Abstract
This study presents a direct comparison of the ligand binding and signaling profiles of a mammalian and non-mammalian mu opioid receptor. Opioid ligand binding and agonist potencies were determined for an amphibian (Rana pipiens) mu opioid receptor (rpMOR) and the human mu opioid receptor (hMOR) in transfected, intact Chinese hamster ovary (CHO) cells. Identical conditions were employed such that statistically meaningful differences between the two receptors could be determined. Identifying these differences is an important first step in understanding how evolutionary changes affect ligand binding and signaling in vertebrate opioid receptors. As expected, the rank of opioid ligand affinity for rpMOR and hMOR were consistent with the ligands’ previously characterized type-selectivity. However, most of the opioid ligands tested had significant differences in affinity for rpMOR and hMOR. For example, the mu-selective agonist, DAMGO ([D-Ala2, N-Me-Phe4, Gly5-ol]-enkephalin), had a 10.9-fold greater affinity for hMOR (Ki = 268 nM) than rpMOR (Ki = 2,914 nM). In addition, differences in signaling between these receptors were found by measuring inhibition of cAMP accumulation by morphine or DAMGO. DAMGO was significantly more potent (13.6-fold) in CHO cells expressing hMOR versus those expressing rpMOR. In addition, a significantly greater maximal inhibition was elicited by both opioid agonists in cells expressing hMOR. In summary, this study supports an ongoing effort to better understand how vertebrate evolution has shaped opioid receptor properties and function.
Keywords: opioid, mu opioid receptor, amphibian, ligand binding, cAMP inhibition
1. Introduction
Opioid analgesics elicit their effects through activation of one or more types (mu, delta, and kappa) of opioid receptor. These receptors belong to the Type A rhodopsin-like family of G protein-coupled receptors and exhibit a typical serpentine, seven-transmembrane topology. Several opioid receptor types have been confirmed by behavioral studies (Martin et al., 1976), radioligand binding (Gillan et al., 1980; Lord et al., 1977), and molecular cloning and characterization in cell lines (Kieffer et al., 1992; Evans et al., 1992; Chen et al. 1993; Wang et al., 1993; Yasuda et al. 1993). Initially, this research was exclusively conducted in mammals, but other studies exploring opioid receptors in non-mammals soon followed.
Early work in our lab demonstrated that opioid receptors mediate antinociceptive effects in non-mammalian species. Specifically, the antinociceptive potency of mu-, delta-, and kappa- selective opioid agonists after systemic (Stevens et al., 1994), intraspinal (Stevens, 1996), or intracerebroventricular (Stevens and Rothe, 1997) administration in amphibians was highly correlated to that observed in mammals and to the relative potency of opioid analgesics in human clinical studies. These results, and other studies over the last two decades, validated the amphibian model as an alternative or adjunct model for pain and analgesia research (Stevens, 2008). However, behavioral studies in Rana pipiens suggested that the selectivity of opioid ligands for the different types of opioid receptors was different in mammalian and non-mammalian species. The mu-, delta-, and kappa- selective opioid antagonists, β-FNA (β-funaltrexamine), naltrindole, and nor-BNI (nor-binaltorphimine), did not show type-selectivity in blocking the antinociceptive effects of selective opioid agonists in amphibians (Stevens and Newman, 1999).
To assess whether distinct opioid receptor types were present in Rana pipiens, radioligand binding studies were conducted in whole brain. Binding analysis of the highly selective opioid agonists, DAMGO (mu-selective; [D-Ala2, N-Me-Phe4, Gly5-ol]-enkephalin), DPDPE (delta-selective; [D-Pen2,5]-enkephalin), and U-69593 (kappa-selective; (+)-(5α,7α,8β)-N-Methyl-N-[7-(1-pyrrolidinyl)-1-oxaspiro[4.5]dec-8-yl]-benzeneacetamide) showed that each bound with high-affinity to a distinct site, implying the possibility of three opioid receptor types (Newman et al., 2002). This finding was supported by previous studies of opioid receptor binding in frog brain which identified multiple opioid binding sites (Benyhe et al., 1990, 1999).
To extend these studies further, we used reverse transcription PCR of frog brain and spinal cord cDNA to isolate distinct opioid receptor sequences which were all fully cloned using rapid amplification of cDNA ends (Stevens et al., 2007). The novel clones were designated rpMOR (Rana pipiens mu opioid receptor), rpDOR (R. pipiens delta opioid receptor), rpKOR (R. pipiens kappa opioid receptor), and rpORL (R. pipiens nociceptin receptor) based on displaying sequence homology with their mammalian counterparts. Besides the frog, several other full-length opioid receptor clones have been identified in non-mammalian species. The earliest example was a mu-like opioid receptor cloned from brain tissue of the white suckerfish (Darlison et al., 1997). Opioid receptors from other non-mammals have included the zebrafish (Barrallo et al., 1998, 2000; Alvarez et al., 2006), and the rough-skinned newt (Bradford et al., 2005, 2006).
In the present study, we characterized the frog mu opioid receptor, rpMOR, since this receptor type is the principal mediator of analgesic drugs in the CNS. In investigating binding and activity of rpMOR, we chose an experimental design to accomplish two goals. First, all studies with rpMOR were done in parallel with its human ortholog, hMOR. Because laboratories often differ in the assay conditions they employ, making statistically meaningful comparisons of receptors from different species is not possible. Thus, the present study characterizes both receptors under identical conditions. Secondly, the reported affinities and potencies of opioid agonists may not be reflective of in vivo values if assay conditions are not physiological. Consequently, the present study characterizes both receptors in intact Chinese hamster ovary (CHO) cells using a physiological buffer. Here, we show for the first time statistically significant differences in the binding and activity of a non-mammalian and mammalian mu opioid receptor.
2. Materials and Methods
2.1 Cell culture and receptor transient transfection
Chinese hamster ovary cells (CHO; American Tissue Bank, McClean, VA) were grown in F-12K growth media (ATCC, Manassas, VA) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 mg/ml streptomycin (Invitrogen, Carlsbad, CA). For transient transfection, CHO cells between passages 5−30 were trypsinized with 0.25% trypsin/0.53mM EDTA (ATCC, Manassas, VA) and plated at 1.50 × 105 cells in 500 μl/well F-12K growth media/10% fetal bovine serum without antibiotics (24-well format). The following day, 1 μg of plasmid DNA/well (pcDNA3.1-rpMOR or –hMOR) was diluted in 50μl/well of Opti-MEM media (Invitrogen, Carlsbad, CA) and incubated at room temperature for 5 min. During the same period, 2 μl/well of Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) was diluted in 50 μl/well of Opti-MEM media (Invitrogen, Carlsbad, CA). The plasmid DNA and the lipofectamine were mixed and incubated at room temperature for 20−30 min. 100 μl of the DNA-lipofectamine complex were added to each well and 24-well plates (Corning, Corning, NY) were mixed by gentle rocking. Five h later, the media was replaced with 50 μl/well F-12K growth media/10% fetal bovine serum without antibiotics. Transfected cells were incubated at 37° C in a 5% CO2 humidified incubator for 24 h before use in binding assays. pcDNA3.1-hMOR was obtained from University of Missouri-Rolla cDNA Resource Center. pcDNA3.1-rpMOR was previously cloned in our lab (GenBank Accession No. AAQ09991).
