Abstract
Normal cell growth is governed by a complicated biological system, featuring multiple levels of control, often deregulated in cancers. The role of microRNAs (miRNAs) in the control of gene expression is now increasingly appreciated, yet their involvement in controlling cell proliferation is still not well understood. Here we investigated the mammalian cell proliferation control network consisting of transcriptional regulators, E2F and p53, their targets and a family of 15 miRNAs. Indicative of their significance, expression of these miRNAs is downregulated in senescent cells and in breast cancers harboring wild-type p53. These miRNAs are repressed by p53 in an E2F1-mediated manner. Furthermore, we show that these miRNAs silence antiproliferative genes, which themselves are E2F1 targets. Thus, miRNAs and transcriptional regulators appear to cooperate in the framework of a multi-gene transcriptional and post-transcriptional feed-forward loop. Finally, we show that, similarly to p53 inactivation, overexpression of representative miRNAs promotes proliferation and delays senescence, manifesting the detrimental phenotypic consequence of perturbations in this circuit. Taken together, these findings position miRNAs as novel key players in the mammalian cellular proliferation network.
Keywords: breast cancer, microarray, miR-106, mir-155, senescence
Introduction
The tumor suppressor p53 is considered a central regulator of cell-fate decisions. Activation of p53 can induce several cellular responses, including cell-cycle arrest, senescence and apoptosis. Thus, absence of functional p53 predisposes cells to neoplastic transformation. Accordingly, mutations of this gene are highly common in human cancers (Hussain and Harris, 1999). p53 is a sequence-specific transcription factor (TF) that exerts many of its downstream effects by activating gene transcription (Ryan et al, 2001). Nevertheless, additional transactivation-independent functions of p53 contribute to its tumor suppressive activity, including protein–protein interactions with additional TFs and other cell-fate regulators. The importance of transcriptional regulation by p53 is exemplified by the fact that most p53 tumor-derived mutants are defective in DNA binding and incapable of transactivation (Kern et al, 1991). In addition to its capability to induce gene transcription, p53 activation results in extensive gene repression (Ginsberg et al, 1991). Direct and indirect transcriptional repression by p53 is considered important for its tumor suppressive functions, such as induction of cell-cycle arrest and apoptosis (Ho and Benchimol, 2003).
microRNAs (miRNAs or miRs in short) are a recently discovered class of small non-coding RNA species that regulate gene expression at the post-transcriptional level. Approximately half of the known miRs are encoded in regions of the genome that are distal to known genes, whereas the remaining reside in introns, or in rare cases in exons, of coding genes, usually in the same orientation as the mRNA. Additionally, some miRs are clustered in the genome and are transcribed as polycistrons that may contain up to ∼50 mature miRs (Bartel, 2004). With the recent identification of miRs that regulate cancer-related processes such as apoptosis, proliferation and differentiation, these RNA species emerge as important regulators of cancer initiation and progression. Accordingly, mutation and transcriptional deregulation of miRs have been linked to cancer (Esquela-Kerscher and Slack, 2006). Deregulated miRs were suggested to exert their function in cancer through silencing of key cell-fate regulators, as shown for let-7 and Ras (Johnson et al, 2005), as well as for miR-106b and p21 (Ivanovska et al, 2008; Petrocca et al, 2008).
In a previous work we suggested that miRs cooperate with certain TFs in the regulation of mutual sets of target genes, allowing the coordinated modulation of gene expression both transcriptionally and post-transcriptionally. Specifically, we found a recurring network motif in which a TF regulates the miR with which it cooperates in regulating a common set of targets, creating a feed-forward loop (FFL). One such case involved E2F and the miR-106b/93/25 polycistron (Shalgi et al, 2007).
