Abstract
DNA-encapsulated silver clusters are readily conjugated to proteins and serve as alternatives to organic dyes and semiconductor quantum dots. Stable and bright on the bulk and single molecule levels, Ag nanocluster fluorescence is readily observed when staining live cell surfaces. Being significantly brighter and more photostable than organics and much smaller than quantum dots with a single point of attachment, these nanomaterials offer promising new approaches for bulk and single molecule biolabeling.
INTRODUCTION
Fluorescent probes can be utilized for highly site-specific labeling to study both bulk and single molecule dynamics. Unfortunately while various methods can be used to mark particular biological processes via either genetic encoding with green fluorescent protein or by incorporating exogenous fluorophores, standard dyes suffer from fast photobleaching and low emission rates (1, 2). Combined with fluorescence intermittency (3) and O2 sensitivity (4), these issues severely limit single molecule experiments (5, 6). One approach to improve sensitivity through increasing fluorophore brightness has been to employ semiconductor quantum dots (QDs) as contrast agents (7). QDs are usually prepared in nonpolar organic solvents, however, and require further modification to improve their solubility in aqueous systems. While there have been numerous applications of QDs in cellular imaging (8-11), the large particle size upon functionalization (∼20nm), tendency to aggregate, and potential toxicity prevent them from being versatile cellular labeling agents (12). The emergence of fluorescent silver clusters that possess advantages of both small size and good brightness opens a complementary path toward biological labeling (13-15). In contrast to QDs, silver clusters protected with short peptides can diffuse through cell membranes into live cells, giving bright cell staining (16). Unfortunately, the fluorescence quantum yield (ΦF) of these initial species was quite low (∼3%), and studies to improve the ΦF are underway. Alternatively, DNA also shows strong affinity for silver (17, 18), but with much higher ΦF’s. Recently, we have produced spectrally pure silver clusters encapsulated in ssDNA, ranging from blue to near IR emission, exhibiting up to 40% fluorescence quantum yields while maintaining small size (19). Herein, we demonstrate the applicability of bright DNA-encapsulated silver clusters to biological imaging in a more specific way through cell surface labeling.
MATERIALS AND METHODS
Materials and cell culture information are reported in accompanying Supplementary Material.
Conjugation of avidin-DNA
The disulfide protected 24mer cytosine (C24, 50 μM) was deprotected with tris(2-carboxyethyl)phosphine (1 mM) at room temperature in phosphate buffered EDTA (PBE, phosphate 100 mM, sodium chloride 137 mM, potassium chloride 2.5 mM, EDTA 5 mM) in a 1000 MWCO dialysis tube (Spectrum Laboratories). The dialysis tube was then suspended in PBE overnight at 4 °C, followed by the addition of avidin (EZ-Link®Maleimide Activated NeutrAvidin™ Protein, Pierce, 270 μM). The mixture was kept at 4 °C for another 8 hrs and the protein was concentrated and washed with deionized water 5 times to remove free DNA in a 10,000 MWCO centrifugal ultrafiltration vial (Vivascience, Stonehouse, UK).
Staining with Avidin-C24-Ag
Cells seeded on coverslips in 12-well-plates were fixed with pre-cooled methanol for 10 min, washed with phosphate buffered saline (PBS, pH 7.9) three times, and some were incubated with biotinylation agent (800 μM in PBS) for 15 min, before being washed with PBS five times. Both the non-biotinylated and the biotinylated cells were incubated with Avidin-C24-Ag (5 μm) for 20 min, then washed with PBS four times and mounted onto slides for microscope. Live cells seeded on coverslips in 6-well-plates were incubated with Sulfo-NHS-LC-Biotin (80 μM) in PBS-Mg (Na2HPO4 20 mM, NaCl 150 mM, MgCl2 5 mM, pH 7.9) at 4 °C for 15 min to maintain maximum cell viability. The biotinylated cells were washed with ice-cooled PBS-Mg and incubated with Avidin-C24-Ag (1 μM) at 4 °C for 10 min, washed with PBS-Mg and mounted onto thick glass slides (Erie Scientific, Portsmouth, NH, USA) with a depression filled with DMEM (Dulbecco’s Modified Eagle’s Medium) to keep the cells alive.
