Abstract
Identifying proteins correctly and with known levels of confidence remain as significant challenges for proteomics. Random or decoy peptide databases are increasingly being used to estimate the false discovery rate (FDR), e.g., from liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses of tryptic digests. We show that this approach can significantly underestimate the FDR, and describe an approach for more confident protein identifications that uses unique partial sequences derived from a combination of database searching and amino acid residue sequencing using high accuracy MS/MS data. Applied to a Saccharomyces cerevisiae tryptic digest, the approach provided 3,132 confident peptide identifications (∼5% modified in some fashion), covering 575 proteins with an estimated zero FDR. The conventional approach provided 3,359 peptide identifications and 656 proteins with 0.3% FDR based upon a decoy database analysis. However, the present approach revealed ∼5% of the 3,359 identifications to be incorrect, and many more as potentially ambiguous, (e.g., due to not considering certain amino acid substitutions and modifications). In addition, 677 peptides and 39 proteins were identified that had been missed by conventional analysis, including non-tryptic peptides, peptides with various expected/unexpected chemical modifications, known/unknown posttranslational modifications, single nucleotide polymorphisms or gene encoding errors, and multiple modifications of individual peptides.
Keywords: precise proteomics, high-precision tandem mass spectrometry, unique sequences, post-translational modifications and mutants, false discovery rate and identification ambiguity
Introduction
Protein identification is fundamental to proteomic studies. Presently, liquid chromatography-tandem mass spectrometry (LC-MS/MS) is widely used to identify proteins from peptides from enzymatic (e.g., trypsin) digestion.1,2 Peptide assignments are typically accomplished using automated database searching algorithms (e.g., SEQUEST,2 MASCOT3, X!Tandem4) that compare tandem mass spectra with in silico generated model spectra derived from candidate peptide sequences, using scoring schemes to determine relative confidence levels.5-7
A currently popular strategy utilizes a comparably sized “decoy” set of “false” peptides to estimate the level of incorrect identifications for a particular set of filtering criteria.5 While low FDRs (e.g.,<1%) have been obtained from conventional precision LC-MS/MS data,5 the accuracy of such estimates is uncertain. The effectiveness of the identification process decreases as the size of the peptide candidates increases,8 and thus proteome coverage is decreased if the FDR is to be held constant. Similar difficulties arise as the candidate list diverges from the actual (i.e., detectable) set of peptides. If the actual FDR is significantly higher than expected, then not only are proteins incorrectly identified, but quantitation also suffers since abundance information from significant numbers of incorrectly identified peptides gets “rolled-up” to the protein level.
The peptide candidate lists are typically generated from genomic data and exclude potential amino acid modifications (or substitutions);9 as a result, both modified and unmodified peptides can be incorrectly (or fail to be) identified. Typically, a large fraction (>50%) of the species detected in MS or MS/MS proteomic measurements do not result in confident peptide identifications, including those from high quality tandem mass spectra;10 and this unidentified fraction increases with proteome complexity. The identification of modified peptides is generally based upon focused searches that consider a limited number of modifications11 and generally fail for peptides that have unknown/unexpected and multiple modifications. Approaches based upon accurate mass and LC retention time data have recently been reported,12 but challenges remain due to proteome complexity. Particularly interesting are so-called “second pass” approaches that use an initial set of identifications to guide a much broader consideration of possible variations and modifications focused on a smaller set of proteins.13 Thus, understanding identification assignment quality and potential ambiguities remain key issues for proteomics.14
In this work we developed and initially applied an approach for broad protein identifications that utilizes initial conventional database searching (to provide a truncated set of candidate sequences) with unambiguous amino acid residue sequencing determination based upon the use of high precision and accuracy LC-MS/MS data. The truncated set of candidate sequences allows a broad set of possible modifications and amino acid sequence variations to be simultaneously considered, in contrast to conventional de novo approaches.15 We demonstrate for yeast Saccharomyces cerevisiae5,16,17 that this unique sequence tag (UStag) method enables higher confidence identification of proteins and their modifications, including chemical artifacts, known and novel post-translational modifications, and amino acid (AA) sequence variations, with more accurately estimated FDR and lower ambiguity.
Results
UStag definition
A UStag is an AA sequence associated with a single protein in a candidate list. Sequence uniqueness generally increases with sequence length (see Supplementary Figure 1 for an in-silico UStags search against the yeast sequence database18), but varies broadly; 4-AA sequences can be unique, while other 50-AA sequences are not (Supplementary Table 1). The UStag concept can be further refined for various purposes by alternatively associating a UStag with a group of similar proteins.
Establishing UStags from high precision LC-MS/MS data
Figure 1 outlines the combined database search and amino acid residue sequencing approach for determining UStags from high precision LC-MS/MS data. The experimental dataset was initially searched against the yeast sequence database with a ±5 u mass tolerance (Supplementary Figure 2), and then with a ±210 u tolerance to generate a sub-dataset that includes potential modifications. The top ten candidates identified by SEQUEST from each tandem mass spectrum were selected for amino acid residue sequencing calculations using accurate masses of the precursor, its fragments, and AA residues (see Methods section and examples shown in Supplementary Figure 3). In addition, spectra were inspected (manually in this work) for “missing” fragment ion peaks, having low intensity and/or non-ideal isotopic envelopes (Supplementary Figure 4), that could join multiple shorter sequences into one larger UStag.
Figure 1.
The UStags process for identifying unique peptide sequences from precise LC-MS and MS/MS experiments. Yeast whole cell tryptic digest was analyzed using an LC-LTQ-Orbitrap platform; resulting LC-MS/MS dataset was searched using SEQUEST against yeast sequence database. Amino acid residue sequencing (see Supplementary Methods) was applied to obtain accurate AA residue assignments and sequences. These initial unique sequences were examined using the residue replacement filter (RRF; see Supplementary Methods), illustrated here for TSGTLEMNLK(1). This sequence was found in a single protein (the number in the parenthesis); however, an isobaric variant sequence SSGTLEMNLK potentially exists in another yeast protein when modifications are considered (i.e., Ser-me-ester has the same mass as Thr). Due to the ambiguity this sequence cannot serve as an UStag (i.e., only sequences remaining after the RRF are considered UStags).