Several studies have implicated p53 in the regulation of miR expression (Xi et al, 2006; Chang et al, 2007; He et al, 2007; Raver-Shapira et al, 2007; Tarasov et al, 2007; Kumamoto et al, 2008). These studies exploited various high-throughput methods to identify p53-regulated miRs in several cellular systems with differential p53 status. Although the resulting candidate lists from each study differed considerably, probably due to differences in the cellular contexts and p53 activation signals, all studies identified members of the miR-34 family as direct transactivation targets of p53. In line with p53 function, induction of miR-34 family members was suggested to mediate cell-cycle arrest, apoptosis and senescence. Importantly, none of these studies focused on miRs, whose expression negatively correlated with p53 activation, and which are likely repressed by this tumor suppressor.
Here, we report the identification of a large set of miRs, the expression of which constitutes a recurring signature in several experiments. The members of this signature are transcriptionally repressed by p53 in primary cells and in human breast cancers. This signature is comprised of known cancer-associated miRs as well as newly proposed ones, and includes the miR-106b/93/25 polycistron. We implicate E2F1 in the p53-dependent repression of these miRs, and demonstrate the oncogenic potential of the miR-106b/93/25 polycistron. Finally, we delineate a network architecture that includes the transcription factor E2F1 and a family of miRs, which co-regulate mutual target genes transcriptionally and post-transcriptionally, thereby enhancing cellular proliferation. This FFL is repressed by p53, possibly to promote senescence and suppress cancer progression.
Results
Identification of p53-regulated miRNAs in primary human cells and in human breast cancers
To identify novel p53-regulated miRs, we established two isogenic cell cultures that differ in their p53 status and analyzed their miRNA profiles both under normal conditions and in contexts involving p53 activation. WI-38 primary human fibroblasts were infected with a retrovirus encoding for the p53-inactivating peptide, GSE56 (Ossovskaya et al, 1996). These cells (GSE) and their active p53 counterparts (con) were treated with the DNA-damaging agent doxorubicin or grown until the onset of replicative senescence (the establishment of the system is depicted in Supplementary Figure S1). Analysis of miRNA expression patterns revealed several expression clusters (see Materials and methods). Notable among these was a cluster populated with miRs, the expression of which was negatively regulated by p53 under normal conditions. The cluster showed additional downregulation in senescent cells, which was attenuated upon p53 inactivation. We named this cluster the ‘p53-repressed miR cluster' (Figure 1A). Notably, doxorubicin treatment, which resulted in a considerable activation of p53 and its mRNA targets (Supplementary Figure S1), did not significantly affect the levels of these miRs.
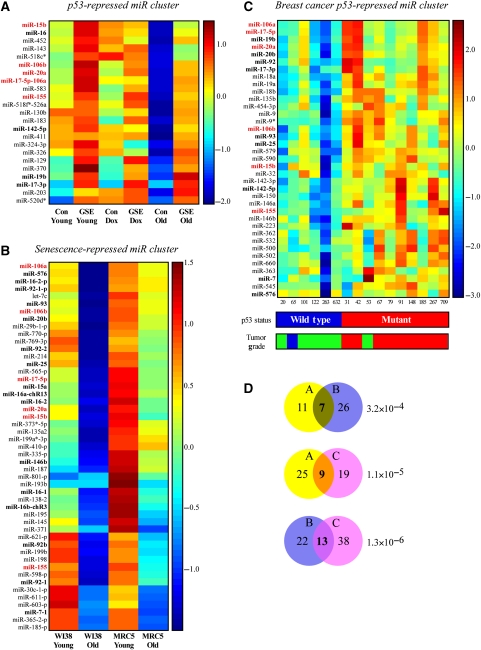
Figure 1.