2.2 Saturation binding assays in whole CHO cells
One day following transfection with the expression plasmid, cells were briefly washed with PBS (Hyclone, Logan, UT). Wells contained a binding volume of 500 μl ice-cold Hank's balanced salts (Invitrogen, Carlsbad, CA) with increasing concentrations (half-log units) of [3H]-naloxone (67 Ci/mmol; Perkin Elmer, Wellesley, MA). 10 μM cold naloxone (Sigma-Aldrich, USA) was used to determine nonspecific binding. Cells were incubated at 4° C for 24 h for convenience and to ensure ligand binding had reached equilibrium. Following the binding reaction, the cells were washed rapidly on ice with 2 × 500 μl/well ice-cold PBS (each well in succession). Cells were lysed with 500 μl/well 0.25 N NaOH and incubated for 30 min at room temperature. Lysates were neutralized with 70 μl/well 2.5 N HCl then removed from the wells and added to 5 ml ScintiVerse (Fisher Scientific, USA) for liquid scintillation counting. Each concentration of radioligand used for specific and nonspecific binding was done in triplicate in 3−4 independent experiments for each mu opioid receptor protein. The total protein in a well was determined using the bicinchoninic acid assay (Pierce, Rockford, IL) as modified for intact cells (Goldschmidt and Kimelberg, 1989).
2.3 Competition binding assays in whole CHO cells
For competition binding in whole cells, we selected [3H]-naloxone as a radioligand due to its lack of membrane permeability under the assay conditions we employed. To verify this, the membrane impermeable, quaternary analog, naloxone methiodide was used to displace [3H]-naloxone binding. More than 94% of the radioligand binding was displaced, and the remaining, non-displaced radioligand was consistent with the range of nonspecific binding observed with several of the ligands employed in this study (data not shown). Following this assessment of radioligand permeability, the selective opioid ligands, DAMGO (mu), morphine (mu), DPDPE (delta), U-50488 (kappa; 3,4-Dichloro-N-methyl-N-[2-(1-pyrrolidinyl)cyclohexyl]-benzeneacetamide), β-funaltrexamine (mu), naltrindole (delta), and nor-binaltorphimine (kappa), were used to displace [3H]-naloxone binding in whole CHO cells transiently expressing human or amphibian mu opioid receptor proteins (all opioid ligands from Sigma-Aldrich, USA). One day following transfection, cells were briefly washed with 2 × 500 μl/well PBS. Wells contained a binding volume of 500 μl ice-cold Hank's balanced salts with a type-selective opioid ligand and 1 nM [3H]-naloxone. A range (in half- or one-third-log units) of concentrations of cold competitor was used such that competition curves had a clear plateau on both ends. Cells were incubated at 4° C for 24 h for convenience and to ensure ligand binding had reached equilibrium. Following the binding reaction, the cells were washed rapidly on ice with 2 × 500 μl/well ice-cold PBS (each well in succession). Cells were lysed with 500 μl/well 0.25 N NaOH and incubated for 30 min at room temperature. Lysates were neutralized with 70 μl/well 2.5 N HCl then removed and added to 5 ml ScintiVerse for liquid scintillation counting (Griffin et al., 2003). Each concentration of radioligand with cold competitor was done in triplicate in 3−4 independent experiments for each mu opioid receptor protein.
2.4 Creation of stable CHO cell lines
The rpMOR and hMOR inserts were cloned using standard techniques into the pIRESneo polycistronic expression vector (Clontech, Mountain View, CA). CHO cells were plated at a concentration of 6.0 × 105 cells in a 35 mm dish containing 2 ml of F-12K growth media/10% fetal bovine serum. The next day, 4 μg of pIRESneo -rpMOR or –hMOR was diluted in 250 μl of Opti-MEM media and incubated at room temperature for 5 minutes. 10 μl of lipofectamine 2000 was diluted in 250 μl of Opti-MEM media, and the DNA and the lipofectamine were mixed and incubated at room temperature for 20 min. The DNA-lipofectamine complex was then added to the dish and mixed gently by rocking. After 5 h, the media was aspirated and replaced with 2 ml of F-12K growth media/10% fetal bovine serum. The next day, the cells were trypsinized with 0.25% trypsin/0.53mM EDTA and plated at a 1/5,000 dilution into 4 × 10 cm plates each containing 15 ml of complete growth media (F-12K and serum with 100 U/ml penicillin and 100 mg/ml streptomycin). The following day, 600 or 800 μg/ml geneticin (Invitrogen, Carlsbad, CA) was added (for rpMOR and hMOR, respectively) to the 10 cm plates. Complete growth media containing geneticin was changed every 2 days. Approximately 8 − 10 days later, 6 mm (outer diameter) cloning rings were used to isolate and trypsinize (20 μl trypsin/ring) colonies, which were then transferred to individual wells (in a 24-well plate) containing 500 μl of complete growth media with geneticin. After several days of growth, colonies were trypsinized and transferred to new wells for maintenance and determination of mu opioid receptor protein expression by intact, whole-cell [3H]-naloxone binding.
2.5 Inhibition of adenylyl cyclase assay
On the day the before the assay, stable rpMOR- or hMOR-CHO cells between passages 14−22 were plated in a 24-well format at 1.50 × 105 cells in 500 μl/well F-12K growth media/10% fetal bovine serum/100 U/ml penicillin/100 mg/ml streptomycin/600−800 μg/ml geneticin. On the day of the experiment, inhibition of adenylyl cyclase assays were conducted as previously described (Griffin et al., 2003). Briefly, cells were washed with 2 × 500 μl/well serum-free F-12K growth media then incubated in 300 μl/well serum-free F-12K media containing 2 μCi [3H]-adenine (27.2 Ci/mmol; Perkin Elmer, Wellesley, MA) and 30 μM adenine (Sigma-Aldrich, USA) for 2 hrs at 37° C in a 5% CO2 humidified incubator. Cells were washed with 2 × 500 μl/well serum-free F-12K media and then pre-incubated with 500 μl/well serum-free F-12K media containing 500 μM IBMX (Sigma-Aldrich, USA) for 12 min at 37° C in a 5% CO2 humidified incubator. The reaction was started by replacing the media with 500 μl/well serum-free F-12K media containing 500 μM IBMX, 10 μM forskolin (Calbiochem, San Diego, CA), and a range of concentrations of morphine or DAMGO and incubating the cells for 12 min at 37° C in a humidified 5% CO2 incubator. The media was then aspirated and replaced with 1 ml/well of ice-cold 9% TCA (Sigma-Aldrich, USA), and the cells were incubated on ice for 30 min. The extracts were then applied to a 1.5 ml Dowex column (AG-50W-X4, 200−400 mesh; Biorad, Hercules, CA) and washed with 2 × 1.5 ml ddiH2O. [3H]cAMP was eluted onto a column of 0.6 g neutral alumina (Sigma-Aldrich, USA) with 5 ml ddiH2O and then eluted into 20 ml scintillation vials with 4 ml 0.1 M imidazole-HCl pH 7.5 (Sigma-Aldrich, USA). 18 ml of Scintiverse were added to each of the vials which were then counted for 5 min.
2.6 Data and statistical analysis
Minimum experiments per plot was N=3 done in triplicate. Opioid ligand equilibrium dissociation constants (Kd), inhibition constants (Ki), and receptor density (Bmax), together with their Hill slopes, were predicted by nonlinear regression curve-fit to a sigmoidal dose-response (variable slope) model using GraphPad Prism software (v. 4.0; Irvine, CA). Calculation of the Ki from competition binding data was derived using the Cheng-Prusoff equation (Cheng and Prusoff, 1973). Opioid agonist potency (IC50) and maximal inhibition (Imax) were predicted by nonlinear regression curve-fit to a one-site competition model using GraphPad Prism software. The mean specific binding from individual experiments was averaged together and plotted in one curve such that error was represented as S.E.M. Differences in values between rpMOR and hMOR were considered statistically significant (P<.05) as determined by a Student's unpaired, two-tailed t-test.
