pregnancy involving a fetus with the said aneuploidy. We set out to test each of these assumptions.
Detection of Fetal DNA in Maternal Plasma.
If MPGS could sequence fetal DNA in maternal plasma, one should be able to detect chrY DNA from plasma of women carrying male fetuses. Plasma samples obtained from four pregnant women carrying euploid fetuses (three males and one female) were processed using the beta ChIP-Seq protocol from Illumina, which included amplification of the adaptor-ligated DNA fragments both before and after (i.e., two rounds of amplification) a gel electrophoresis-based size fractionation step as described in supporting information (SI) Text.
The clinical information and sequenced counts for these four samples are shown in Table S1. The total number of sequences obtained from each sample was ≈9 × 106. The total U0–1-0–0 counts ranged from ≈1.8 × 106 to 2.0 × 106 per case. The percentages of the U0–1-0–0 counts mapped to each chromosome are shown in Fig. S1. For the three pregnancies with male fetuses, i.e., cases 3009, 3034, and 3143, the absolute and fractional (in parentheses) U0–1–0–0 counts mapped to chrY were 636 (0.032%), 858 (0.048%), and 1,054 (0.056%), respectively. However, it was unexpected that 177 (0.009%) sequences were also mapped to chrY in the sample involving a female fetus. Real-time PCR for the SRY gene (8) was negative for this latter plasma sample. We next considered that contamination from male sequences might occur during the gel electrophoresis.
Sequencing Protocol for Plasma DNA.
We developed a new protocol to prepare plasma DNA samples for MPGS whereby the gel electrophoresis and second amplification steps were omitted. The new and original protocols were compared and denoted as protocols A and B, respectively. To minimize the chance of bias in the sequencing results caused by low DNA input, 100 ng of DNA were extracted from three plasma samples. Half (50 ng) of each plasma sample was processed by either protocol and sequenced in the same manner. The tested plasma samples included one from a pregnant woman carrying a female fetus, one from a pregnant woman carrying a male fetus, and one that was a mixture of plasma from two male individuals. A mixture was required for the last sample so that 100 ng of DNA could be obtained. The three samples were named samples 1, 2, and 3, respectively.
The clinical details and sequencing counts for each sample and each protocol are shown in Table S2. The total U0–1–0–0 counts ranged from 2.0 × 106 to 2.2 × 106. The absolute and fractional U0–1–0–0 counts (in parentheses) mapped to chrY for samples 1, 2, and 3 using the new protocol were 184 (0.009%), 1,444 (0.066%), and 3,523 (0.175%), respectively. The corresponding numbers for the original protocol were 218 (0.011%), 1,615 (0.077%), and 3,468 (0.169%), respectively. Thus, contamination attributable predominantly to the gel purification and the second amplification steps could not be substantiated.
We next explored if there might be a bioinformatic explanation. We used the Basic Local Alignment Search Tool (BLAST) to analyze each of the U0–1–0–0 sequences mapped to chrY for each of the three samples and for both protocols. We assessed the proportion of those DNA sequences that could genuinely be aligned just to chrY using BLAST. The proportion of sequences aligned uniquely to chrY by BLAST was comparable for both the new and the original protocols (Table S3). For the plasma sample obtained from the pregnancy with a female fetus, only ∼30% of sequences mapped to chrY by ELAND were confirmed to map just to chrY by BLAST. This was in contrast to samples 2 and 3, where >90% of the sequences mapped to chrY by ELAND could be confirmed by BLAST. Nonetheless, the chrY sequences detected in the plasma sample from a pregnancy with a male fetus confirmed that fetal DNA in maternal plasma could be sequenced by MPGS.
To confirm that there was little mapping error among the U0–1–0–0 sequences aligned by the ELAND software, we performed a BLAST analysis on 120 randomly selected U0–1–0–0 sequences for each of the other chromosomes for the three plasma DNA samples processed by the new protocol. As shown in Table S4, among the selected test sequences, > 99% of U0–1–0–0 sequences mapped by ELAND to the autosomes were confirmed to align only to the corresponding chromosome by BLAST. All 120 chrX sequences mapped by ELAND were confirmed by BLAST in sample 1, which was composed of female DNA only. More than 97% of chrX sequences mapped by ELAND were confirmed by BLAST in samples 2 and 3, which contained male DNA. These data suggested that U0–1–0–0 sequences mapped by the ELAND software were generally accurate with chrY being the exception.