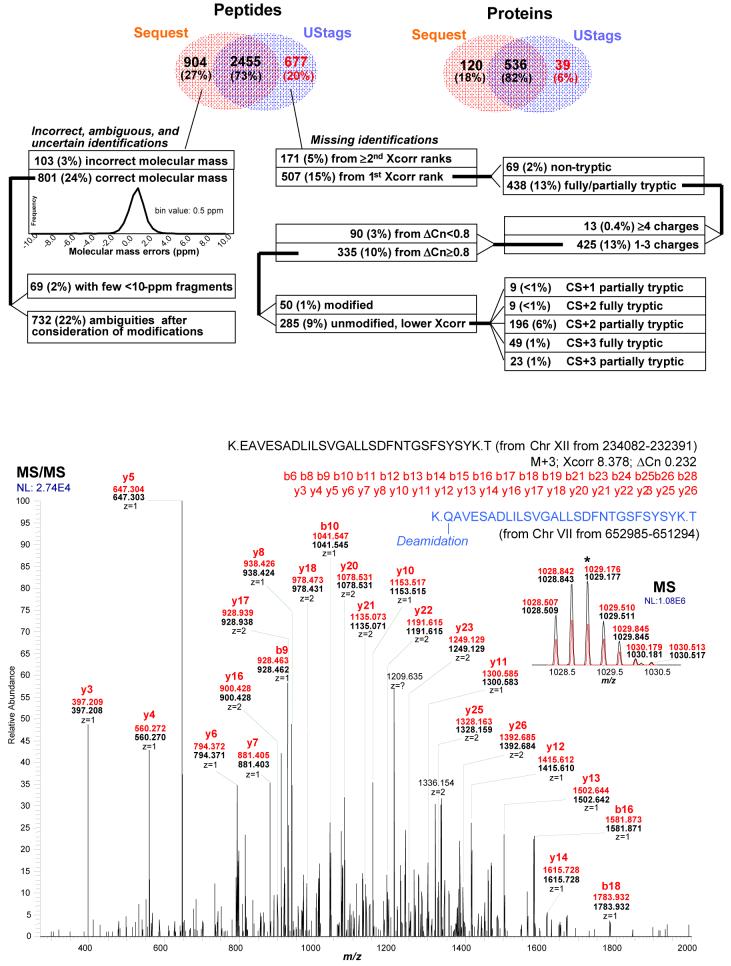
We developed a residue replacement filter (RRF) that considers all AA substitutions and a broad set of possible modifications9 to construct a candidate list for further consideration. The RRF is made up of a broadly inclusive list of AA (or modified AA) sequence combinations that could potentially explain the mass differences observed in a MS/MS spectrum. A peptide is not considered as unambiguously identified if a replacement of same mass segments or modification(s) for the others in the database leads to generating the same mass sequence for each residue as the peptide to be identified. For example, the peptide R.SAYLAAVPLAAILIK.T (from YCL040W) cannot be distinguished from R.SAYLAAVPIA AILIK.T (from YDR516C) due to I and L being isobaric. Similarly, the peptide S.SSANK.L (from YKR092) cannot be distinguished from D.SSAGGK.Q (from YJL012C) due to the isobaric segment GG for N as one cannot get guarantee the MS/MS spectrum would reveal the G-G bond cleavage (however, D.SSAGGK.Q can be distinguished from S.SSANK.L using Ustag, as the UStag method effectively requires sequencing each individual amino acid in the tag; the replacement was operated from GG to N, not from N to GG; the same situation was for AG/GA and Q). Additionally, the peptide K.EAVESADLILSVGALLSDFNTGSFSYSYK.T (from YLR044C) cannot be distinguished from K.Q(Deamidation)AVESADLILSVGALLSDFNTGSFSYSY.K (from YGR087C) due to the modification (additional details and examples are given below). It is well established that even MSn approaches cannot typically distinguish such isobaric differences. The LC retention time might distinguish some peptides containing such isobaric residues.19
The initial putative unique sequences from the first pass candidate peptide list search and the amino acid sequence were additionally filtered by the RRF to exclude ambiguous AA combinations from the resulting UStag set. Supplementary Table 2 lists the ambiguous AA combinations and modifications having mass differences that need high mass measurement accuracy (MMA) for differentiation. In this study, we used an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific) that generally provided <5 ppm MMA for parent and fragment ions and ≤0.003 u for AA residues (i.e., sequencing precision) (Figure 1). Approximately 5,700 putative unique sequences (from ∼650 different yeast proteins) were identified after the initial database search and the sequence determination; ∼5,200 putative unique sequences (from ∼630 yeast proteins) remained after the RRF eliminated isomeric combinations of AA residues from ambiguous masses, and ∼4,400 UStags (from 575 proteins) remained after the RRF eliminated ambiguous masses arising from potential modifications. Supplementary Figure 5 shows the MMA distributions obtained for identified peptides, and their corresponding fragment ions and AA residues. We also searched the 34,261 MS/MS spectra in this dataset against reversed and scrambled yeast sequence databases using the same procedure. Importantly, both scrambled and reversed database searches resulted in a zero FDR (i.e., no “UStags” were found).
Unambiguous identification of yeast proteins and their modifications
UStag identified peptides from the yeast tryptic digest analysis varied in length (5-45 AA residues) and mass (450-5000 u) (Supplementary Figure 6). UStags identified 575 yeast proteins: 442 of these proteins (77%) from unmodified peptides, 129 (22%) from both unmodified and modified peptides, and 4 (<1%) from only modified peptides (Supplementary Tables 3 and 4). Figure 2 shows the classification of identified proteins according to their associated molecular function. Less defined yeast proteins [e.g., unclassified proteins, interaction with the environment (systematic) and transposable elements, viral, and plasmid proteins] were not found, even when only a single UStag was required for identification. The ability to identify modifications to a specific AA was evaluated by alkylating reduced cysteines with iodoacetamide (Supplementary Methods) and using an appropriate mass tolerance (210 u) in a SEQUEST search. Using UStags, we identified alkylation of not only Cys, but also of 13 other N-terminal residues (Supplementary Table 3), indicating artifactual overalkylation20 during sample preparation (see Supplementary Methods).