3. Results
3.1 [3H]-Naloxone saturation in rpMOR- and hMOR-transfected CHO cells
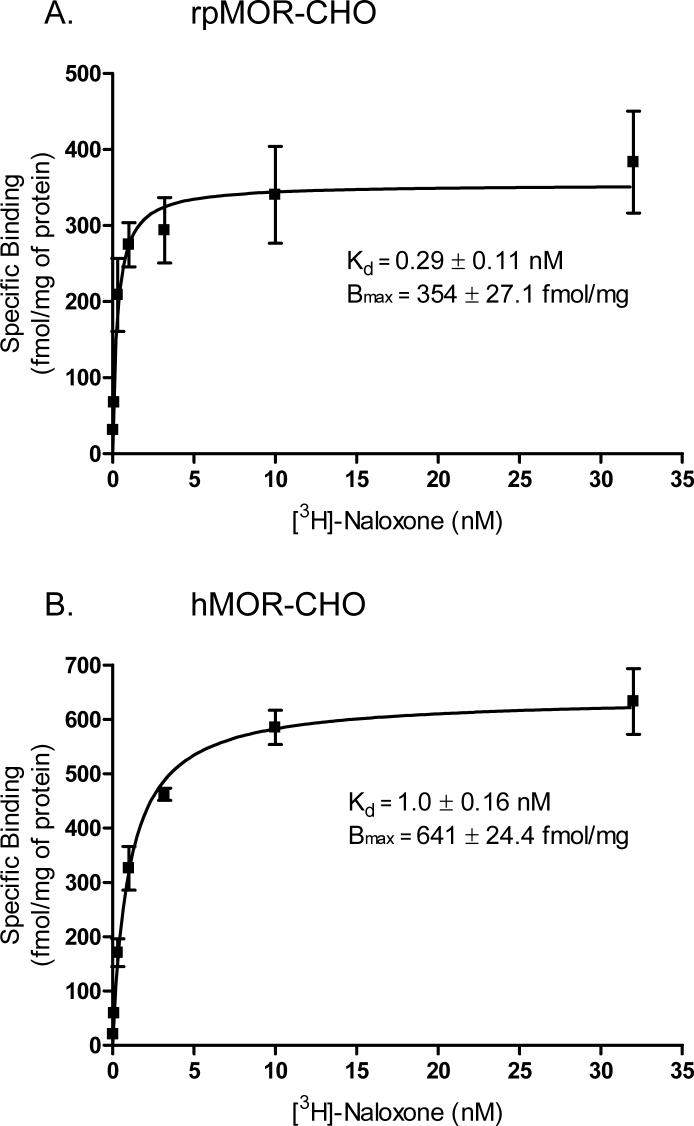
To determine the expression of rpMOR and hMOR in intact, transfected CHO cells, saturation binding assays were performed in parallel for both receptors using increasing, log-spaced amounts of [3H]-naloxone (Fig. 1). In rpMOR-transfected CHOs, the maximum receptor density (Bmax) was 354 ± 27.1 fmol/mg of protein, while the Bmax for hMOR-transfected CHOs was 641 ± 24.4 fmol/mg of protein. Although rpMOR was sufficiently expressed for determining its pharmacological properties, its Bmax was significantly lower (almost 2-fold) than that for hMOR. In addition to differences in receptor density, the equilibrium dissociation constant (Kd) also differed significantly for the two receptors. [3H]-naloxone bound with high affinity to both receptors, but the affinity was 3-fold greater for rpMOR (Kd = 0.29 ± 0.11 nM) versus hMOR (Kd = 1.0 ± 0.16 nM).
Fig. 1.
[3H]-Naloxone saturation binding in intact, whole CHO cells transiently expressing rpMOR (A) or hMOR (B). In rpMOR-CHO, [3H]-naloxone bound to a single site with a Kd of 0.29 nM and a Bmax of 354 fmol/mg of protein. In hMOR-CHO, [3H]-naloxone bound to a single site with a Kd of 1.0 nM and a Bmax of 641 fmol/mg of protein. Specific binding for each concentration of [3H]-naloxone was determined in triplicate in 3−4 independent experiments (error bars indicate S.E.M.).
3.2 [3H]-Naloxone displacement by type-selective opioid agonists in rpMOR- and hMOR-transfected CHO cells
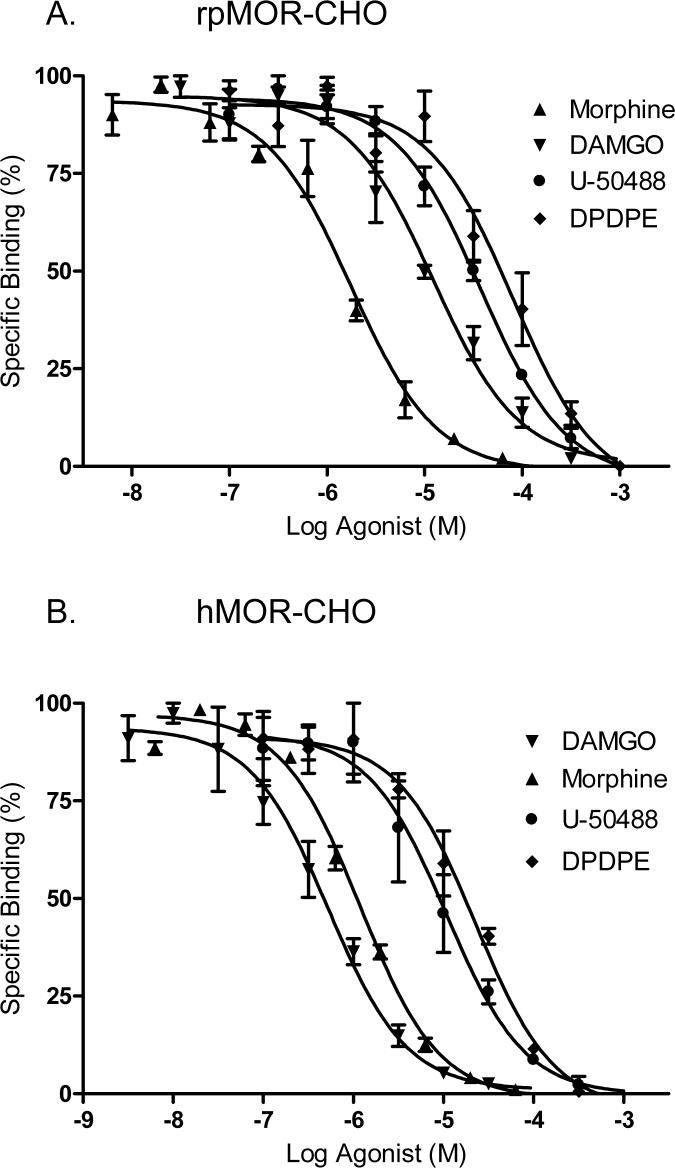
In order to characterize rpMOR and hMOR type-selectivity, parallel experiments were conducted where 1 nM [3H]-naloxone was displaced by type-selective opioid agonists in intact, transfected CHO cells. As predicted from its high amino acid sequence identity (∼80%) with mammalian mu opioid receptors, rpMOR displayed a mu receptor binding profile with the following rank order of agonist affinities: morphine (mu-selective) > DAMGO (mu-selective) > U-50488 (kappa-selective) > DPDPE (delta-selective; Fig. 2A). Inhibition constants (Ki) are reported for each agonist in Table 1. As expected, [3H]-naloxone displacement in hMOR-transfected CHOs also followed a mu receptor binding profile (Fig. 2B); however, the rank order of agonist affinities for hMOR was slightly different than that for rpMOR: DAMGO > morphine > U-50488 ≥ DPDPE. Notably, the affinities of morphine and DAMGO are significantly different for both receptors. Morphine affinity for rpMOR is 1.6-fold greater than its affinity for hMOR, and DAMGO affinity for hMOR is 10.9-fold greater than that determined for rpMOR.
Fig. 2.
[3H]-Naloxone displacement by multiple opioid agonists in intact, whole CHO cells transiently expressing rpMOR (A) or hMOR (B). Agonists displaced [3H]-naloxone and bound to a single site. The Ki and Hill slope for each is shown in Table 1. Percent specific binding of [3H]-naloxone at each concentration of competitor was determined in triplicate in 3−4 independent experiments (error bars indicate S.E.M.).