Distribution of Maternal Plasma DNA Sequences Among the Human Chromosomes.
The percentage contributions of U0–1–0–0 count by each chromosome among the total U0–1–0–0 sequences were calculated for samples 1, 2, and 3. To investigate if maternal plasma DNA sequences were evenly distributed across the human genome, we compared the plasma DNA data with the expected genomic contribution of each chromosome. Our main goal was to analyze maternal plasma DNA in which the predominant DNA background was female. Thus, we calculated the relative genomic representation, i.e., size, of each chromosome, on the basis of the nucleotide content of each chromosome within a repeat-masked haploid reference human genome of a female. The relative size of each chromosome was plotted alongside the percentage of chromosomal contribution of U0–1–0–0 sequences of the sequenced plasma DNA samples.
As shown in Fig. 2, aliquots of plasma DNA processed by the new protocol, i.e., samples 1A, 2A, and 3A, bore closer resemblances to the expected genomic representation of each human chromosome than the corresponding aliquot processed by the original protocol, i.e., samples 1B, 2B, and 3B. We performed linear regression analyses to compare the % U0–1–0–0 per chromosome obtained from both the new and the original protocols against the expected genomic representation of each chromosome in the human genome. As shown in Fig. S2, the slopes of the lines obtained from samples 1A, 2A, and 3A were >0.95, while those for samples 1B, 2B, and 3B were 0.755, 0.795, and 0.859, respectively. R2 was >0.980 for samples 1A, 2A, and 3A but was 0.803, 0.840, and 0.910 for samples 1B, 2B, and 3B, respectively. These data objectively confirmed that the DNA processing protocol with just one PCR amplification step and the omission of the gel electrophoresis procedure produced a quantitative profile of sequences that better resembled the genomic content of each human chromosome than the original protocol. More importantly, these data suggested that the overall distribution of DNA molecules in maternal plasma (inclusive of maternal and fetal DNA) across the human genome was quite even. The chromosomal distribution of DNA molecules in the maternal plasma samples (1A and 2A) was also similar to that of adult male plasma (sample 3A). This observation suggested that it would be unlikely for the maternal and fetal DNA sequences in maternal plasma to bear significant discrepancies among their genomic distributions. Otherwise, if the genomic representation of the maternal DNA differed substantially from that of fetal DNA, one would expect the overall genomic representation to be discrepant from that of a nonpregnant human plasma DNA sample.
Fig. 2.
Bar chart of % U0–1–0–0 sequences per chromosome for a maternal plasma sample involving a female fetus (sample 1), a maternal plasma sample involving a male fetus (sample 2), and a mixture of plasma from two adult males (sample 3) processed using the new (protocol A) and original (protocol B) protocols. The percentage of genomic representation of each chromosome as expected for a repeat-masked reference haploid female genome was plotted as a reference (black bars).
Fetal Trisomy 21 Detection from Maternal Plasma.
We proceeded to test if fetal chromosomal aneuploidy would lead to quantitative perturbations in the percentage contribution in aligned sequences for the aneuploid chromosome. Plasma samples were obtained in the first and second trimesters of pregnancies from 14 women each pregnant with a euploid fetus and 14 women each pregnant with a T21 fetus. The chromosomal status of the fetuses was confirmed by full karyotyping. Plasma DNAs from the 28 pregnancies (median gestational age: 14.1 weeks) were processed by the new protocol and sequenced. The clinical details and sequencing counts for each sample are shown in Table S5. The 28 samples were processed as two batches on dates 6 weeks apart and sequenced in four flow cells.
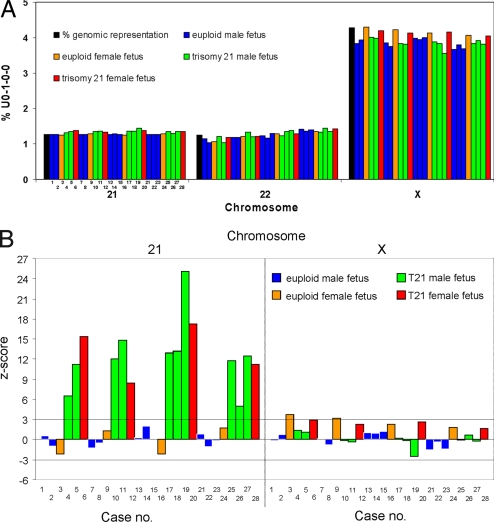
The mean number of sequence reads generated per sample was 10.8 × 106. The mean U0–1–0–0 count was 2.5 × 106. The percentage contributions of U0–1–0–0 sequences to each chromosome were plotted against the percentage of genomic representation per chromosome of the human genome as described above and are shown in Fig. S3. The data for chr21 and chrX are further shown in Fig. 3A. The percentage of U0–1–0–0 sequences aligned to chr21 was slightly higher for all T21 than for euploid cases. The % chrX was much higher and the % chrY was much lower for all female than male fetuses.