Conjugation of anti-HS ab C24
(a) Anti-Heparin/Heparan Sulfate (anti-HS, clone T320.11, Millipore, MAB204, 0.1 mL) was added with Sulfo-SMCC(0.6 mg, Sigma) dissolved in 0.06 mL PBS (pH 7.2, 10 mM) and stored at 4 °C for 2 hrs, and then purified over a Sephadex G100 column with nitrogen-degassed PBS buffer as eluant. (b) 9 mg TCEP and 1600 nmol thiolated C24 DNA (IDT) were mixed in 0.6 mL borate buffer (pH8.5) and stored at 4 °C for 4 hrs, and then purified over a Sephadex G50 column with nitrogen-degassed PBS buffer as eluant. (c) The above two products were mixed in the presence of an extra 2 mg of TCEP and then reacted at 4 °C for 48 hrs, followed by concentration with 5k mwco membrane centrifugal tubes (Vivaspin) and purified over a Sephadex G100 column. Anti-HS ab C24-Ag was prepared in a manner identical to nanocluster formation in free ss-DNA(17, 18), but using the Anti-HS ab C24 conjugate as the scaffold. Briefly, protein-C24 conjugate and silver nitrate were mixed at a molar ratio of two bases of DNA to one silver nitrate and reduced with aqueous sodium borohydride.
RESULTS AND DISCUSSION
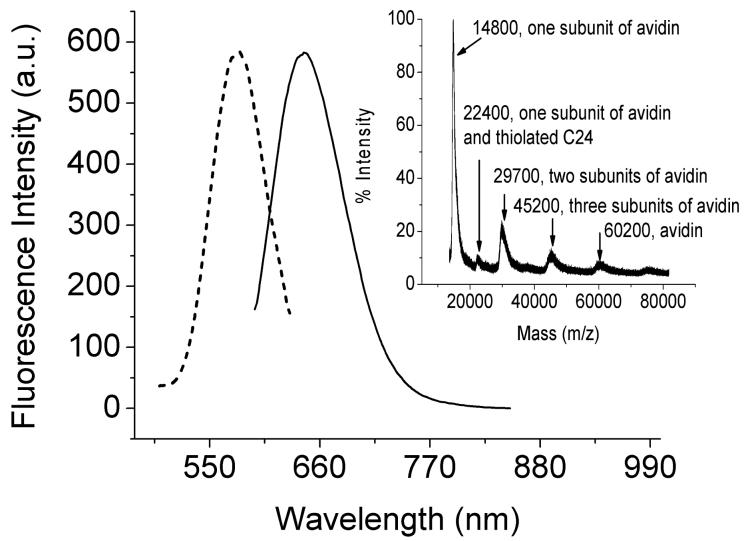
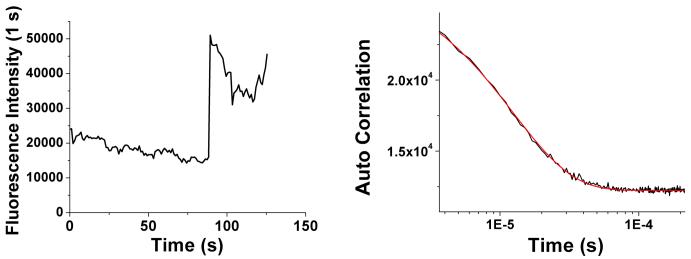
Modified proteins, either genetically or chemically, are widely applied for specific staining, yet for long term studies or those involving low copy numbers in the presence of high background, available labels limit experimental observations. The ssDNA scaffold affords a single point of attachment, while simultaneously stabilizing the few-atom, strongly emissive nanoclusters. Since the DNA chain can be easily modified with a thiol group, this thiolated DNA is conveniently covalently conjugated to any protein activated with maleimide. Capitalizing on the strong biotin-avidin interaction (20), an avidin-C24 conjugate was prepared through maleimide coupling (EZ-Link®Maleimide Activated NeutrAvidin™ Protein, Pierce, 50 μM) to disulfide-protected 24mer oligocytosine (5′-/5ThioMC6-D/CCCCCCCCCCCCCCCCCCCCCCCC-3′, Integrated DNA Technologies (IDT), Coralville, IA, USA, abbreviated as C24, 50 μM), after deprotection with tris(2-carboxyethyl)phosphine (TCEP) (21). Labeling was intentionally kept low to ensure <1 label/avidin tetramer. Creation of avidin-C24 conjugate was confirmed with MALDI-TOF mass spectrometry (Figure 1, inset). The avidin-C24 conjugate was then mixed with silver nitrate and chemically reduced as reported in unconjugated DNA to prepare fluorescent silver clusters (Avidin-C24-Ag) (14). The silver clusters show similar photophysics as those protected with C24 alone (C24-Ag), i.e., a major emissive species with emission maximum at 634 nm and an excitation spectrum centered at 580 nm is observed (Figure 1) with a fluorescence lifetime of 2.86 +/- 0.01 ns. Without conjugated ssDNA, we see no emission from avidin when subjected to identical cluster creation conditions. The photophysical similarity of silver clusters