Table 1.
Affinities of opioid competitors in rpMOR- and hMOR-transfected CHO cells. The Ki for each competitor is reported in nM, and, for each competition curve, Hill slopes were determined using the sigmoidal dose-response (variable slope) curve-fitting program, GraphPad Prism (v.4.0).
| rpMOR | Ki (nM) | S.E.M. (nM) | Hill Slope | S.E.M. (nM) | hMOR | Ki (nM) | S.E.M. (nM) | Hill Slope | S.E.M. (nM) | Ratioa |
|---|---|---|---|---|---|---|---|---|---|---|
| Agonists | Agonists | |||||||||
| Morphine | 390 | ± 67.0 | −1.20 | ± 0.18 | Morphine | 611 | ± 48.0 | −1.12 | ± 0.09 | 1.6# |
| DAMGO | 2,914 | ± 615 | −0.85 | ± 0.13 | DAMGO | 268 | ± 65.0 | −0.85 | ± 0.15 | 0.1# |
| DPDPE | 19,130 | ± 8,242 | −0.93 | ± 0.24 | DPDPE | 11,690 | ± 2,886 | −0.97 | ± 0.20 | 0.6 |
| U-50488 | 8,421 | ± 1,509 | −1.00 | ± 0.15 | U-50488 | 5,561 | ± 1,985 | −0.97 | ± 0.28 | 0.7 |
| Antagonists | Antagonists | |||||||||
| Naloxone | 0.25 | ± 0.06 | −1.19 | ± 0.13 | Naloxone | 0.76 | ± 0.21 | −1.40 | ± 0.15 | 3.0# |
| β-FNA | 0.63 | ± 0.15 | −0.67 | ± 0.11 | β-FNA | 1.85 | ± 0.15 | −1.01 | ± 0.09 | 2.9# |
| Naltrindole | 2.96 | ± 0.56 | −0.96 | ± 0.15 | Naltrindole | 6.10 | ± 1.00 | −0.99 | ± 0.14 | 2.1 |
| nor-BNI | 14.4 | ± 3.35 | −1.02 | ± 0.20 | nor-BNI | 67.4 | ± 19.6 | −0.99 | ± 0.23 | 4.7# |
The ratio of the competitor Ki hMOR-transfected CHOs over that in rpMOR-transfected CHOs was determined
annotated where p<0.05 (unpaired, two-tailed Student's t-test).
3.3 [3H]-Naloxone displacement by type-selective opioid antagonists in rpMOR- and hMOR-transfected CHO cells
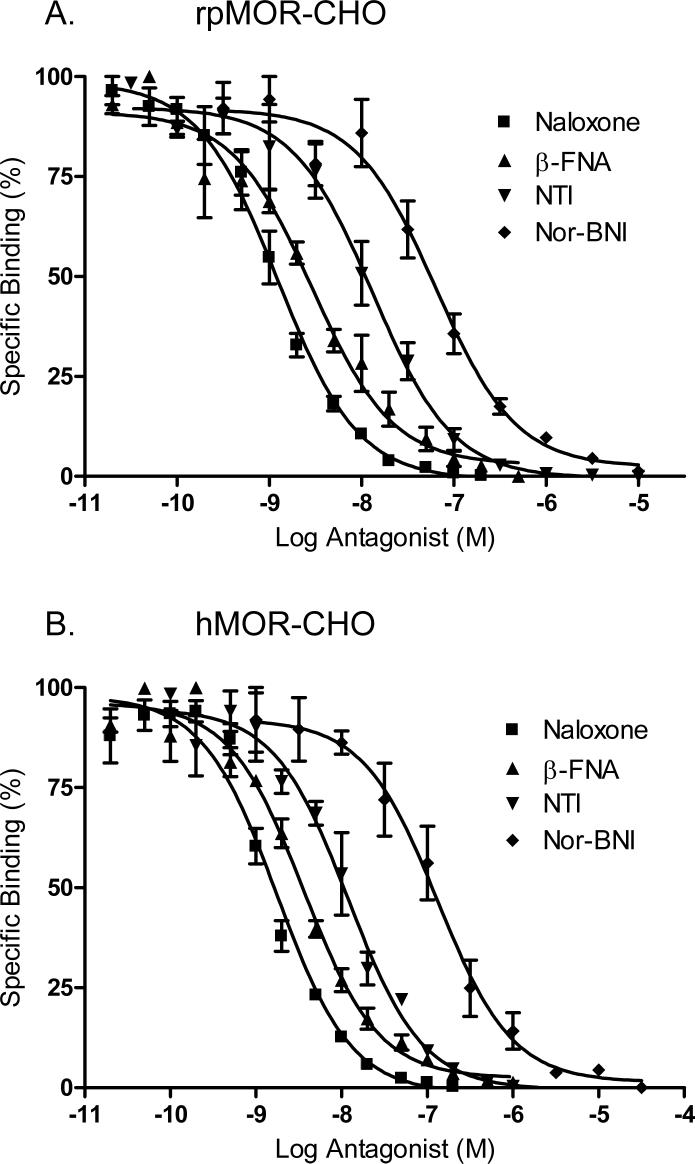
To further characterize the selectivity of rpMOR and hMOR, type-selective opioid antagonists were used in parallel experiments to displace 1 nM [3H]-naloxone in transfected CHO cells. The Ki for each antagonist tested in shown in Table 1. Similar to the agonist data, rpMOR again displayed a mu receptor binding profile with the following rank order of antagonist affinities: naloxone (non-selective) > β-FNA (mu-selective) > NTI (delta-selective) > nor-BNI (kappa-selective; Fig. 3A). In hMOR-transfected CHOs, the rank order of affinities of the selective antagonists was the same as that for rpMOR (Fig. 3B). When comparing the opioid antagonist affinities for the two receptors, all with the exception of NTI had a significantly greater affinity (ranging from 2.9- to 4.7-fold) for rpMOR than hMOR.
Fig. 3.
[3H]-Naloxone displacement by multiple opioid antagonists in intact, whole CHO cells transiently expressing rpMOR (A) or hMOR (B). Antagonists displaced [3H]-naloxone and bound to a single site. The Ki and Hill slope for each is shown in Table 1. Percent specific binding of [3H]-naloxone at each concentration of competitor was determined in triplicate in 3−4 independent experiments (error bars indicate S.E.M.).