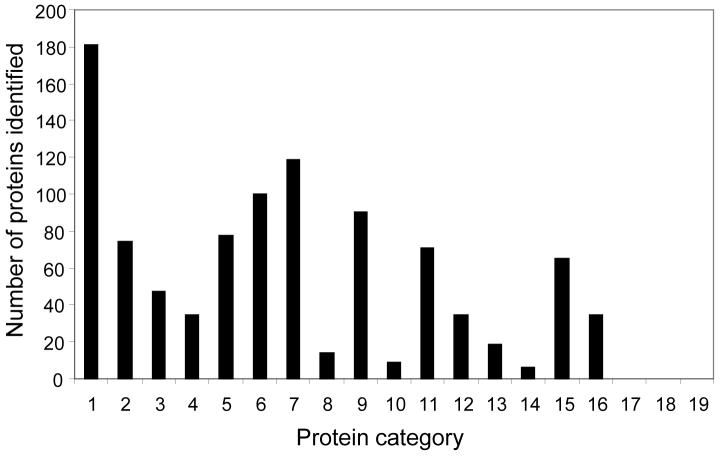
Figure 2.
The classification of unambiguously identified proteins according to the molecular function they are associated. Protein function category: 1 Metabolism, 2 Energy, 3 Cell cycle and DNA processing, 4 Transcription, 5 Protein synthesis, 6 Protein fate, 7 Protein with binding function or cofactor requirement, 8 Protein activity regulation, 9 Cellular transport, transport facilitation and transport routes, 10 Cellular communication/signal transduction mechanism, 11 Cell rescue, defense and virulence, 12 Interaction with the cellular environment, 13 Cell fate, 14 Development (systemic), 15 biogenesis of cellular components, 16 Cell type differentiation, 17 Interaction with the environment (systemic), 18 Transposable elements, viral and plasmid proteins, 19 Unclassified proteins. Note that proteins annotated as unclassified, interaction with the environment (systematic), transposable elements, and viral and plasmid proteins (a total of 129 database entries) were not found using the UStag method with this cellular lysate tryptic dataset.
Modifications that could not be assigned to a specific AA residue based upon a UStag-derived sequence were determined from the mass differences between the precursor mass and the candidate mass calculated, the MS/MS information for parts of the peptide not contained in the UStag sequence, and the mass contributions from modifications. Many examples of glycylation and lysylation were found (Supplementary Figure 7). Acetylation was especially prevalent on Ser, indicating cell-directed posttranslational modification9 as opposed to random chemical derivation of hydroxyl-containing residues (e.g., Asp and Glu). Deamidation of both Asn and Gln was found, but Asn deamidation was much greater than Gln (35 vs. 2), which is consistent with a previous report.21 Artifactual carbamylation from the breakdown of urea was observed on peptide N-termini, indicating modification either during or after the trypsin digestion, while Pyro-glu (17 cases) and Hse_lact (2 cases) were observed on the peptide N-terminal Glu and Met, in agreement with previous reports.22,23 Oxidation observed for Met and Trp occurs before the separation and electrospray ionization (ESI) as the peptide and its oxidized version were detected at different LC separation times (Supplementary Figure 8).
Unexpected sequence modifications, i.e., AA mutations or genome sequence errors, were also identified using UStags. The UStag SSIFDASAGI (see Figure 3A) identified the peptide D.SHSSIFDASAGIR and not the sequence D.SHSSIFDASAGIQ expected from yeast glyceraldehyde-3-phosphate dehydrogenase 3. The data indicates the precursor ion mass was shifted either +28.042 u or -113.082 u (not shown in Fig 3A) from the mass of the peptide sequence identified. The negative mass shift was excluded since no single and/or combined modifications on Gln and Leu9 corresponds to such mass shift. The mass positive shift of 28.042 u was attributed to Gln to Arg substitution. More complex, multiple modifications were also observed that could not be identified by conventional methods. Figure 3B shows a peptide from yeast killer toxin-resistance protein 5 that has both Gln deamidation and a Val to Tyr substitution.
Figure 3.
UStags for characterization of unexpected modifications. (A) UStag SSIFDASAGI measured from b ions (Figure 3A) led to the identification of the peptide D.SHSSIFDASAGIQL... predicted from the yeast protein Chr VII. The precursor ion mass (survey MS) was shifted by +28.042 u corresponding to the (C)(H4)(N2)(-O) change in elemental composition, which could readily be explained by a Gln to Arg switch. Measured values are denoted in black; theoretical values derived from the sequence database are given in red, and theoretical values for corrected sequence are given in blue. (B) An example showing a case where both, deamidation on Gln and Val to Tyr substitution were determined to provide correct interpretation of the spectrum. Measured values and theoretical values for corrected sequence are denoted in blue; theoretical values derived for deamidated sequence are given in green.
Peptides from multiplexed spectra and unexpected digestion products identified by UStags
Conventional approaches generally identify a single peptide per tandem mass spectrum (i.e., the “top hit”); however, simultaneous dissociation of multiple precursor ions (i.e., multiplexed MS/MS) has been well documented. The UStag approach identifies multiple peptides from tandem mass spectrum by piecing together various b- and y-ion series into multiple sequences. For example, two peptides were identified using UStags GFTFSFPA from yeast hexokinase-2 and INFLTE from yeast RIM1 (Figure 4A). The selection of the peptide precursor ion at m/z =1073 (selected with a 3 m/z “window”) yielded two partially overlapping isotopic envelopes and two UStag-identified peptides. ∼5% of the UStags identified in this study were from multiplexed tandem mass spectra (Supplementary Tables 3 and 4). It is noted that the multiple UStags identified in a single MS/MS spectrum were required to have their corresponding peptide molecular masses (with a mass error of <10 ppm) in the precursor MS scan.