protected with avidin-C24 conjugate or C24 alone further indicates that the silver ions are predominantly bound to DNA even in the presence of covalently linked protein, promising wide applicability of silver nanocluster biolabeling. As observed for free DNA-encapsulated nanoclusters, those conjugated to avidin exhibit excellent photostability and brightness (∼20,000 counts/s/molecule excited at 568-nm CW laser with an excitation intensity of 2 kW/cm2, Figure 2), in line with the regular bright, stable, essentially non-blinking ssDNA-encapsulated silver clusters (22). Single molecule intensity autocorrelations yield a fast decay (∼12 μs) characteristic of the very short-lived dark state of silver nanocluster emitters (22), indicating the independence of nanocluster photophysics upon protein conjugation. Detected single molecule emission rates easily exceed 40,000 counts/sec with a further 2-fold excitation intensity increase.
Figure 1.
Excitation and emission spectra of aqueous avidin-C24 silver clusters solution. Emission (solid) was excited at 580 nm, and excitation (dotted) was detected at 640 nm. Inset: MALDI mass spectrum of avidin-C24 conjugate.
Figure 2.
Emission rate of single avidin-C24 silver nanocluster. Left: fluorescence intensity trajectory of a single avidin-C24 silver cluster excited at 568-nm CW with an excitation intensity of 2.0 kW/cm2 for the first 75 sec and 4.1 kW/cm2 thereafter. Diminution of intensity with time results from slight defocusing of the microscope. Right: autocorrelation trace and fit of the lower intensity level of the left time trace showing a 12 μs decay.
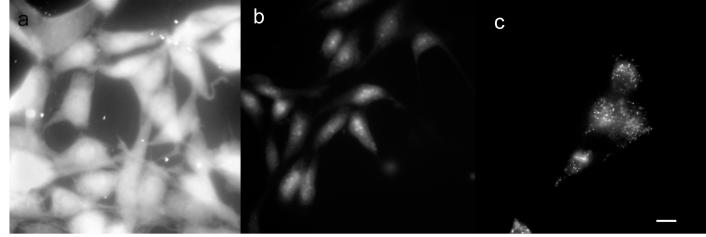
The avidin-biotin interaction has been widely used for molecular targeting and supramolecular assembly (23, 24). Biotin can be conjugated to targets via chemical reactions or specifically targeted using biotin ligase (25). Here sulfosuccinimidyl-6-(biotinamido) hexanoate (EZ-Link®Sulfo-NHS-LC-Biotin, Pierce) was used to react with exposed primary amines (lysine residues) to biotinylate cellular proteins. This reaction was first tested on methanol fixed NIH 3T3 cells. As shown in Figure 3, the fixed cells exhibit quite different fluorescence images, in which the biotinylated cells (Figure 3a) show much higher fluorescence intensity than the non-biotinylated cells (Figure 3b) when both are stained with Avidin-C24-Ag (5 μM). The application of Sulfo-NHS-LC-Biotin to fixed cells (800 μM) was too harsh for live cells, therefore the concentration was lowered to 80 μM to maintain cell viability. Biotinylated live cells loaded with Avidin-C24-Ag were examined at room temperature shortly after preparation, as shown in Figure 3c. The silver clusters were bound to the cell surface yielding diffuse labeling with some bright aggregates, possibly resulting from endocytic uptake. However, the non-biotinylated live cells showed only weak emission, indicative of autofluorescence alone.