3.5 Inhibition of forskolin-stimulated cAMP accumulation in stable rpMOR- and hMOR-CHO cells
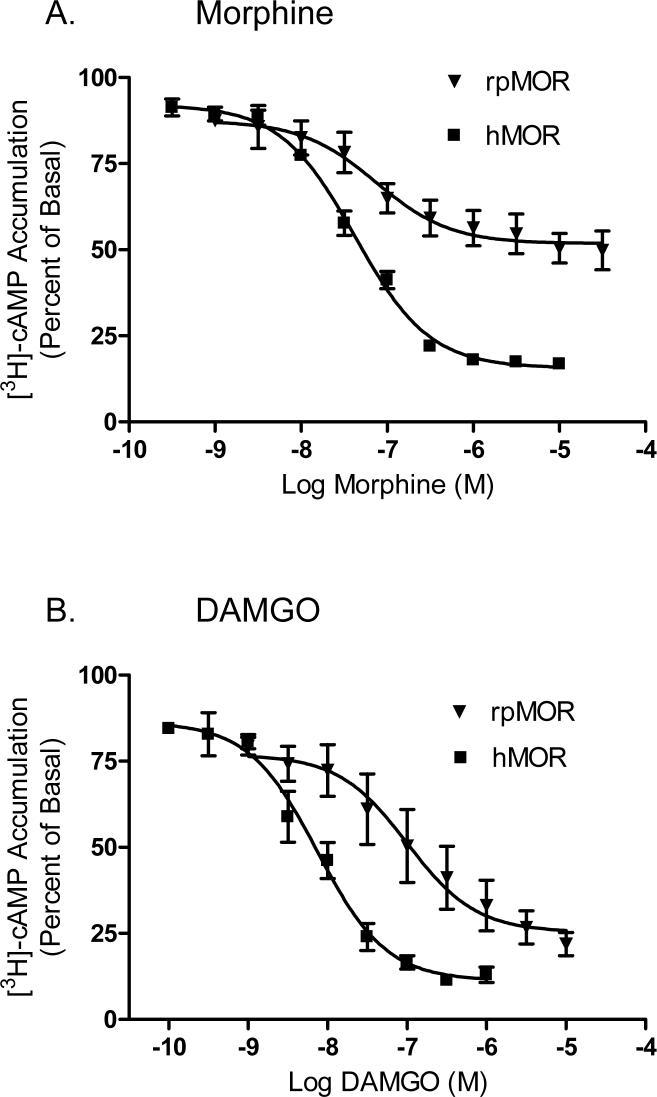
In order to demonstrate the functionality of rpMOR, we measured the ability of morphine and DAMGO to inhibit forskolin-simulated cAMP accumulation. In addition, hMOR activity was determined so that our species comparative model could be extended to receptor function. Because both opioid agonists were unable to inhibit cAMP accumulation in rpMOR and hMOR transiently-transfected CHO cells, we chose to construct stable CHO cell lines. Several stable clones for both receptors were obtained and matched for expression. For the clones used in the assays, the rpMOR stable clone had a Bmax of 980 ± 26.8 fmol/mg of protein while the Bmax for the hMOR stable clone was 1,167 ± 28.2 fmol/mg of protein (data not shown). Both receptors were activated by the opioid agonists, but the potencies and maximal inhibition were different. With regard to potency, DAMGO had significantly greater potency (13.6-fold) in CHO cells expressing hMOR (Fig. 4B) than those expressing rpMOR (Fig.4A). Individual values for both receptors are shown in Table 2. With respect to maximal inhibition, a significantly greater inhibitory effect for both opioid agonists was observed in hMOR- versus rpMOR-expressing cells. The Imax for morphine was 1.8-fold greater in hMOR versus rpMOR-expressing cells, while the Imax for DAMGO was 1.2-fold greater in hMOR versus rpMOR-expressing cells. Interestingly, both agonists had a similar Imax value in hMOR-expressing cells; but in rpMOR-expressing cells, morphine had a significantly lower Imax (1.5-fold) than that elicited by DAMGO.
Fig. 4.
Inhibition of [3H]cAMP accumulation by morphine (A) or DAMGO (B) in CHO cells stably expressing rpMOR or hMOR. A range of opioid agonist concentrations were used in order to determine the concentration for 50% inhibition (IC50) and maximal inhibition (Imax). Individual values are reported in Table 2. [3H]-cAMP accumulation is plotted as percent versus that in cells treated with 10 μM forskolin only. For each condition, 3−5 independent experiments were performed in triplicate (error bars indicate S.E.M.).
Table 2.
Potency and maximal effect data for cAMP inhibition experiments. For each condition of rpMOR or hMOR treatment with morphine or DAMGO, the maximum percent inhibition (Imax) is shown as the greatest percent of inhibition of [3H]-cAMP accumulation elicited by opioid agonist in forskolin-stimulated cells. In addition, the potency of inhibition (IC50) is noted for each condition. Both parameters were predicted by a best-fit to a one-site competition curve using GraphPad Prism software (v. 4.0).
| rpMOR | IC50 (nM) | S.E.M. (nM) | Imax (%) | S.E.M. (nM) | hMOR | IC50 (nM) | S.E.M. (nM) | Imax (%) | S.E.M. (nM) | IC50 Ratioa | Imax Ratiob |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Agonists | Agonists | ||||||||||
| Morphine | 73.9 | ± 49.0 | 48.2 | ± 2.49 | Morphine | 42.1 | ± 4.26 | 84.4 | ± 1.13 | 0.6 | 0.3# |
| DAMGO | 101 | ± 65.2 | 74.7 | ± 4.30 | DAMGO | 7.42 | ± 1.67 | 88.8 | ± 2.37 | 0.07# | 0.4# |
The ratios of the IC50a and Imaxb values for rpMOR over those for hMOR were determined and #annotated where p<0.05 (unpaired, two-tailed Student's t-test).
4. Discussion
This study, for the first time, presents a direct comparison of the ligand binding and signaling profiles of a non-mammalian and a mammalian mu opioid receptor. On an evolutionary timescale, the genes for these receptors are separated by about 360 million years (Kumar and Hedges, 1998). Over this period, several major vertebrate classes have arisen including, Amphibia, Reptilia, Aves, and Mammalia. The present study adopts a comparative evolutionary approach in order to provide an initial characterization of how changes in these receptors over millions of years have resulted in significant differences in their opioid ligand binding and signaling properties.
Both the amphibian and human mu opioid receptors (rpMOR and hMOR, respectively) were investigated in parallel, and, as a result, we were able to demonstrate significant differences in the pharmacology and signaling activity of these receptors. Moreover, in the binding assays, a physiological approach was adopted whereby whole CHO cells and a physiological binding buffer were employed. This allowed us to determine opioid ligand affinity and type-selectivity under conditions that would better reflect values as they would occur under in vivo conditions. Consequently, these data suggested that physiological conditions can exert a large impact on opioid binding.
Not surprisingly, the competition binding experiments revealed binding profiles consistent with a mu-type opioid receptor for both rpMOR and hMOR. The rank order of agonist and antagonist affinities indicated that the mu-selective ligands had the greatest affinities for both receptor clones (Figs. 1 and 2, Table 1). On the other hand, several significant differences were observed in opioid binding. Most notable, the mu-selective agonist, DAMGO, displayed a significantly lower affinity (10.9-fold) for rpMOR versus hMOR. Several studies using opioid receptor chimeras and site-directed mutagenesis have found individual amino acid sites important for DAMGO binding in mammalian mu opioid receptors. In particular, sites in the first and third extracellular loop domains were shown to discriminate DAMGO binding between the mu opioid receptor and the delta and kappa opioid receptors, respectively (Fukuda, et al., 1995; Minami, et al., 1996; Wang, et al., 2000; Xue, et al., 1995; Seki, et al., 1998). A comparison of these amino acid residues in rpMOR and hMOR revealed that all of the previously identified sites were conserved between the two receptors. However, 17 total amino acids in the extracellular loops of rpMOR and hMOR are different (Fig. 5) and may account for the differences observed with DAMGO affinity. Further study will be needed to ascertain the relative importance of individual sites in DAMGO binding.
Fig. 5.

ClustalW alignment of rpMOR and hMOR amino acid sequences. Alignment was done using default values (Mega v. 4.0, www.megasoftware.net). Identical sites are noted by white text with black background, conservative substitutions are noted by grey background, and different sites are boxed on the rpMOR sequence. .
The observation that DAMGO displays reduced binding for rpMOR is consistent with previous reports characterizing the pharmacology of cloned, non-mammalian mu opioid receptors. Studies in both the white suckerfish (Darlison, et al., 1997) and the rough-skinned newt (Bradford, et al., 2006) reported DAMGO affinities that were one to two orders of magnitude lower than those typically observed for mammalian mu opioid receptors (Wang, et al., 1993; Raynor, et al., 1994; Raynor, et al., 1995). The binding data for non-mammalian mu opioid receptors may not be not be entirely unanticipated since DAMGO is a synthetic, enkephalin derivative developed for its high affinity and specificity for mammalian mu opioid receptors (Handa et al., 1981). In addition, the degree of amino acid sequence divergence between mammalian and non-mammalian opioid receptors may play a role. Non-mammalian opioid receptor types, on the whole, have greater protein identity than their mammalian counterparts (Stevens et al., 2007). Consequently, some highly selective opioid ligands, such as DAMGO, might be expected to show a reduced capacity to discriminate between receptor types with fewer amino acid sequence differences.