Fig. 3.
Plot of (A) % U0–1–0–0 counts and (B) z-scores for chromosome 21 and chromosome X for 28 maternal plasma samples. The sample numbers correspond to the cases described in Table S5.
To objectively quantify the degree of overrepresentation in chr21 sequences of the T21 fetuses, we used the data from the 10 euploid male fetuses as a reference population to calculate the mean and SD in % U0–1–0–0 per chromosome. The reference population was restricted to euploid male fetuses so that an expected increase in % chrX could also be explored in female fetuses. Using these reference values, we calculated the z-scores for each of the chromosomes, except the Y chromosome, for each of the 28 cases, as shown in Fig. S4. The z-scores for chr21 and chrX are further shown in Fig. 3B. All of the T21 cases had a z-score of >3 (range 5.03–25.11) for chr21, i.e., at 3 standard deviations above the reference established from the euploid male fetuses. The cases with female fetuses had a z-score >1.67 for chrX. All of the other chromosomes had z-scores within ±3 for all 28 cases.
Reproducibility of Measuring Percentage of Chromosome Representation.
Among the 28 tested maternal plasma samples, we expected a difference in % chr21 representation between the T21 and euploid fetuses and the % chrX representation between the female and male fetuses. However, it was interesting to observe a small absolute difference in % chr21 representation, which translated to a large z-score difference but a large absolute difference in % chrX representation that translated to a less impressive z-score difference among the respective cases (Fig. 3). The absolute differences in chrX counts between female and male fetuses were expected to be much larger than the difference in chr21 counts between T21 and euploid fetuses. This was because there was a 2-fold increase in the dosage of chrX for a female than a male individual, but just a 1.5-fold increase in chr21 dosage for a T21 than a euploid individual. Furthermore, chrX is much larger than chr21 and contributed to a mean of 9.5 × 104 U0–1–0–0 counts in male fetuses compared with a mean of 3.2 × 104 for chr21 in all samples.
As the z-score reflects the extent of differences in a measurement expressed as the number of SDs from the mean of a reference data set, we postulated that the SD was small for the measurement of % chr21 but large for the measurement of % chrX. As the SD of a data set was in fact reflecting the precision of its measurement, we used the data from the 10 euploid male fetuses to calculate the coefficient of variation (CV = SD/mean × 100%) of measuring the percentage of representation of each chromosome. As shown in Table S6, chr21 had the third lowest CV (0.54%) among all chromosomes while the CV for the % chrX measurement was 3.10%. As the absolute number of U0–1–0–0 sequences counted for chrX was threefold higher than that for chr21, the number of sequences counted could not explain the variation in the precision. We therefore explored the relationship between the CV in % U0–1–0–0 counts and the GC content of each chromosome (Fig. S5). Human chromosomes can be distributed into five groups with different levels of GC content (20). Group I chromosomes have the lowest levels while group V chromosomes have the highest levels of GC content. Interestingly, there was a statistically significant difference (P < 0.001, ANOVA) in the CVs for the five groups of chromosomes. A Bonferroni t-test further identified that the CV for group V was significantly higher (P < 0.05) than that for the other four groups. The CV for group IV and group I was each significantly higher (P < 0.05) than for both groups II and III.
Discussion
We have demonstrated that MPGS can be used as a diagnostic tool in noninvasive prenatal diagnosis. We have shown that differences in amounts of chr21 DNA sequences in maternal plasma contributed by T21 fetuses compared with euploid fetuses can be unambiguously detected. Absolute differences in amounts of chrX and chrY DNA sequences in maternal plasma contributed by male fetuses compared with female fetuses can also be observed robustly. The ability of MPGS to differentiate small quantitative perturbations in genomic distributions of chromosomes lies in the very large number of molecules analyzed, which minimizes the imprecision of the quantitative measurement. As no specific gene locus was targeted, all plasma DNA fragments together provide an unprecedented number of molecules analyzed per plasma sample.