Figure 4.
The UStags identifies peptides that challenge conventional proteomics approaches. (A) UStags allow identification of peptides arising from multiplexed spectra. Two UStags: GFTFSFPA from protein Chr VII and INFLTE from protein Chr III were measured in a single spectrum. The selection of the precursor ion at m/z =1073 u (with selection window 3 m/z units wide) yielded two partially overlapping isotopic envelopes and two UStag-identified proteins. Measured values are denoted in black; theoretical values derived from the two UStag-identified peptides are denoted in red and blue, respectively. (B) UStags allow identification of non-tryptic peptides. An UStag was measured using both b- or y ion series for protein Chr VII and no modification(s) were found that could make the sequence has Arg/Lys terminus and still support the measured UStag, precursor mass and fragment masses. Measured values are denoted in black; theoretical values derived from the UStag-identified peptide are denoted in red.
While search candidate lists typically include partially or fully tryptic peptides (i.e., cleavage at least at one C-terminus of Arg or Lys),5 even non-tryptic peptides associated with endogenous protease activities can be confidently identified by UStags, as illustrated for yeast glyceraldehyde-3-phosphate dehydrogenase 3 (Figure 4B). In this example, no modification(s) were found to be consistent with Arg/Lys termini and measured precursor and fragment masses; however, intracellular degradation by endogenous proteases24 (e.g., yeast chymotrypsin-like proteases and caboxypeptidases) could cleave Tyr-Thr and Ala-Ser bonds to generate the observed peptide. Approximately 3% of the identified peptides were associated with proteolytic activity other than trypsin (i.e., peptides determined to have C-termini other than Arg or Lys).
UStags vs. conventional database searching for peptide and protein identification
Using previously suggested criteria for obtaining <1% peptide level FDR for yeast5 (0.3% determined for the present data), the use of SEQUEST identified 3,359 peptides from this dataset (see Supplementary Methods). Figure 5 (top) compares numbers of SEQUEST and UStag identified peptides and proteins. While the two methods had 2,455 peptides in common, 904 additional peptides were identified solely by SEQUEST. Importantly, all 904 could be shown to be erroneous or to have arguably ambiguous assignments.
Figure 5.
The comparison between SEQUEST and UStags identified peptides. Top: While the two methods had 2455 peptides in common, 904 peptides were identified only using SEQUEST and 678 only by UStag method. Measured precursor masses for 103 out of 904 SEQUEST identified peptides had unacceptably high errors (>10 ppm) and the remaining 801 peptides with accurately measured precursor masses (histogram shown as an inset) included 69 peptides identified with few fragments when MMA <10 ppm requirement was applied and 732 peptides that could not be uniquely assigned after consideration of isomeric AA combinations and modifications. 678 UStag-identified peptides that were missed by SEQUEST included multiply modified peptides, multiplexed spectra, non-tryptic peptides, sequence polymorphisms, low SEQUEST scores, etc. Xcorr and ΔCn are SEQUEST cross correlation and relative Xcorr values; CS: peptide charge state. Bottom: An example illustrating that even a peptide identified with high scores using database search (e.g., Xcorr >8 and ΔCn > 0.2) can harbor inaccuracy or ambiguity. (Precursor and fragment measured masses of SEQEST identified peptide are denoted in red and alternative peptide sequence consistent with MS/MS data n blue.)
The 103 peptides were considered incorrect due to high mass errors (i.e., ≫10 ppm) indicated by the precursor MS. It is unlikely that the 103 incorrect molecular masses arise due to missing matches to the correct peaks or from “multiplexed spectra” as we matched the wide m/z range (i.e., 400-2000, see Methods) of the precursor MS spectrum, not the narrow m/z range around the precursor, and also considered theoretical isotopic peak envelopes and possible charge states for the parent peptide. The failure to observe correct molecular masses means that the peptide molecular formats (or chemical composition) of these identifications derived from the conventional SEQUEST database search must be incorrect. Peptide modifications may be one reason for the observed incorrect molecular masses.
Another 69 peptides having <10 ppm precursor mass error but also had few fragments (see examples in Supplementary Table 5) such that a nearly 100% FDR would result from a reverse database search (e.g., >200 such peptides were obtained from searching the reverse database and amino acid sequence determination of search candidates with <10 ppm mass errors). These incorrect peptide identifications can be easily removed through use of accurate MS/MS data.
The remaining 732 peptides having <10 ppm parent mass error (Figure 5) were ambiguous if isomeric AA combinations or possible modifications were considered. Table 1 gives some examples that had high SEQUEST scores among these 732 peptides and Figure 5 (bottom) shows such a case. As an example, the peptides R.SAYLAAVPLAAILIK.T (from YCL040W) cannot be distinguished from R.SAYLAAVPIAAILIK.T (from YDR516C) as they have the same mass sequence due to I and L being isobaric. Use of mass spectrometric information to derive amino acid sequence information needs to exclude the isobaric amino acids and other isobaric assignments (e.g., modified amino acids) to achieve effectively unambiguous peptide identifications (i.e., unique explanation of spectra for specific peptides). This goal has been partially accomplished in conventional database search methods; for example, SEQUEST sets ΔCn = 0 for the above two peptides and conventional database search methods require ΔCn > 0 (e.g., 0.08 or 0.1) for peptide identification.5,16 However, conventional database search methods omit the consideration of isobaric segments generated e.g. from various possible modifications (see examples shown in Table 1 and Figure 5) for peptide identification, which makes the methods in currently used inconsistent with obtaining unambiguous peptide identifications (i.e. incorrect assignments are possible due to the failure to consider such alternatives). Thus, a key goal of the UStag method is to greatly reduce or eliminate the possibility of incorrect assignments that result from such assumptions as to the set of possible peptides that can exist in the proteome mixture as well as the modifications that may exist for these peptides. To accomplish this, the approach removes such ambiguous peptides (i.e., the 732 peptides) from the identification list. Therefore, the approach effectively sacrifices the subset of correct identifications (e.g. with high SEQUEST score) because they are ambiguous when the broader set of possibilities is considered.