Figure 3.
Fluorescence images of NIH 3T3 cells stained with Avidin-C24-Ag. a, Fixed cells, biotinylated; b, Fixed cells, non-biotinylated; c, Live cells, biotinylated. All cells were stained with Avidin-C24-Ag before imaging. Images a to c were recorded on Zeiss Axiovert 200 microscope with a CoolSNAP CCD camera (Roper Scientific). Scale bar 30 μM.
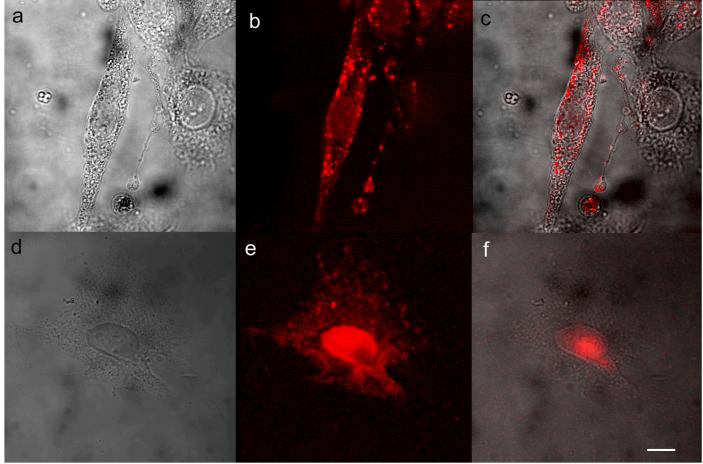
Although the cell surface was readily stained with Avidin-C24-Ag under these conditions, the somewhat indiscriminate biotinylation agent was still rather toxic to NIH 3T3. The affinity to and permeability of NIH 3T3 cells to DNA are low, resulting in poor staining of cells by C24-Ag alone (supplementary information). Targeting membrane-bound components enables effective cell surface labeling and uptake via receptor mediated endocytosis (26). Heparin sulfate (HS) is a linear, polysulfated, and highly negatively-charged polysaccharide attached to the cell surface and extracellular matrix proteins (27). Cellular uptake of a wide variety of extracellular ligands such as fibroblast growth factor and cell penetrating peptides are facilitated through initial electrostatic interaction between the negatively-charged HS and positively-charged ligands, therefore HS proteoglycan is recruited as a plasma membrane carrier (28). To better maintain cell viability, we conjugated C24 ssDNA to a heparin sulfate antibody and labeled with emissive Ag nanoclusters. In this paper, cells incubated with anti-HS-C24-Ag (4 μM) at 4 °C for 15 min showed staining of only the cell surface (Fig 4a - 4c), with no internalization. While most labeling was diffuse, some aggregation on the cell surface was observed, resulting in a few bright spots. Longer incubation time and higher antibody concentration induces denser and more even surface staining. Silver clusters were quickly internalized when the above cells were incubated at 37°C and emission is concentrated in the nuclei (Fig 4d - 4f). Likely, the antibody silver cluster conjugate first bonds to cell surface HS followed by endocytosis (29), as further indicated by the lack of observed fluorescence from cells incubated with C24-Ag only at either 4°C or 37°C. The internalization of silver clusters illustrates that silver clusters can be applied not only to label cell surface proteins to investigate extracellular dynamics, but also potentially as a reporter of endocytic uptake and vesicular transport.
Figure 4.