This approach is in marked contrast to previous methods that quantified only DNA molecules that could serve as templates for locus-specific PCR assays, for example, SRY on chrY (8). The gene locus-specific DNA templates represent only an extremely small proportion of DNA fragments present in maternal plasma. In fact, MPGS is such a powerful tool for quantifying the relative genomic representation of plasma DNA molecules that only an amount corresponding to just a representative fraction of the human genome would need to be sequenced. For example, ∼10 million 36-bp reads were generated for each plasma sample, which was equivalent to just one-tenth of the human genome. Furthermore, in this study, only the U0–1–0–0 sequences, representing just ≈20% of all of the reads sequenced from each plasma DNA sample, were used to generate a quantitative profile of chromosomal distribution. Thus, this is quite unlike some previously described sequencing-based methods for quantitative nucleic acid profiling that relied on sequencing at high fold coverage (21), for example, to determine the relative abundance of RNA species in transcriptome analysis (21). On the contrary, our present method simply sequences a random representative fraction of the human genome. The majority of DNA fragments are sequenced once, if at all. The relative chromosome size is then deduced by counting the relative number of sequences aligned to the chromosome. Each of the counted DNA fragments would be of a different nucleotide sequence. In fact, the pool of DNA sequenced for a sample would vary from run to run.
Despite the randomness of the sequencing, the quantitative estimation of % chr21 sequences was so precise and robust that the z-scores for chr21 of the T21 pregnancies were markedly different from the mean of a reference euploid sample set. In this study, the median gestational age of the T21 pregnancies (14.1 weeks) was comparable with the median of the euploid group (15.4 weeks). All samples from the euploid group were collected before any invasive procedure in the present pregnancy. Blood samples from 11 of the T21 pregnancies were collected immediately before pregnancy termination at a median of 6 days (range: 2–22 days) after invasive prenatal diagnostic procedure. Our previous study (22) indicated that there would be no substantial difference in the fetal DNA concentration in samples collected days after amniocentesis. Nonetheless, blood samples from 3 T21 pregnancies, cases 17, 19, and 25 (Table S5), were collected in the first trimester before chorionic villus sampling. Increases in their z-scores for chr21 were readily identifiable (Fig. 3B).
Theoretically, the determination of the presence of quantitative perturbations in any particular chromosome could be achieved more precisely, for example, by taking into account the fetal DNA concentration to estimate the expected degree of chromosomal perturbation. Fetal DNA concentrations can be readily measured using either fetal epigenetic markers (23) or paternally inherited polymorphic markers (24). In this study, the fetal DNA concentration of each case was not required to derive cutoff values for determining the disease status of each case. First, according to Table S6, chr21 is one of the chromosomes whose percentage of representation could be measured at very low imprecision with our current protocol. Second, when compared to methods like digital RCD whereby disease cutoff values related to the fetal DNA concentration were required (15), many more chr21 sequences were measured by sequencing. For digital RCD, we reported that for a sample with 25% fetal DNA concentration, 7,680 digital PCRs would need to be performed to achieve a correct classification rate of 97%. Our previous data also showed that ≈20% of the total number of digital PCRs analyzed, equal to 1,536 chr21 molecules for a 7,680-well experiment, would contain only the chr21 gene target and hence be counted as informative. Thus, the number of chr21 molecules (mean: 3.2 × 104) analyzed by the sequencing method is ∼20-fold that of the digital RCD method. Hence, the measurement would be significantly more precise than the present scale of digital PCR analyses. However, by taking the fetal DNA concentration into account, the measurement of the percentage of chromosomal representation could be made more precisely for some of the other chromosomes or across batches and hence minimize false diagnoses.