miRNA clusters derived from three different datasets. The figure depicts three different expression matrices for miRNA clusters that originated from three microarray experiments. miRs appearing in all three clusters are indicated in red bold font. miRs appearing in two clusters are indicated in black bold font. (A) The ‘p53-repressed miR cluster.' Primary WI-38 cells that were infected with the p53-inactivating peptide GSE56 (GSE) and their empty vector control counterparts (Con) were analyzed for miRNA expression at early passage (Young), after doxorubicin treatment (0.2 μg/ml, 24 h) of early passage cells (Dox), and at the onset of replicative senescence (Old). A cluster of miRs that were repressed by p53 at normal conditions and in senescent cells is presented. (B) The ‘Senescence-repressed miR cluster.' Primary WI-38 and MRC5 cells were analyzed for miRNA expression at early passage (Young) and at the onset of replicative senescence (Old). A cluster of miRs that were repressed upon senescence in both cell types is presented. (C) The ‘breast cancer p53-repressed miR cluster.' Human primary breast cancers described by Sorlie et al (2006) and Naume et al (2007) were analyzed for their miRNA profiles. A cluster of miRs, the expression of which was anticorrelated with the presence of a wild-type p53 in the tumor is presented. p53 status was determined using TTGE and sequencing of exons 2–11. Grading was performed using histopathological evaluation according to the modified Scarff–Bloom–Richardson method and is represented by blue for grade 1, green for grade 2 and red for grade 3. (D) Venn diagrams depicting the overlaps between cluster pairs. The values in each circle represent the number of miRs from the indicated cluster that was detected by the array corresponding to the second cluster. The values in the circle overlapping regions represent the number of miRs that are shared between the two clusters. Hypergeometric P-values on the size of the overlaps are provided.
Interestingly, a significant number of miRs from this cluster were also clustered together in a similar experiment, in which miR expression was profiled in young and senescent human embryonic fibroblasts (WI-38 and MRC5). This cluster is termed ‘senescence-repressed miR cluster' (Figure 1B). The significant overlap between the clusters (P-value=3.2 × 10−4) is interesting as the second experiment was not designed to discover p53-regulated miRs, but rather to identify a general signature of miRs that are altered upon replicative senescence. However, p53 activity was increased in both senescent fibroblast cultures (data not shown). Strikingly, clustering analysis of miR expression data derived from a set of breast cancer tumors with differing p53 status also resulted in a cluster highly overlapping the ‘p53-repressed miR cluster' (Figure 1C; P-value=1.11 × 10−5) (samples, p53 status and histological grading were described by Naume et al (2007) and Sorlie et al (2006), and detailed description of the mutation status is listed in Supplementary Table S1). The ‘breast cancer p53-repressed miR cluster' was comprised of miRs, the expression of which was negatively correlated with the presence of a wild-type p53 in the tumors. Additionally, the miRNA expression and p53 status partially correlated with tumor grade, as almost all cancer samples that contained a mutant p53 and expressed high levels of the miRs were derived from high-grade tumors.
We thus revealed a recurring signature of miRs that are coordinately regulated both in primary human cells in vitro and in human breast tumors in vivo. We suggest that these miRs are repressed by wild-type p53 during both normal growth and cancer progression.
The presented clusters contain families of paralogous cancer-related miRNAs
Interestingly, 15 miRs represented in the three clusters (Figure 1) are transcribed from three homologous genomic loci, reported earlier to be paralogs that evolved from a common evolutionary origin (Tanzer and Stadler, 2004). These include miRs-106b/93/25, which reside within an intron of the cell-cycle gene ‘minichromosome maintenance protein 7' (MCM7); miRs-17/18a/19a/20a/19b-1/92a-1 (miR-17-92 polycistron), which are transcribed as the non-coding RNA c13orf25, and miRs-106a/18b/20b/19b-2/92-2 (miR-106a-92 polycistron), which are clustered on chromosome X. Our data indicate that not only were these miRNA sequences and genomic organization conserved during evolution but also was their transcriptional regulation. Additional well-represented miRs in the clusters include the miR-15b/16 polycistron and miR-155.
Many members of the clusters are overexpressed in various tumors, consistent with the frequent p53 loss of function in cancer, and some were shown to possess oncogenic functions. For example, miR-92, miR-106a, miR-17-5p, miR-20a and miR-155, which appear in at least two expression clusters, were reported to be overexpressed in solid tumors (Volinia et al, 2006). Members of the miR-17-92 polycistron are overexpressed in lymphomas and in lung and colorectal carcinomas (He et al, 2005; Schetter et al, 2008), and were shown to accelerate tumor growth (O'Donnell et al, 2005). Interestingly, the MCM7 gene that contains three of the clusters' miRs in its intron (miRs-106b/93/25) is amplified or overexpressed in diverse types of cancers (Ren et al, 2006), as are its resident miRs (Petrocca et al, 2008).