Besides the difference observed in DAMGO binding between rpMOR and hMOR, significant differences in binding were also observed with the opioid agonist, morphine, as well as all of the opioid antagonists tested. In contrast to DAMGO binding, the fold-differences in the affinities of these opioid ligands were more modest and ranged from 1.6-fold for morphine to between 2.1- to 4.7-fold for the opioid antagonists (Figs. 1 and 2, Table 1). Similar to the studies localizing important binding determinants for DAMGO, several groups have also identified binding sites important for morphine and all of the opioid antagonists employed in this study (Surratt, et al., 1994; Chen, et al., 1996; Mansour, et al., 1997; Xu, et al., 1999). In these studies, amino acid residues residing in or adjacent to the transmembrane domains of mammalian mu opioid receptors were investigated. However, as was the case with the previously identified DAMGO binding determinants, all the amino acid sites implicated in these studies were found to be conserved between rpMOR and hMOR.
In addition to the extracellular loops and transmembrane domains, other mu opioid receptor domains have also been implicated in opioid binding. For instance, the N-terminal domains of rpMOR and hMOR are highly divergent (Stevens et al., 2007), and at least one study has demonstrated the importance of this receptor domain in ligand-binding to the mu opioid receptor (Chaturvedi, et al., 2000). A 64 amino acid N-terminal domain deletion resulted in decreased binding for several opioid ligands, including naloxone, morphine, and DAMGO. Hence, future studies exploring the opioid binding determinants responsible for the differences observed between rpMOR and hMOR should not be limited to the extracellular loops and transmembrane domains.
As mentioned above, the current binding studies were also conducted using physiological conditions. As a result, the values for opioid ligand affinity and type-selectivity reported in this study should better reflect values as that might occur in an in vivo setting. Specifically, a whole-cell model and a physiological buffer were employed since these variables have been previously shown to influence opioid binding. For example, the presence of guanyl nucleotides has been shown to reduce opioid agonist binding in brain and neuroblastoma cell membranes (Blume, 1978; Werling, et al., 1988). In the current study, guanyl nucleotides were preserved since the binding assays utilized an intact, whole-cell preparation. In addition, several studies have also demonstrated that cations, such as sodium, regulate opioid ligand binding in brain membranes (Pert and Snyder, 1974; Paterson, et al., 1986; Werling, et al., 1986). Consequently, a binding buffer containing physiological concentrations of cations was used in the present study. These conditions were in contrast to most opioid binding studies which employ broken cell membranes and binding buffers low in or devoid of sodium ions. A comparison of the hMOR binding data in the current study with a previous study characterizing opioid binding in CHO cells expressing hMOR (Toll, et al., 1998) illustrated the large degree to which physiological conditions can impact opioid binding (Table 3). Not only were opioid ligand affinities generally lower in the current study, but a comparison of the degree of type selectivity of the mu-selective agonist, DAMGO, and the mu-selective antagonist, β-FNA, revealed that these opioid ligands also displayed lower selectivity for hMOR in the present study. Although future studies will be necessary to make a direct statistical comparison of the effects of physiological versus non-physiological conditions on opioid binding, the comparison with the Toll et al. study suggests that opioid ligand affinity and type-selectivity may be substantially reduced under physiological conditions.
Table 3.
Comparison of type-selectivity of opioid ligands for hMOR in present study with previously reported type-selectivities for hMOR in Toll et al. 1998 study. The degree of type-selectivity was determined by calculating the ratios of opioid Ki values for delta- vs. mu-selective and kappa- vs. mu-selective ligands.
| Type-Selective Opioids | Present Studya | Toll et al. 1998b | ||
|---|---|---|---|---|
| Ki (nM) | Ratio | Ki (nM) | Ratio | |
| Agonists | ||||
| DPDPE/DAMGO (delta/mu) | 11,690/268 | 44 | 504/0.5 | 1,008 |
| U-50,488/DAMGO (kappa/mu) | 5,561/268 | 21 | 290/0.5 | 580 |
| Antagonists | ||||
| NTI/β-FNA (delta/mu) | 6.10/1.85 | 3.2 | 6.3/0.3 | 21 |
| Nor-BNI/β-FNA (kappa/mu) | 67.4/1.85 | 35 | 21/0.3 | 70 |
Opioid ligand binding was conducted in intact cells in Hank's balanced salts buffer at pH 7.4
Opioid ligand binding was conducted using cell membrane homogenates in 50mM Tris-HCl pH 7.5.
Besides the comparison of rpMOR and hMOR opioid binding, receptor function was also compared by conducting inhibition of adenylyl cyclase assays. In general, hMOR signaling was more robust than that of rpMOR. Notably, DAMGO displayed a 13.6-fold greater potency in CHO cells expressing hMOR (Fig. 4, Table 2). This likely resulted, at least in part, from the significantly reduced binding DAMGO has for rpMOR. In addition to the potency data, both morphine and DAMGO were significantly more efficacious in cells expressing hMOR, although differences were more modest (1.8- and 1.2-fold, respectively). Like opioid binding, several receptor domains and specific amino acids have been identified which are critical for opioid receptor signaling. Synthetic peptides representing portions of the second and third intracellular loops have been used to demonstrate the importance of these domains for opioid receptor activation (Merkouris et al., 1996; Georgoussi et al., 1997). Both regions are identical in rpMOR and hMOR except for one amino acid (Met in rpMOR, Leu in hMOR) that differs at the very proximal end of the third intracellular loop (Fig. 5). In addition to the second and third intracellular loops, the C-terminal domain of the mu opioid receptor has also been shown to be important for signaling. One study demonstrated that truncation of the C-terminal tail by 33 amino acids blocked the ability of DAMGO, but not morphine, to inhibit adenylyl cyclase (Surratt et al., 1994). Interestingly, mammalian mu opioid receptors all have an additional 11 amino acids at the end of their C-terminal domains which are not present in non-mammalian mu opioid receptors (Fig. 5). Although the absence of 11 amino acids in the non-mammalian mu opioid receptors is much less than a truncation of 33 residues, one might expect a similar or partial effect with regard to activation of rpMOR by morphine and DAMGO. However, as noted above, the maximal inhibition elicited by DAMGO was only modestly lower (1.2-fold) in CHO cells expressing rpMOR. While the observed differences were not as great as those in the truncation study, the conspicuous differences in the C-terminal tails of non-mammalian and mammalian mu opioid receptors makes this receptor domain a good candidate for future studies exploring its role in opioid receptor signaling.
This study represents an important first step in understanding how changes in two evolutionary distant receptor proteins can result in significant differences in their opioid binding and signaling profiles. One of the ultimate goals of our lab is to utilize an alternative approach by which the evolution of receptor domains in non-mammalian vertebrates is used to inform which individual residues become functionally important in humans. Comparison and characterization of opioid binding and signaling in a comprehensive fashion lays the groundwork for studies in progress evaluating rates of evolution for individual amino acid sites and their relative importance in opioid binding and receptor signaling.