In fact, the precision and accuracy of MPGS for determining the genomic representation of maternal plasma DNA could be further improved by a number of postsequencing analysis strategies. For example, sequences occurring in regions of known copy number variations (25) could be adjusted for so that the reference range for euploid pregnancies might be even tighter. Sequences other than U0–1–0–0, for example, with one or two mismatches to the reference genome that, in some instances, may represent a polymorphic difference between the tested sample and the reference human genome, may also be used to increase the number of usable sequence counts. We have also shown that the reproducibility of measuring the percentage contribution of plasma DNA sequences varied between chromosomes and the GC content of the chromosome may partly explain this variability. Thus, the amount of sequencing to be done per sample could be varied to ensure that measurements could be made precisely enough for the detection of quantitative perturbations of each of the other chromosomes. In fact, we predict that if adequate numbers of plasma DNA fragments are counted, MPGS may be precise enough to detect quantitative aberrations involving regions less than a whole chromosome.
We have found a discrepancy in the accuracy of ELAND mapping for chrY when compared with other chromosomes. This may be attributable to the known presence of many repetitive sequences in chrY (26). Even after repeat masking, much of the remaining chrY sequences are composed of low-copy-number repeat sequences, which increases the difficulty of aligning such sequences accurately. Nonetheless, for women carrying female fetuses, there still appeared to be a small number of sequences genuinely mapped to chrY. We had confirmed that these plasma samples were negative for male DNA using a SRY real-time PCR assay that was widely used in the field (8). The MPGS approach is potentially much more sensitive than the real-time PCR approach. Technically, it might mean that an extremely low level of carryover contamination would be unavoidable in a laboratory environment that has been optimized for the less sensitive real-time PCR system. Alternatively, the sequences that appeared to be uniquely mapped to chrY by both ELAND and BLAST may in fact be mappable to the gap regions of the human genome whose sequence has not been uncovered. Yet, it is still obvious that one can clearly distinguish the male from the female fetuses just from the % chrY counts in maternal plasma (Fig. S3F).
We have demonstrated the principle of the plasma DNA sequencing approach using one of the available MPGS platforms. The same approach can also be used for the other platforms (16, 27). The main limitation of the method described here is the relatively high costs. Sequencing reagents alone cost $700 for each sample. A high capital outlay is involved in setting up the instrumentation that for the Illumina Solexa platform at present has a throughput of 16 samples per week per instrument. However, it is expected that such technology will rapidly become more affordable over the next few years (17). In the interim period, one could potentially reduce the cost by barcoding individual patients' samples such that one sequencing reaction could generate diagnostic information for multiple cases. Alternatively, sequencing could be focused just on the chromosomes of interest by array capture before random sequencing of DNA fragments originating from those chromosomes. In fact, other non-sequencing-based methods of single-molecule analyses may be suitable for our application and may ultimately prove to be more cost effective (28). To contain costs of a noninvasive prenatal diagnostic program, one could also potentially combine the sequencing approach with for example, the RNA–SNP allelic ratio approach (11) such that fetuses that are not heterozygous and thus uninformative for the RNA–SNP approach could be analyzed by the more expensive MPGS approach.
Finally, the data reported in this study need to be confirmed by large-scale clinical trials. Ultimately it is hoped that noninvasive prenatal diagnosis will make future prenatal testing safer for pregnant women and their fetuses. In this work, we demonstrate the use of MPGS for quantitative genomic sequencing in the form of a diagnostic tool. We expect that the massively parallel plasma DNA sequencing strategy described in this article could also be used to analyze the various pathological conditions associated with aberrations in plasma nucleic acids, e.g., cancer.
While this article was under review, Fan et al. also reported the use of Solexa sequencing for fetal chromosomal aneuploidy diagnosis (29). These authors analyzed 18 maternal plasma samples, of which 17 were collected 15–30 min after amniocentesis or chorionic villus sampling. The median gestational age of the T21 group (18 weeks) was in fact older than that of the euploid group (12 weeks). As fetal DNA release into the maternal circulation increases significantly within the immediate period of invasive procedures (30) and with pregnancy progression, the potential confounding effects of these factors need to be considered. Nonetheless, the report by Fan et al. and our study independently demonstrate the feasibility of MPGS for noninvasive prenatal diagnosis.
Materials and Methods
Details are in the SI Text.
Massively Parallel Genomic Sequencing.
For plasma DNA sequencing, 11–50 ng of plasma DNA were used for DNA library construction by the beta Chromatin Immunoprecipitation Sequencing (ChIP-Seq) sample preparation kit (Illumina) according to the manufacturer's instructions except when specifically noted below. For the first experiment described in Results, the enriched adapter-ligated DNA fragments in the range of 150–300 bp were size selected using 2% agarose electrophoresis. The selected DNA libraries were then additionally amplified using a 15-cycle PCR. After the first experiments, all other experiments in this study followed the protocol with the omission of the last two steps. The adapter-ligated DNA was purified directly using spin columns provided in a QIAquick PCR purification kit (Qiagen).