Representative miRNAs show p53-dependent repression during senescence in many cell types
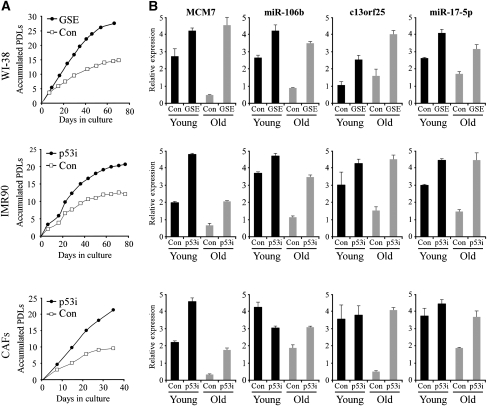
To further validate our data, we generated two additional human isogenic cell culture pairs from the IMR90 lung primary fibroblasts and from prostate-cancer-associated fibroblasts. Each culture was infected with a retrovirus encoding for either a small hairpin RNA (shRNA) targeting p53 (p53i) or a control shRNA (con), and grown until the onset of replicative senescence. p53 knockdown, which significantly reduced the mRNA and protein levels of both p53 and its target p21, delayed the onset of senescence by approximately 10 population doublings (PDLs) (Figure 2A; Supplementary Figure S2C). For these cell types, as well as for the WI-38 cells, we compared the levels of representative miRs that appear in the three expression clusters using TaqMan miRNA assays. Analyses of miR-106b and miR-17-5p, as well as their host transcripts MCM7 and c13orf25, respectively, revealed transcriptional repression upon replicative senescence in all three tested cell cultures in a manner that was partially or completely p53 dependent (Figure 2B). Additionally, the non-coding RNA BIC and its resident miR-155 were also transcriptionally repressed in a p53-dependent manner upon replicative senescence (Supplementary Figure S2D).
Figure 2.
Validation of microarray data. Inactivation of p53 by the GSE56 peptide (GSE) or shRNA (p53i) in three different human primary fibroblasts delays replicative senescence and attenuates the repression of miRs and their hosts upon senescence. (A) Growth curves for the human primary fibroblasts WI-38 and IMR90 and for the prostate cancer-associated fibroblasts (CAFs) PF179. PDLs, population doublings. (B) QRT–PCR for miR-106b and miR-17-5p, and their host transcripts MCM7 and c13orf25, respectively, in early passage (Young) versus late passage (Old) fibroblasts. Data are represented as mean±s.d.
Co-clustering of miRNA and mRNA expression data from human breast cancers reveals two distinct groups of p53-repressed miRNAs
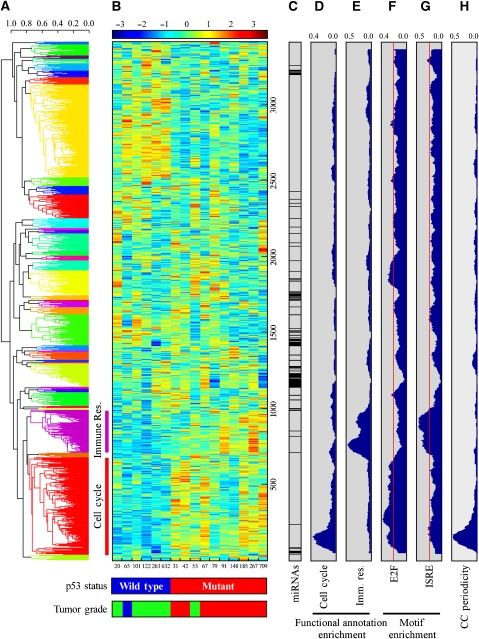
To gain further insights into the regulation and function of the identified miRNA signature, we exploited previously published mRNA profiling (Sorlie et al, 2006; Naume et al, 2007), performed on the same set of breast cancer specimens from which the ‘breast cancer p53-repressed miR cluster' was derived. The mRNA and miRNA array data were combined into one set of expression profiles, and were clustered into 40 co-clusters; each may consist of both miRs and mRNAs (Figure 3). Interestingly, the members of the ‘breast cancer p53-repessed miR cluster' were separated into two distinct co-clusters with dissimilar characteristics, as revealed by functional annotation using DAVID (Dennis et al, 2003) and by promoter motif analyses using AMADEUS (Linhart et al, 2008) of the mRNAs from each