Table 1.
Examples showing potential ambiguities in the identification of peptides from MS/MS when isobaric AA substitutions and a broad set of AA modifications are considered.*
| Peptide without modification | Xcorr | ΔCn | CS | ORF | Possible modified peptide | ORF |
|---|---|---|---|---|---|---|
| K.IEGVATPQEAQFYLGK.R | 4.959 | 0.538 | 2 | YOR234C | K.IEGVATPQD(-methylation)AQFYLGK.R* | YPL143W |
| V.TPSFVAFTPEER.L | 3.358 | 0.260 | 2 | YDL229W | V.TPSFVAFTPQ(-Deadmid)ER.L | YNL209W |
| R.VVNEPTAAALAYGLEK.S | 5.037 | 0.553 | 3 | YJR045C | R.VVNEPTAAALAYGLD(-methylation)K.S | YEL030W |
| K.GGNIPMIPGWVMEFPTGK.E | 4.313 | 0.357 | 2 | YFR053C | K.GGNIPMIPGWVMD(-Methylation)FPTGK.E | YGL253W |
| K.SPIKVVGLSTLPEIYEK.M | 6.046 | 0.442 | 2 | YOL086C | K.SPIKVVGLSS(-Methylation)LPEIYEK.M | YMR303C |
| K.GILFVGSGVSGGEEGAR.Y | 5.735 | 0.484 | 2 | YHR183W | Q.GILFVGSGVSGGE(-Methylation)DGAR.F | YGR256W |
| K.AVYAGENFHHGDK.L | 4.643 | 0.463 | 2 | YHR174W | N.AVF(-Hydroxyl)AGENFHHGDK.L | YGR254W |
| K.VVGLSTLPEIYEK.M | 3.831 | 0.427 | 2 | YOL086C | K.VVGLSS(-Methylation)LPEIYEK.M | YMR303C |
| R.MPELIPVLSETMWDTKK.E | 3.229 | 0.218 | 2 | YLR249W | R.MPELIPVLSES(-Methylation)MWDTKK.G | YNL014W |
| K.EAVESADLILSVGALLSDFNT GSFSYSYK.T | 8.378 | 0.232 | 3 | YLR044C | K.Q(-Deamid)AVESADLILSVGALLSDFN TGSFSYSY.K | YGR087C |
The sequences underlined were identified based upon accurate mass MS/MS (i.e., <10 ppm mass errors), but cannot be unambiguously distinguished by MS present methods, (see alternatives (right), and are rejected by the UStag method for use in protein identification.
Thus, an important aspect of the present approach is the initial consideration of an expanded set of possible peptides. We note that the UStag method identified 677 peptides missed by the SEQUEST analysis due to the various modifications not considered, as well as the consideration of multiplexed data, unexpected digest products, etc (Figure 5). The support for the identification of these 677 peptides included the spectral similarity (i.e., their inclusion in the top 10 candidates of the SEQUEST database search, see Methods section), correct molecular masses (i.e., <10 ppm errors) and consistent isotopic envelope patterns, consistent fragment ion masses (i.e., <10 ppm errors), correct residue mass differences (i.e., <0.005 u errors), and the uniqueness in the database for the sequences constructed from the consecutive fragments and the residues measured after the consideration of various possible modifications. The number of peptide false negative identifications was additionally reduced compared to the conventional SEQUEST analysis by the acceptance of identifications excluded by the SEQUST filters applied (see Supplementary Methods); e.g., UStags confidently identified some peptides having SEQUEST Xcorr <1.000 and ΔCn=0; see Supplementary Figure 9).
UStags vs. peptide sequence tag-based approaches
Various “peptide sequence tag”-based approaches, ranging from early heuristic to recent probabilistic versions,25-28 have been proposed for interpretation of tandem mass spectra. These approaches typically use short AA sequences and fragment locations together with a set of rules/filters such as (in its most simplistic version) a fully tryptic peptide requirement.25 Figure 6 shows an example of peptide identifications based on 2-9 AA residue “peptide sequence tags” approach for the same dataset. 3,368 peptides from 782 proteins were obtained when 2-AA residue tags were used for tryptic peptide identification.25 Of these 3,368 peptides, ∼60% (2,102 peptides) contain the UStags, while the remaining ∼40% are ambiguous (i.e., having alternative explanations for their tandem mass spectra due to modifications and/or AA substitutions). Longer sequences improved the identification specificity, but specificity comparable to the UStag approach could not be achieved, as even long sequence tags (e.g., >10-AA residues) may not be unique (see Figure 5). Also, lengthening the tag to 7-8 AA residues reduced the number of identified peptides (e.g., to ∼1,500) below that obtained with the UStag approach (i.e., 3,132 peptides). Increasing the number of the short “peptide sequence tags” (i.e., multiTag26) in individual peptides improves the probability of correct identifications, but similarly does not ensure unambiguous identifications. (Figure 6 shows an ambiguous identification even given 2 short AA segments: GQ and SLLL). Allowing partially- and non-tryptic peptides increases the number of identifications to 7,009 (from 2,186 proteins), but results in significantly lower confidence (>50% of identified peptides are ambiguous). The reliance on the precursor (and fragments) masses also limits the ability of the “peptide sequence tag” approach to identify unexpected/unknown modifications; e.g., see Figure 3.
Figure 6.
Comparison of peptide identifications based on measured peptide sequence tags containing 2-9 AA residues with UStags-derived peptide identification. Both peptide fragment measured masses are filtered using a 10 ppm MMA cutoff. Top: ○ and ◇ indicate the number of peptides and proteins, respectively, identified with 2-9 AA-residues; ○ and ◆ indicate the number of peptides and proteins, respectively, that overlap between the “peptide sequence tag” and “UStag” sets of identifications. Venn diagrams compare peptides and proteins identified using “peptide sequence tags” with ≥2 AA residues requirement (mass (MS) and sequencing error (MS/MS) histograms from are shown in the upper right corner) and UStags with unconstrained peptide C-termini. Bottom: An example of ambiguous peptide identification using the “peptide sequence tag” approach, even with two AA segments GQ and SLLL (labeled in blue), which was unambiguously identified using UStag approach.