Sequence Alignment.
All 36-bp sequence reads were aligned to the repeat-masked human genomic reference sequences (NCBI Build 36, version 48) downloaded from the Ensembl Genome Browser (http://www.ensembl.org), using the ELAND program in the GAPipeline-0.2.2.5 software package provided by Illumina. A result output file was generated after running ELAND, in which code U0 on the third field of the output file indicated that the best match found was a unique match in the repeat-masked human reference genome. A sequence with codes 1 in the fourth, 0 in the fifth, and 0 in the sixth fields (hence U0–1–0–0) indicated that it had just a single exact match in the repeat-masked human reference genome without any nucleotide mismatch. U0–1–0–0 sequences in each chromosome were sorted and counted using the awk utility in Linux, and the sorted sequences were used for further analysis.
Calculation of the Genomic Representation of Each Chromosome in the Reference Human Genome.
Except for the genomic representation values used as reference in Fig. S2C (sample 3), the expected chromosome size for a haploid female genome was calculated as described here. The reference sequences (NCBI Build 36, version 48) for each human chromosome except chrY were downloaded from the Ensembl Genome Browser (http://www.ensembl.org). All sequences were subjected to repeat masking and the number of remaining nucleotides was counted per chromosome. The expected percentage of representation of each chromosome was obtained by dividing the repeat-masked nucleotide counts per chromosome by the total repeat-masked nucleotide counts of all chromosomes without including chrY. The reference values used in Fig. S2C were derived by including the repeat-masked nucleotide counts of chrY to obtain the total repeat-masked genome size.
Supplementary Material
Acknowledgments.
We thank Rebecca Chan, Macy Heung, Yongjie Jin, Yu Kwan Tong, and Dana Tsui for technical assistance. We thank the Information Technology Services Center of The Chinese University of Hong Kong and the Center for High Performance Computing at Virginia Commonwealth University for data processing. This study was supported by the University Grants Committee of the Government of the Hong Kong Special Administration Region, China, under the Areas of Excellence Scheme (AoE/M-04/06) and a sponsored research agreement with Sequenom. Y. M. D. Lo was supported by the Chair Professorship scheme of the Li Ka Shing Foundation.
Footnotes
Conflict of interest statement: R.W.K.C., K.C.A.C., N.B.Y.T., F.M.F.L., B.C.Y.Z., C.R.C., and Y.M.D.L. have filed patent applications on the detection of fetal nucleic acids in maternal plasma for noninvasive prenatal diagnosis. Part of this patent portfolio has been licensed to Sequenom. C.R.C. is Chief Scientific Officer of and holds equities in Sequenom. Y.M.D.L is a consultant to and holds equities in Sequenom.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810641105/DCSupplemental.
References
- 1.Tabor A, et al. Randomised controlled trial of genetic amniocentesis in 4606 low-risk women. Lancet. 1986;1:1287–1293. doi: 10.1016/s0140-6736(86)91218-3. [DOI] [PubMed] [Google Scholar]
- 2.Malone FD, et al. First-trimester or second-trimester screening, or both, for Down's syndrome. N Engl J Med. 2005;353:2001–2011. doi: 10.1056/NEJMoa043693. [DOI] [PubMed] [Google Scholar]
- 3.Bianchi DW, et al. Isolation of fetal DNA from nucleated erythrocytes in maternal blood. Proc Natl Acad Sci USA. 1990;87:3279–3283. doi: 10.1073/pnas.87.9.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung MC, Goldberg JD, Kan YW. Prenatal diagnosis of sickle cell anaemia and thalassaemia by analysis of fetal cells in maternal blood. Nat Genet. 1996;14:264–268. doi: 10.1038/ng1196-264. [DOI] [PubMed] [Google Scholar]
- 5.Bianchi DW, et al. Fetal gender and aneuploidy detection using fetal cells in maternal blood: analysis of NIFTY I data. National Institute of Child Health and Development Fetal Cell Isolation Study. Prenat Diagn. 2002;22:609–615. doi: 10.1002/pd.347. [DOI] [PubMed] [Google Scholar]