co-cluster. Of 37 miRs, 31 were co-clustered (Figure 3A, red cluster; Supplementary Table S2) with genes highly enriched for cell-cycle-related annotations (Figure 3D; enrichment P-value for ‘cell-cycle' annotation=8.5 × 10−20, see Supplementary Table S3A for other enriched annotations) and for regulatory binding motifs of known cell-cycle-related TFs such as E2F (Figure 3F; P-value=2.5 × 10−16). All members of the three paralogous polycistrons described above were included in this ‘cell-cycle-associated co-cluster,' as were their hosts MCM7 and c13orf25. Supporting the notion that this cluster consists mainly of cell-cycle-related genes, we compared the mRNA dataset to a previously published list of genes expressed in a cell-cycle periodic manner (Whitfield et al, 2002), and found a significant enrichment of periodically expressed genes in this cluster (Figure 3H; P-value=4.9 × 10−17). Another co-cluster, which contained six miRs (Figure 3A, purple cluster; Supplementary Table S2), was comprised of genes highly enriched for immune response-related functions (Figure 3E; P-value for ‘immune response' annotation=1.2 × 10−48, see Supplementary Table S3B for other enriched annotations). The promoters of these genes were enriched for immune response-related motifs such as interferon-stimulated-responsive element (Figure 3G; P-value=1.62 × 10−12) as well as for the IRF and NF-κB motifs (data not shown). Importantly, both mRNAs and miRNAs of both co-clusters were downregulated in tumors that harbor wild-type p53, suggesting that our miR signature belongs to a larger transcriptional program that mediates p53-dependent gene repression of both RNA types.
Figure 3.
Co-clustering of miRNA and mRNA expression data from human breast cancers. (A) A dendrogram for the expression data based on hierarchical clustering and average linkage. Data were clustered into 40 clusters, which are indicated by different colors of the dendrogram. miRs from the ‘breast cancer p53-repressed miR cluster' were mapped to the red (cell cycle) and purple (immune response) clusters. (B) Expression matrix of the mRNAs and miRNAs analyzed. For p53 status and tumor grade analyses, see Figure 1 legend. Breast cancer samples are indicated by numbers below the matrix. (C) The bar indicates the position of miRs along the expression matrix. (D, E) Functional annotation analysis for ‘cell cycle' (C) and ‘immune response' (Imm. Res.) (D) terms. The plots represent the density (from 0 to 1) of mRNAs corresponding to each annotation term in windows of 100 genes. (F, G) Density plots for the appearance of the E2F and ISRE motifs, the most enriched elements in the cell cycle and immune response co-clusters, respectively. Red lines indicate the background levels of each motif, calculated as the fraction of genes in the genome containing the motif. (H) Density plot for cell-cycle periodic genes as defined by Whitfield et al (2002).