Discussion
A key issue for proteomics is the quality (e.g. FDR) of the sets of peptides and proteins ultimately identified. The peptide identification process is conventionally dependent upon the quality of the set of possible peptides sequences in the database being used (e.g. for spectrum matching). The ideal database should contain peptides from all protein sequences, including those predicted from genome translations, as well as possibly unpredictable posttranslational modifications, and artifact products from proteomic sample manipulation. Since this ideal database is not achievable (and if achieved by greatly expanding the set of possibilities considered, such a database would significantly challenge computational throughput), and conventional approaches will lead to either significant sets of false positive identifications or some level of false negative identifications, and generally both. The present UStag approach aims to effectively eliminate false positive identifications and simultaneously takes steps to reduce false negative identifications by the consideration of an expanded set of possible peptides. Thus, this approach provides insights into the effectiveness of conventional approaches, and can be directly contrasted with conventional approaches by the detailed evaluation of MS/MS spectra to determine if incorrect or ambiguous assignments have been made. We note that this approach differs fundamentally from the conventional approaches that provide probability scores for identifications (of unknown accuracy, due to the issues noted above), since all the resulting identifications are both unambiguous and highly confident. Thus, by definition, probability scores for these identifications cannot be assigned.
This work demonstrates that LC-MS/MS proteomics based on conventional database searching/scoring algorithms can result in much higher levels of erroneous or ambiguous identifications than previously indicated due to not considering alternative assignments. For instance, ∼5% of the peptides identified in this work were modified, and the FDR in the example studied was underestimated by more than an order of magnitude (∼5% rather than the calculated ∼0.3%). Contributions of incorrect and ambiguous identifications from conventional approaches arise from both mass measurement accuracy limitations (i.e., for both peptide precursors and fragment ions) and limitations of the candidate list (discussed above). The former issue29,30 can be largely eliminated using accurate mass measurements, but the latter issue challenges current approaches (Table 1 and the spectrum in Figure 5) and FDR evaluations. Many approaches do not fully exploit the high MMA achievable with newer instrumentation and produce substantially greater levels of incorrect and ambiguous peptide identifications than indicated by conventional FDR analysis due to factors that include not considering possible sequence modifications.
Perfection in terms of simultaneously optimizing both false positive and false negative identifications is effectively unachievable in proteomics (due to unavoidable imperfections in the set of possible peptides being considered), and present approaches and methods effectively seek some compromise between the two. A key question that can be raised for this approach is the extent of false negative peptide identifications. This work shows that the UStag approach provides a significant overall improvement in the quality of the resulting set of peptide identifications. Importantly, this work has also shown that the UStag method (Figure 1) allows identification of approximately comparable numbers of unique peptides and proteins along with an essentially zero FDR and elimination of ambiguity in identifications. This finding is probably related to the approach used in this work to in the first pass definition of the set of peptide possibilities (the top 10 SEQUEST matches). It is clear that a significant number of the present unambiguous peptide identifications are derived from peptides that do not achieve “top” scores, and suggests further refinement of the scoring algorithms may significantly improve performance.
The more effective identification of non-tryptic protein degradation products by the UStag approach significantly enhances chance for identification of, e.g., secreted and low molecular weight proteins from human biofluids, missed by conventional approaches. The UStag method should also be useful for “top-down”31 approaches, and to more broadly characterize PTMs to identify unexpected (but biologically significant) proteins stemming from sequence polymorphisms, alternative splicing, and programmed frame shifts. In addition, the use of alternative search engines (e.g., MASCOT, X!Tandem) and de novo algorithms for generating more complete initial sets of peptide candidates may further improve the coverage without affecting the specificity. Also, we note that when extending the UStag to the study of mammalian (e.g., human) proteomes, the database typically contains redundant protein products (e.g. from the same gene) that need to be grouped into one entry (e.g. according to the gene) in order to minimize the false negative peptide identifications. Finally, we note that this approach does not rule out the use of lower confidence and more ambiguous MS and MS/MS identifications (and indeed the assignment of probability scores for such cases). Along with the higher quality identifications, such lower confidence identifications can be more effectively recognized and used properly.
In addition to the extremely low FDR discussed above for identification of tryptic digests of the cellular extracts, the UStag method has other attractive characteristics for peptide identification that include: 1) effective independence of peptide terminal properties, 2) independence of the peptide sequence length and its charge states, and 3) the ability to probe for unknown mutations and modifications. We are now using the first two characteristics to study proteolytic posttranslational modifications of proteins, intracellular proteases’ activity and specificity, and polypeptides (including intact proteins) and the third for study of amino acid mutations.
Methods
High-resolution capillary LC coupled with high precision MS/MS analyses
LC separations were performed using a 20K psi LC system32 equipped with a 90 cm × 100 μm i.d. capillary column packed with 2 μm C18-bonded porous (120 Å pores) silica particles (Phenomenex, Terrence, CA). FT-MS and FT-MS/MS data were collected using an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific) and AGC targets of 1×106 and 2×105, respectively. Spectra were acquired at 30K resolution, using a survey scan with 400≤m/z≤ 2000 followed by FT- MS/MS of the 5 most intense ions from the survey scan. FT-MS/MS employed an isolation window of 3 m/z units and 35% normalized collision energy. Dynamic exclusion was enabled with no repeat count and using a mass window of ±1.5 m/z units and duration of 25 sec. Mass calibration was performed according to the method provided by the instrument manufacturer.