Fluorescence images of live NIH 3T3 cells stained with anti-HS-C24-Ag. Live cells incubated with anti-HS-C24-Ag at 4 °C for 20 min (a, bright field; b, silver clusters; c, merge), or at 37 °C for 6 min (d, bright field; e, silver clusters; f, merge). Images a to f were recorded on Zeiss LSM 10 confocal microscope. The fluorescence images were taken at 543 nm excitation. Scale bar 25 μm.
In summary, the oligoDNA protected silver clusters are readily covalently conjugated to proteins such as avidin, and primary antibodies without significant interference of either biological function or nanocluster photophysics. As silver cluster label size is significantly less than 10% of labeled protein molecular weight, and overall isolated label size is quite small (19), proteins are likely minimally perturbed by cluster-based labeling. Employing either avidin-biotin or antibody/antigen interactions, these silver clusters can stain the cell surface and be internalized, possibly enabling studies of transmembrane transport or drug delivery. Because silver nanocluster-based labels simultaneously exhibit small size, excellent photostability, and bright emission, they offer new opportunities in high sensitivity biolabeling.
Supplementary Material
ACKNOWLEDGMENTS
The authors gratefully acknowledge financial support from Invitrogen Corp., NIH R01GM68732, and NIH P20GM072021. We also thank Prof. D. F. Doyle for use of his cell culture facility, Prof. C. J. Fahrni for equipment loan, and acknowledge productive discussions with Prof. C. K. Payne.
Footnotes
This invited paper is part of the Series: Applications of Imaging to Biological and Photobiological Systems
REFERENCES
- 1.Eggeling C, Widengren J, Rigler R, Seidel CAM. Photobleaching of fluorescent dyes under conditions used for single-molecule detection: Evidence of two-step photolysis. Anal. Chem. 1998;70:2651–2659. doi: 10.1021/ac980027p. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt T, Kubitscheck U, Rohler D, Nienhaus U. Photostability data for fluorescent dyes:an update. single mol. 2002;3:327. [Google Scholar]
- 3.Hoogenboom JP, Hernando J, van Dijk E, van Hulst NF, Garcia-Parajo MF. Power-law blinking in the fluorescence of single organic molecules. ChemPhysChem. 2007;8:823–833. doi: 10.1002/cphc.200600783. [DOI] [PubMed] [Google Scholar]
- 4.Lill Y, Hecht B. Single dye molecules in an oxygen-depleted environment as photostable organic triggered single-photon sources. Appl. Phys. Lett. 2004;84:1665–1667. [Google Scholar]
- 5.Oparka KJ, Roberts AG, SantaCruz S, Boevink P, Prior DAM, Smallcombe A. Using GFP to study virus invasion and spread in plant tissues. Nature. 1997;388:401–402. [Google Scholar]
- 6.Gronemeyer T, Godin G, Johnsson K. Adding value to fusion proteins through covalent labelling. Curr. Opin. Biotechnol. 2005;16:453–458. doi: 10.1016/j.copbio.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Michalet X, Kapanidis AN, Laurence T, Pinaud F, Doose S, Pflughoefft M, Weiss S. The power and prospects of fluorescence microscopies and spectroscopies. Annu. Rev. Biophys. Biomolec. Struct. 2003;32:161–182. doi: 10.1146/annurev.biophys.32.110601.142525. [DOI] [PubMed] [Google Scholar]
- 8.Chan WCW, Nie SM. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 9.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao XH, Yang LL, Petros JA, Marshal FF, Simons JW, Nie SM. In vivo molecular and cellular imaging with quantum dots. Curr. Opin. Biotechnol. 2005;16:63–72. doi: 10.1016/j.copbio.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nat. Biotechnol. 2003;21:47–51. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- 12.Howarth M, Takao K, Hayashi Y, Ting AY. Targeting quantum dots to surface proteins in living cells with biotin ligase. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng J, Dickson RM. Individual water-soluble dendrimer-encapsulated silver nanodot fluorescence. J. Am. Chem. Soc. 2002;124:13982–13983. doi: 10.1021/ja028282l. [DOI] [PubMed] [Google Scholar]