Acknowledgments
The first author would like to thank Crystal Shults for her patience and assistance in the laboratory and also the OSU—Center for Health Sciences Biomedical Sciences Graduate Program for their generous stipend support. Research funding was supported in part by the Oklahoma Center for the Advancement of Science and Technology, Health Research Contract HR02-122R and NIH grant DA12248 to CWS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez FA, Rodriguez-Martin I, Gonzalez-Nunez V, de Velasco EM, Gonzalez SR, Rodriguez RE. New kappa opioid receptor from zebrafish Danio rerio. Neurosci. Lett. 2006;405:94–99. doi: 10.1016/j.neulet.2006.06.028. [DOI] [PubMed] [Google Scholar]
- Barrallo A, González-Sarmiento R, Porteros A, Gracia-Isídoro M, Rodríguez RE. Cloning, molecular characterization, and distribution of a gene homologous to delta opoid receptor from zebrafish (Danio rerio). Biochem. Biophys. Res. Commun. 1998;245:544–548. doi: 10.1006/bbrc.1998.8496. [DOI] [PubMed] [Google Scholar]
- Barrallo A, González-Sarmiento R, Alvar F, Rodriguez RE. ZFOR2, a new opioid receptor-like gene from the teleost zebrafish (Danio rerio). Mol. Brain Res. 2000;84:1–6. doi: 10.1016/s0169-328x(00)00152-2. [DOI] [PubMed] [Google Scholar]
- Benyhe S, Varga E, Hepp J, Magyar A, Borsodi A, Wollemann M. Characterization of kappa1 and kappa2 opioid binding sites in frog (Rana esculenta) brain membrane. Neurochem. Res. 1990;15:899–904. doi: 10.1007/BF00965909. [DOI] [PubMed] [Google Scholar]
- Benyhe S, Monory K, Farkas J, Toth G, Guerrini R, Salvadori S, Orosz G, Wollemann M, Borsodi A. Nociceptin binding sites in frog (Rana esculenta) brain membranes. Biochem. Biophys. Res. Commun. 1999;260:592–596. doi: 10.1006/bbrc.1999.0907. [DOI] [PubMed] [Google Scholar]
- Blume AJ. Interaction of ligands with the opiate receptors of brain membranes: regulation by ions and nucleotides. Proc. Natl. Acad. Sci. U.S.A. 1978;75:1713–1717. doi: 10.1073/pnas.75.4.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford CS, Walthers EA, Searcy BT, Moore FL. Cloning, heterologous expression and pharmacological characterization of a kappa opioid receptor from the brain of the rough-skinned newt, Taricha granulosa. J. Mol. Endocrinol. 2005;34:809–823. doi: 10.1677/jme.1.01711. [DOI] [PubMed] [Google Scholar]
- Bradford CS, Walthers EA, Stanley DJ, Baugh MM, Moore FL. Delta and mu opioid receptors from the brain of a urodele amphibian, the rough-skinned newt, Taricha granulosa: Cloning, heterologous expression, and pharmacological characterization. Gen. Comp Endocrinol. 2006;146:275–290. doi: 10.1016/j.ygcen.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Chaturvedi K, Christoffers KH, Singh K, Howells RD. Structure and regulation of opioid receptors. Biopoly. 2000;55:334–346. doi: 10.1002/1097-0282(2000)55:4<334::AID-BIP1006>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Chen Y, Mestek A, Liu J, Hurley J, Yu L. Molecular cloning and functional expression of a mu opioid receptor from rat brain. Mol Pharmacol. 1993;44:8–12. [PubMed] [Google Scholar]
- Chen C, Yin J, de Riel JK, DesJarlais RL, Raveglia LF, Zhu J, Liu-Chen L-Y. Determination of the amino acid residue involved in [3H] β-funaltrexamine covalent binding in the cloned rat mu opioid receptor. J Biol Chem. 1996;271:21422–21429. doi: 10.1074/jbc.271.35.21422. [DOI] [PubMed] [Google Scholar]
- Cheng Y-C, Prusoff WH. Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50 per cent inhibition (IC50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- Darlison MG, Greten FR, Harvey RJ, Kreienkamp H, Stuhmer T, Zwiers H, Lederis K, Richter D. Opioid receptors from a lower vertebrate (Catostomus commersoni): Sequence, pharmacology, coupling to a G-protein-gated inward-rectifying potassium channel (GIRK1), and evolution. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8214–8219. doi: 10.1073/pnas.94.15.8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CJ, Keith DE, Morrison H, Magendzo K, Edwards RH. Cloning of a delta opioid receptor by functional expression. Science. 1992;258:1952–1955. doi: 10.1126/science.1335167. [DOI] [PubMed] [Google Scholar]
- Fukuda K, Terasako K, Kato S, Mori K. Identification of the amino acid residues involved in selective agonist binding in the first extracellular loop of the delta- and mu-opioid receptors. FEBS Lett. 1995;373:177–181. doi: 10.1016/0014-5793(95)01034-c. [DOI] [PubMed] [Google Scholar]
- Georgoussi Z, Merkouris M, Mullaney I, Megaritis G, Carr C, Zioudrou C, Milligan G. Selective interactions of mu opioid receptors with pertussis toxin-sensitive G proteins: involvement of the third intracellular loop and the c-terminal tail in coupling. Biochim Biophys Acta. 1997;1359:263–274. doi: 10.1016/s0167-4889(97)00097-9. [DOI] [PubMed] [Google Scholar]
- Gillan MG, Kosterlitz HW, Paterson SJ. Comparison of the binding characteristics of tritiated opiates and opioid peptides. Br. J. Pharmacol. 1980;70:481–490. doi: 10.1111/j.1476-5381.1980.tb08727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt RC, Kimelberg HK. Protein analysis of mammalian cells in monolayer culture using the bicinchoninic assay. Anal. Biochem. 1989;177:41–45. doi: 10.1016/0003-2697(89)90010-9. [DOI] [PubMed] [Google Scholar]
- Griffin MT, Hsu JC, Shehnaz D, Ehlert FJ. Comparison of the pharmacological antagonism of M2 and M3 muscarinic receptors expressed in isolation and in combination. Biochem. Pharmacol. 2003;65:1227–1241. doi: 10.1016/s0006-2952(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Handa BK, Lane AC, Lord JAH, Morgan BA, Rance MJ, Smith CFC. Analogues of beta-LPH 61−64 possessing selective agonist activity at mu opiate receptors. Eur J Pharmacol. 1981;70:531–540. doi: 10.1016/0014-2999(81)90364-2. [DOI] [PubMed] [Google Scholar]
- Kieffer BL, Befort K, Gaveriaux-Ruff C, Hirth CG. The delta opioid receptor: Isolation of a cDNA by expression cloning and pharmacological characterization. Proc Natl Acad Sci U.S.A. 1992;89:12048–12052. doi: 10.1073/pnas.89.24.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Lord JAH, Waterfield AA, Hughes J, Kosterlitz HW. Endogenous opioid peptides: multiple agonists and receptors. Nature. 1977;267:495–499. doi: 10.1038/267495a0. [DOI] [PubMed] [Google Scholar]
- Mansour A, Taylor LP, Fine JL, Thompson RC, Hoversten MT, Mosberg HI, Watson SJ, Akil H. Key residues defining the mu-opioid receptor binding pocket: A site-directed mutagenesis study. J. Neurochem. 1997;68:344–353. doi: 10.1046/j.1471-4159.1997.68010344.x. [DOI] [PubMed] [Google Scholar]
- Martin WR, Eades CG, Thompson JA, Huppler RE, Gilbert PE. The effects of morphine-and nalorphine-like drugs in the nondependent and morphine-dependent chronic spinal dog. J. Pharmacol. Exp. Ther. 1976;197:517–532. [PubMed] [Google Scholar]
- McFadyen IJ, Metzger TG, Paterlini MG, Ferguson DM. Exploring the unique pharmacology of a novel opioid receptor, ZFOR1, using molecular modeling and the ‘message-address’ concept. Protein Engineering. 2001;14:953–960. doi: 10.1093/protein/14.12.953. [DOI] [PubMed] [Google Scholar]