Preparations of the yeast trypsin digest
Saccharomyces cerevisiae (ATCC 26108, Lot 137504) was grown in a batch shaker flask at 37°C on yeast nitrogen base without amino acids (Y0626, Sigma Aldrich, St. Louis MO). Two different media were prepared, one with the addition of 5 g/L of glucose and the other, with 5 g/L fructose. Cells were harvested at mid- logarithmic phase and stationary phase by centrifugation at 3200 rcf for 10 min and combined in a 1:3 ratio of stationary to mid- logarithmic phase cells. 50 mM Tris buffer with 10 mM MgCl2 (pH 7.5) was added to the cell pellet at a 10:1 (v/v) ratio of buffer to pellet volume. Lysis was accomplished by bead beating the cell mixture with 0.1 mm zirconia/silica beads in a minibeadbeater (Biospec, Bartlesville, OK) at 5500rcf for 90 s. The lysate was ultracentrifuged at 355,040 rcf with a Beckman (Fullerton, CA) Optima TL ultracentrifuge for 10 min. The supernatant was collected and placed immediately on ice to inhibit proteolysis. The sample was denatured in a solution of 7 M urea, 50 mM ammonium bicarbonate (pH 7.8), and 5 mM TCEP (Pierce, Rockford, IL), and then incubated at 60°C for 30 min and diluted 10-fold with 50 mM ammonium bicarbonate. 1 μL of 1 M CaCl2 was added with sequencing grade modified trypsin in a protease-to-sample protein ratio of 1:50 and incubated at 37°C for 5 h. Iodoacetamide (36 mg/mL) was added to a final concentration of 10 mM and incubated at room temperature for 30 min. The resultant peptides were desalted using Supelco Supelclean C-18 tubes (St. Louis, MO). Peptide concentration was determined by BCA protein assay (Pierce, Rockford, IL), and the sample concentration was adjusted to 1.0 mg/mL with water.
Processing high-accuracy MS/MS data
FT MS/MS data were processed using SEQUEST (Thermo Fisher Scientific), using tolerances of ±5 u and ±210 u in two separate database searches. The top 10 candidates from each database search were used as a starting point for the amino acid residue sequencing evaluation of peptide fragments. In-house developed software ICR2LS (http://ncrr.pnl.gov) was used to generate the theoretical isotopic envelopes for each peptide candidates from the database searches and match the raw spectra with the charge states, a mass tolerance of 10 ppm, and the quality of the isotopic envelope (or pattern): the correct molecular mass must be matched with at least the 3 most abundant isotopic peaks. Peptide ion peak candidate b and y fragments were searched with a mass tolerance of 10 ppm and used to establish a frame for sequencing (reading) consecutive fragments. The amino acid residues were determined from the distance of the consecutive b or y fragments assigned for the peptide candidates from database searches with a mass tolerance of 0.005 u; sequence fragments were then constructed according to the residues determined. The obtained sequenced fragments were searched against the yeast sequence database to establish sequence uniqueness using the developed residue replacement filter. Measurement errors were reported separately for fragment and amino acid residue masses. Supplementary Figure 10 outlines steps for identification of the UStags.
Conventional processing using SEQUEST search
Data were searched against the yeast sequence database using SEQUEST with a 5 u mass tolerance and fixed alkylation (on Cys), and the following previously suggested criteria for yeast proteome analysis:5 1) top hits with ΔCn ≥0.08 and Xcorr values ≥2.0, ≥1.5, and ≥3.3 for +1, +2, +3 charged fully tryptic peptides (having two termini ended with Arg/Lys prior to the cleavage points), respectively; and 2) top hits with ΔCn ≥0.08 and Xcorr values ≥3.0, ≥4.0 for +2, +3 charged partial tryptic peptides (having one terminus ended with Arg/Lys prior to the cleavage point), respectively. Only unique peptides, i.e., associated with a single protein, were counted and used for protein identification.
Supplementary Material
ACKNOWLEDGMENTS
Portions of this research were supported by the NIH NIAID (AGRY1-AI-4894) and NIH National Center for Research Resources (RR18522). Work was performed in the Environmental Molecular Science Laboratory, a U. S. Department of Energy (DOE) national scientific user facility located on the campus of Pacific Northwest National Laboratory (PNNL) in Richland, Washington. PNNL is a multiprogram national laboratory operated by Battelle Memorial Institute for the DOE under contract DE-AC05-76RLO-1830.
REFERENCES
- 1.Hunt EF, Yates JR, III, Shabanowitz J, Winston S, Hauer CR. Protein sequencing by tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 1986;84:620–623. doi: 10.1073/pnas.83.17.6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eng JK;, Mccormack AL, Yates JR. III an approach to correlate tandem mass spectral data of peptides with amino acid sequence in a protein database. J. Am. Soc. Mass spectrum. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 3.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification searching sequence database using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Craig R, Beavis RC. A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun. Mass Spectrom. 2003;17:2310–2316. doi: 10.1002/rcm.1198. [DOI] [PubMed] [Google Scholar]
- 5.Peng J, et al. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]; Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identification by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 6.Weatherly DB, et al. A heuristic method for assigning a false-discovery rate for protein identifications from Mascot database search results. Mol. Cell. Proteomics. 2005;4:762–772. doi: 10.1074/mcp.M400215-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Higgs RE, et al. Estimating the statistical significance of peptide identifications from shotgun proteomics experiments. J. Proteome Res. 2007;6:1758–1767. doi: 10.1021/pr0605320. [DOI] [PubMed] [Google Scholar]
- 8.Erisson J, Fenyö D. The statistical significance of protein identification results as a function of the number of proteins sequences searched. J. Proteome. Res. 2004;3:979–982. doi: 10.1021/pr0499343. [DOI] [PubMed] [Google Scholar]
- 9.Information for various modifications. http://www.unimod.org/ and http://www.abrf.org/index.cfm/dm.home.
- 10.More than 100,000 different species are typically detectable, for example for a tryptic digest of yeast lysate [Shen Y, et al. High-throughput proteomics using high-efficiency multiple-capillary liquid chromatography with on-line high-performance ESI FTICR mass spectrometry. Anal. Chem. 2001;73:3011–3021. doi: 10.1021/ac001393n.], using high resolution LC-high accuracy FTICR MS; however, yeast proteomics studies (ref. 5) limit the number of different peptides identified significantly less than 50,000; this situation becomes more significant in our human plasma analysis.