- 14.Petty JT, Zheng J, Hud NV, Dickson RM. DNA-templated Ag nanocluster formation. J. Am. Chem. Soc. 2004;126:5207–5212. doi: 10.1021/ja031931o. [DOI] [PubMed] [Google Scholar]
- 15.Peyser-Capadona L, Zheng J, Gonzalez JI, Lee TH, Patel SA, Dickson RM. Nanoparticle-free single molecule anti-stokes Raman spectroscopy. Phys. Rev. Lett. 2005;94 doi: 10.1103/PhysRevLett.94.058301. art. no. 058301. [DOI] [PubMed] [Google Scholar]
- 16.Yu J, Patel SA, Dickson RM. In vitro and intracellular production of peptide-encapsulated fluorescent silver nanoclusters. Angew. Chem.-Int. Edit. 2007;46:2028–2030. doi: 10.1002/anie.200123456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luk KFS, Maki AH, Hoover RJ. Studies of heavy-metal binding with polynucleotides using optical detection of magnetic-resonance-silver(I) binding. J. Am. Chem. Soc. 1975;97:1241–1242. doi: 10.1021/ja00838a047. [DOI] [PubMed] [Google Scholar]
- 18.Ritchie CM, Johnsen KR, Kiser JR, Antoku Y, Dickson RM, Petty JT. Ag nanocluster formation using a cytosine oligonucleotide template. J. Phys. Chem. C. 2007;111:175–181. doi: 10.1021/jp0648487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards CI, Choi S, Hsiang JC, Antoku Y, Vosch T, Bongirono A, Tzeng YL, Dickson RM. Oligonucleotide-stablized Ag nanocluster fluorophores. J. Am. Chem. Soc. 2008;130:5038–5039. doi: 10.1021/ja8005644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilchek M, Bayer EA. Avidin biotin technology 10 years on - has it lived up to its expectations. Trends Biochem.Sci. 1989;14:408–412. doi: 10.1016/0968-0004(89)90289-2. [DOI] [PubMed] [Google Scholar]
- 21.Visser CC, Voorwinden LH, Harders LR, Eloualid M, Van Bloois L, Crommelin DJA, Danhof M, De Boer AG. Coupling of metal containing homing devices to liposomes via a maleimide linker: Use of TCEP to stabilize thiol-groups without scavenging metals. J. Drug Target. 2004;12:569–573. doi: 10.1080/10611860400010689. [DOI] [PubMed] [Google Scholar]
- 22.Vosch T, Antoku Y, Hsiang JC, Richards CI, Gonzalez JI, Dickson RM. Strongly emissive individual DNA-encapsulated Ag nanoclusters as single-molecule fluorophores. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12616–12621. doi: 10.1073/pnas.0610677104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao Y, Kopplow K, Liu GY. In-situ immun-PCR to detect antigens. Lancet. 2000;356:1002–1003. doi: 10.1016/S0140-6736(00)02696-9. [DOI] [PubMed] [Google Scholar]
- 24.Niemeyer CM, Burger W, Peplies J. Covalent DNA - streptavidin conjugates as building blocks for novel biometallic nanostructures. Angew. Chem.-Int. Edit. 1998;37:2265–2268. doi: 10.1002/(SICI)1521-3773(19980904)37:16<2265::AID-ANIE2265>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 25.Chen I, Howarth M, Lin WY, Ting AY. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat. Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 26.Haucke V. Cargo takes control of endocytosis. Cell. 2006;127:35–37. doi: 10.1016/j.cell.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 27.Iozzo RV. Matrix proteoglycans: from molecular design to cellular function. Annu. Rev. Biochem. 1998;67:609–652. doi: 10.1146/annurev.biochem.67.1.609. [DOI] [PubMed] [Google Scholar]
- 28.Belting M. Heparan sulfate proteoglycan as a plasma membrane carrier. Trends Biochem.Sci. 2003;28:145–151. doi: 10.1016/S0968-0004(03)00031-8. [DOI] [PubMed] [Google Scholar]
- 29.Payne CK, Jones SA, Chen C, Zhuang XW. Internalization and trafficking of cell surface proteoglycans and proteoglycan-binding ligands. Traffic. 2007;8:389–401. doi: 10.1111/j.1600-0854.2007.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.