- 6.Lo YMD, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 7.Lo YMD, Chiu RWK. Prenatal diagnosis: progress through plasma nucleic acids. Nat Rev Genet. 2007;8:71–77. doi: 10.1038/nrg1982. [DOI] [PubMed] [Google Scholar]
- 8.Lo YMD, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62:768–775. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng EKO, et al. mRNA of placental origin is readily detectable in maternal plasma. Proc Natl Acad Sci USA. 2003;100:4748–4753. doi: 10.1073/pnas.0637450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oudejans CB, et al. Detection of chromosome 21-encoded mRNA of placental origin in maternal plasma. Clin Chem. 2003;49:1445–1449. doi: 10.1373/49.9.1445. [DOI] [PubMed] [Google Scholar]
- 11.Lo YMD, et al. Plasma placental RNA allelic ratio permits noninvasive prenatal chromosomal aneuploidy detection. Nat Med. 2007;13:218–223. doi: 10.1038/nm1530. [DOI] [PubMed] [Google Scholar]
- 12.Chim SSC, et al. Systematic search for placental epigenetic markers on chromosome 21: towards noninvasive prenatal diagnosis of fetal trisomy 21. Clin Chem. 2008;54:500–511. doi: 10.1373/clinchem.2007.098731. [DOI] [PubMed] [Google Scholar]
- 13.Old RW, Crea F, Puszyk W, Hulten MA. Candidate epigenetic biomarkers for non-invasive prenatal diagnosis of Down syndrome. Reprod Biomed Online. 2007;15:227–235. doi: 10.1016/s1472-6483(10)60713-4. [DOI] [PubMed] [Google Scholar]
- 14.Tong YK, et al. Noninvasive prenatal detection of fetal trisomy 18 by epigenetic allelic ratio analysis in maternal plasma: theoretical and empirical considerations. Clin Chem. 2006;52:2194–2202. doi: 10.1373/clinchem.2006.076851. [DOI] [PubMed] [Google Scholar]
- 15.Lo YMD, et al. Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc Natl Acad Sci USA. 2007;104:13116–13121. doi: 10.1073/pnas.0705765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuster SC. Next-generation sequencing transforms today's biology. Nat Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- 18.Dear PH. One by one: single molecule tools for genomics. Brief Funct Genomic Proteomic. 2003;1:397–416. doi: 10.1093/bfgp/1.4.397. [DOI] [PubMed] [Google Scholar]
- 19.Chan KCA, et al. Size distributions of maternal and fetal DNA in maternal plasma. Clin Chem. 2004;50:88–92. doi: 10.1373/clinchem.2003.024893. [DOI] [PubMed] [Google Scholar]
- 20.Kel-Margoulis OV, et al. Composition-sensitive analysis of the human genome for regulatory signals. In Silico Biol. 2003;3:145–171. [PubMed] [Google Scholar]
- 21.Reinartz J, et al. Massively parallel signature sequencing (MPSS) as a tool for in-depth quantitative gene expression profiling in all organisms. Brief Funct Genomic Proteomic. 2002;1:95–104. doi: 10.1093/bfgp/1.1.95. [DOI] [PubMed] [Google Scholar]
- 22.Lo YMD, et al. Increased fetal DNA concentrations in the plasma of pregnant women carrying fetuses with trisomy 21. Clin Chem. 1999;45:1747–1751. [PubMed] [Google Scholar]
- 23.Chan KC, et al. Hypermethylated RASSF1A in maternal plasma: a universal fetal DNA marker that improves the reliability of noninvasive prenatal diagnosis. Clin Chem. 2006;52:2211–2218. doi: 10.1373/clinchem.2006.074997. [DOI] [PubMed] [Google Scholar]
- 24.Lun FMF, et al. Microfluidics digital PCR reveals a higher than expected fraction of fetal DNA in maternal plasma. Clin Chem. 2008;54:1664–1672. doi: 10.1373/clinchem.2008.111385. [DOI] [PubMed] [Google Scholar]
- 25.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skaletsky H, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 27.Harris TD, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 28.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 29.Fan HC, et al. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci USA. 2008;42:16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samura O, et al. Cell-free fetal DNA in maternal circulation after amniocentesis. Clin Chem. 2003;49:1193–1195. doi: 10.1373/49.7.1193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.