- 11.For SEQUEST™ algorithm, 5 modifications can be probed for each database search and Mascot allows 9 modifications in each search.
- 12.Savitsk MM, Nielsen ML, Zubarev RA. ModifiComb, a new proteomic tool for mapping substoichiometric post-translational modifications, finding novel types of modifications, and fingerprinting complex protein mixtures. Mol. Cell. Proteomics. 2006;5:935–948. doi: 10.1074/mcp.T500034-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Startkweather R, et al. Virtual polymorphism: finding divergent peptide matches in mass spectrometry data. Anal. Chem. 2007 doi: 10.1021/ac0703496. ASAP. DOI: 10.1021/ac0703496. [DOI] [PubMed] [Google Scholar]
- 14.Carr S, et al. The need for guidelines in publication of peptide and protein identification data: working group on publication guidelines for peptide and protein identification data. Mol. Cell. Proteomics. 2004;3:531–533. doi: 10.1074/mcp.T400006-MCP200. [DOI] [PubMed] [Google Scholar]; Taylor CF, et al. The minimum information about a proteomics experiment (MIAPE) doi: 10.1038/nbt1329. http://www.nature.com/nbt/consult/index.html. [DOI] [PubMed]; Mischak H, et al. Clinical proteomics: a need to define the filed and to begin to set adequate standards. Proteomics Clin. Appl. 2007;1:148–156. doi: 10.1002/prca.200600771. [DOI] [PubMed] [Google Scholar]
- 15.Frank AM, et al. De novo peptide sequencing and identification with precision mass spectrometry. J. Proteome Res. 2007;6:114–123. doi: 10.1021/pr060271u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Washburn MP, Wolters D, Yates JR. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 17.Huh W-K, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]; Ghaemmagham S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]; Gavin A-C, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:477–483. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]; Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:627–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 18.Yeast. ftp://genome-ftp.stanford.edu/pub/yeast/data_download/sequence/GenBank/ _2004-08- 27.
- 19.Petritis K, et al. Use of artificial neural networks for the accurate prediction of peptide liquid chromatography elution times in proteome analysis. Anal. Chem. 2003;75:1039–1048. doi: 10.1021/ac0205154. [DOI] [PubMed] [Google Scholar]
- 20.Boja ES, Fales HM. Overalkylation of a protein digest with lodoacetamide. Anal. Chem. 2001;73:3576–3582. doi: 10.1021/ac0103423. [DOI] [PubMed] [Google Scholar]
- 21. http://www.ionsource.com/
- 22.Miller DL, et al. Peptide composition of the cerebrovascular and senile plaque core amyloid deposits of Alzheimer’s disease. Arch. Biochem. Biophy. 1993;301:41–52. doi: 10.1006/abbi.1993.1112. [DOI] [PubMed] [Google Scholar]
- 23.Jones MD, et al. Peptide mass analysis of recombinant human granulocyte colony stimulating factor: elimination of methionine modification and nonspecific cleavages. Anal. Bioanal. 1994;216:135–146. doi: 10.1006/abio.1994.1017. [DOI] [PubMed] [Google Scholar]
- 24.Bochtler M, et al. The proteasome. Annu. Rev. Biophys. Biomol. Struct. 1999;28:295–317. doi: 10.1146/annurev.biophys.28.1.295. [DOI] [PubMed] [Google Scholar]; Huang W-P;, Klionsky DJ. Autophagy in yeast: a review of the molecular machinery. Cell. Struct. Funct. 2002;27:409–420. doi: 10.1247/csf.27.409. [DOI] [PubMed] [Google Scholar]
- 25.Mann M, Wilm M. Error-tolerance identification of peptides in sequence databases by peptide sequence tags. Anal. Chem. 1994;66:4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- 26.Sunyaev S, et al. MultiTag: Multiple error-tolerant sequence tag search for the sequence-similarity identification of proteins by mass spectrometry. Anal. Chem. 2003;75:1307–1315. doi: 10.1021/ac026199a. [DOI] [PubMed] [Google Scholar]
- 27.Tabb DL, Saraf A, Yates JR. GutenTag: High-throughput sequence tagging via an empirically derived fragmentation model. Anal. Chem. 2003;75:6415–6421. doi: 10.1021/ac0347462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frank A, et al. Peptide sequence tags for fast database search in mass spectrometry. J. Proteome Res. 2005;4:1287–1295. doi: 10.1021/pr050011x. [DOI] [PubMed] [Google Scholar]
- 29.Yates JR, et al. Performance of a linear ion trap-Orbitrap hybrid for peptide analysis. Anal. Chem. 2006;78:493–500. doi: 10.1021/ac0514624. [DOI] [PubMed] [Google Scholar]
- 30.Zubarev R, Mann M. On the proper use of mass accuracy in proteomics. Mol. Cell. Proteomics. 2007;6:377–381. doi: 10.1074/mcp.M600380-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Kelleher NL. Top-down proteomics. Anal. Chem. 2004;76:197A–203A. [PubMed] [Google Scholar]; Zabrouskov V, et al. New and automated MSn approaches for top-down identification of modified proteins. J. Am. Soc. Mass Spectrom. 2005;16:2027–2038. doi: 10.1016/j.jasms.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Patrie SM, et al. Top-down mass spectrometry of <60-kDa proteins from Methanosarcina acetivorans using quadrupole FTMS with automated octopole collisionally activated dissociation. Mol. Cell. Proteomics. 2006;5:14–25. doi: 10.1074/mcp.M500219-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Shen Y, et al. Automated 20 kpsi RPLC-MA and MS/MS with chromatographic peak capacities of 1000-1500 and capabilities in proteomics and metabolomics. Anal. Chem. 2005;77:3090–3100. doi: 10.1021/ac0483062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.