- Merkouris M, Dragatsis I, Megaritis G, Konidakis G, Zioudrou C, Milligan G, Georgoussi Z. Identification of the critical domains of the delta opioid receptor involved in G protein coupling using site-specific synthetic peptides. Mol. Pharmacol. 1996;50:985–993. [PubMed] [Google Scholar]
- Minami M, Onogi T, Nakagawa T, Katao Y, Aoki Y, Katsumata S, Satoh M. DAMGO, a mu-opioid receptor selective ligand, distinguishes between mu- and kappa-opioid receptors at a different region from that for the distinction between mu- and delta-opioid receptors. FEBS Lett. 1995;364:23–27. doi: 10.1016/0014-5793(95)00340-f. [DOI] [PubMed] [Google Scholar]
- Minami M, Nakagawa T, Seki T, Onogi T, Aoki Y, Katao Y, Katsumata S, Satoh M. A single residue, Lys108, of the delta-opioid receptor prevents the mu-opioid-selective ligand [D-Ala2, N-MePhe4, Gly-ol5]enkephalin from binding to the delta-opioid receptor. Mol. Pharmacol. 1996;50:1413–1422. [PubMed] [Google Scholar]
- Newman LC, Sands SS, Wallace DR, Stevens CW. Characterization of mu, kappa, and delta opioid binding in amphibian whole brain tissue homogenates. J. Pharmacol. Exp. Ther. 2002;301:364–370. doi: 10.1124/jpet.301.1.364. [DOI] [PubMed] [Google Scholar]
- Onogi T, Minami M, Katao Y, Nakagawa T, Aoki Y, Toya T, Katsumata S, Satoh M. DAMGO, a mu-opioid receptor selective agonist, distinguishes between mu- and delta-opioid receptors around their first extracellular loops. FEBS Lett. 1995;357:93–97. doi: 10.1016/0014-5793(94)01341-w. [DOI] [PubMed] [Google Scholar]
- Paterson SJ, Robson LE, Kosterlitz HW. Control by cations of opioid binding in guinea pig brain membranes. Proc. Natl. Acad. Sci. U.S.A. 1986;83:6216–6220. doi: 10.1073/pnas.83.16.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pert CB, Snyder SH. Opiate receptor binding of agonist and antagonists affected differentially by sodium. Mol. Pharmacol. 1974;10:868–879. [Google Scholar]
- Puttfarcken P, Werling LL, Brown SR, Cote TE, Cox BM. Sodium regulation of agonist binding at opioid receptors. I. Effects of sodium replacement on binding at mu- and delta-type receptors in 7315c and NG108−15 cells and cell membranes. Mol. Pharmacol. 1986;30:81–89. [PubMed] [Google Scholar]
- Raynor K, Kong H, Chen Y, Yasuda K, Yu L, Bell GI, Reisine T. Pharmacological characterization of the cloned mu-, delta-, and kappa-opioid receptors. Mol. Pharmacol. 1994;45:330–334. [PubMed] [Google Scholar]
- Raynor K, Kong H, Mestek A, Bye LS, Tian M, Liu J, Yu L, Reisine T. Characterization of the cloned human mu opioid receptor. J. Pharmacol. Exp. Ther. 1995;272:423–428. [PubMed] [Google Scholar]
- Rodríguez RE, Barrallo A, Garcia-Malvar F, McFadyen IJ, González-Sarmiento R, Traynor JR. Characterization of ZFOR1, a putative delta-opioid receptor from the teleost zebrafish. Neurosci. Lett. 2000;288:207–210. doi: 10.1016/s0304-3940(00)01239-8. [DOI] [PubMed] [Google Scholar]
- Seki T, Minami M, Nakagawa T, Ienaga Y, Morisada A, Satoh M. DAMGO recognizes four residues in the third extracellular loop to discriminate between mu- and kappa-opioid receptors. Eur. J. Pharmacol. 1998;350:301–310. doi: 10.1016/s0014-2999(98)00240-4. [DOI] [PubMed] [Google Scholar]
- Stevens CW, Klopp AJ, Facello JA. Analgesic potency of mu and kappa opioids after systemic administration in amphibians. J. Pharmacol. Exp. Ther. 1994;269:1086–1093. [PubMed] [Google Scholar]
- Stevens CW. Relative analgesic potency of mu, delta and kappa opioids after spinal administration in amphibians. J. Pharmacol. Exp. Ther. 1996;276:440–448. [PubMed] [Google Scholar]
- Stevens CW, Brasel CM, Mohan S. Cloning and bioinformatics of amphibian mu, delta, kappa, and nociceptin opioid receptors expressed in brain tissue: Evidence for opioid receptor divergence in mammals. Neurosci. Lett. 2007;419:189–194. doi: 10.1016/j.neulet.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens CW, Rothe KS. Supraspinal administration of opioids with selectivity for mu, delta, and kappa opioid receptors produces analgesia in amphibians. Eur. J. Pharmacol. 1997;331:15–21. doi: 10.1016/s0014-2999(97)01026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens CW. Non-Mammalian Models for the Study of Pain. In: Conn M, editor. Sourcebook of Models for Biomedical Research. Humana Press; Towata, NJ: 2008. pp. 341–352. [Google Scholar]
- Stevens CW, Newman LC. Spinal administration of selective opioid antagonists in amphibians: evidence for an opioid unireceptor. Life Sci. 1999;64:PL125–PL130. doi: 10.1016/s0024-3205(99)00013-2. [DOI] [PubMed] [Google Scholar]
- Surratt CK, Johnson PS, Moriwaki A, Seidleck BK, Blaschak CJ, Wang JB, Uhl GR. Mu opiate receptor. Charged transmembrane domain amino acids are critical for agonist recognition and intrinsic activity. J. Biol. Chem. 1994;269:20548–20553. [PubMed] [Google Scholar]
- Toll L, Berzetei-Gurske IP, Polgar WE, Brandt SR, Adapa ID, Rodriguez L, Schwartz RW, Haggart D, O'Brien A, White A, Kennedy JM, Craymer K, Farrington L, Auh JS. Standard binding and functional assays related to medications development division testing for potential cocaine and opiate narcotic treatment medications. NIDA Res. Monogr. 1998;178:440–466. [PubMed] [Google Scholar]
- Wang JB, Imai Y, Eppler CM, Gregor P, Spivak CE, Uhl GR. Mu opiate receptor: cDNA cloning and expression. Proc. Natl. Acad. Sci. U.S.A. 1993;90:10230–10234. doi: 10.1073/pnas.90.21.10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WW, Shahrestanifar M, Jin J, Howells RD. Studies on mu and delta opioid receptor selectivity utilizing chimeric and site-mutagenized receptors. Proc Natl Acad Sci U.S.A. 2000;92:12436–12440. doi: 10.1073/pnas.92.26.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling LL, Brown SR, Puttfarcken P, Cox BM. Sodium regulation of agonist binding at opioid receptors. II. Effects of sodium replacement on opioid binding in guinea pig cortical membranes. Mol. Pharmacol. 1986;30:90–95. [PubMed] [Google Scholar]
- Werling LL, Puttfarcken PS, Cox BM. Multiple agonist-affinity states of opioid receptors: regulation of binding by guanyl nucleotides in guinea pig cortical, NG108−15, and 7315c cell membranes. J. Pharmacol. Exp. Ther. 1988;33:423–431. [PubMed] [Google Scholar]
- Xu W, Ozdener F, Li JG, Chen C, de Riel JK, Weinstein H, Liu-Chen LY. Functional role of the spatial proximity of Asp114(2.50) in TMH 2 and Asn332(7.49) in TMH 7 of the mu opioid receptor. FEBS Lett. 1999;447:318–324. doi: 10.1016/s0014-5793(99)00316-6. [DOI] [PubMed] [Google Scholar]
- Xue JC, Chen C, Zhu J, Kunapuli S, DeRiel JK, Yu L, Liu-Chen LY. Differential binding domains of peptide and non-peptide ligands in the cloned rat κ-opioid receptor. J. Biol. Chem. 1994;269:30195–30199. [PubMed] [Google Scholar]
- Yasuda K, Raynor K, Kong H, Breder C, Takeda J, Reisine T, Bell GI. Cloning and functional comparison of kappa and delta opioid receptors from mouse brain. Proc Natl Acad Sci U.S.A. 1993;90:6736–6740. doi: 10.1073/pnas.90.14.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]