The miRNAs from the ‘cell-cycle-associated co-cluster' are associated with p53 and E2F in a proliferation-related regulatory network
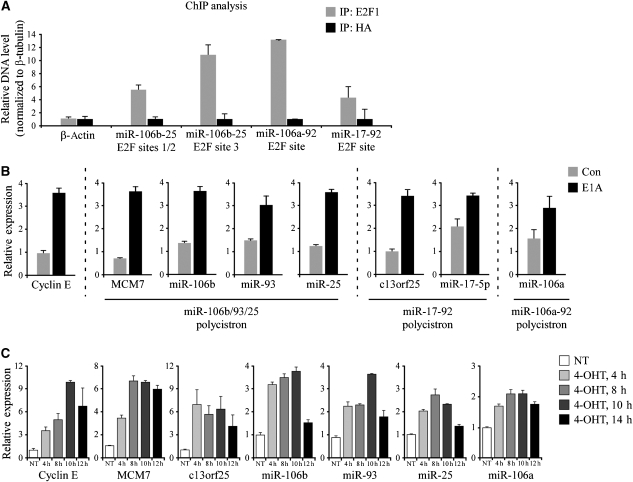
We have reported earlier the identification and characterization of an mRNA cluster termed the ‘proliferation cluster' that consists mainly of cell-cycle-related genes (Milyavsky et al, 2005). This cluster emerged from mRNA profiling of an in vitro transformation process, in which primary WI-38 cells were gradually transformed into tumorigenic cells. Importantly, the ‘proliferation cluster' is one of the most prominent expression signatures revealed when tumors are compared to normal tissues or when highly proliferating cells are compared to slow growing cells, and contains many cell-cycle periodic genes (Whitfield et al, 2006). The expression pattern of the ‘proliferation cluster' is highly similar to that of the ‘p53-repressed miR cluster'; i.e. the ‘proliferation cluster' mRNAs display p53-dependent downregulation. The similarity in expression patterns prompted us to hypothesize that both clusters share a common transcriptional program. It was shown earlier that the p53-mediated repression of the ‘proliferation cluster' was mediated through E2F (Tabach et al, 2005). Providing further support, the promoters of the mRNAs from the ‘cell-cycle-associated co-cluster' are highly enriched with E2F-binding motifs (Figure 3F), and in particular, a conserved E2F-binding site is found upstream of the three miRNA polycistrons (Supplementary Figure S3A and Supplementary Table S4). Confirming these predictions, chromatin immunoprecipitation analysis revealed that E2F1 binds to its conserved motifs upstream of each of the three polycistrons (Figure 4A). Similarly, it was shown recently that the miR-17-92 and the miR-106b/93/25 polycistrons are transcriptionally activated by E2F family members (O'Donnell et al, 2005; Sylvestre et al, 2007; Woods et al, 2007; Petrocca et al, 2008).
Figure 4.
E2F binds the promoters of the paralogous miRNA polycistrons and induces their transcription. (A) E2F1 binds conserved E2F sites upstream of each of the paralogous miRNA polycistrons. ChIP analysis was performed on U2OS cells with an anti-E2F1 antibody (IP: E2F1) and a control antibody against HA (IP: HA). The precipitated DNA was measured using QRT–PCR. The β-actin gene serves as a negative control for E2F1 binding. Values were normalized to the levels of β-tubulin. For schematic representation of the polycistrons' genomic organization and corresponding E2F sites, see Supplementary Figure S3A. (B) E2F activation by E1A induces the polycistronic miRNAs. WI-38 cells were infected with the oncoprotein E1A or a control vector (Con) and selected with puromycin. QRT–PCR revealed upregulation of the known E2F1 target, Cyclin E, as well as of host transcripts and miRNAs, representatives of the three paralogous polycistrons (miRs-106b/93/25, miR-17-92 and miR-106a-92). (C) E2F1 activation results in rapid induction of the polycistronic miRNAs. WI-38 cells were stably infected with ER-E2F1 and treated with 4-OHT (300 nM) for the indicated time periods. QRT–PCR analysis was performed to measure the levels of miRNAs and mRNAs. QRT–PCR data are represented as mean±s.d.
In view of the above, it appears conceivable that the miRs from the ‘cell-cycle-associated co-cluster' are transcriptionally activated by E2F, and that p53 exerts its repression through E2F inhibition. In agreement with the observed downregulation of the ‘p53-repressed miR cluster' in senescence, it was shown that E2F activity is significantly downregulated in senescent cells (Campisi and d'Adda di Fagagna, 2007). In addition, the miRs presented here are proposed to be novel members of the well-established ‘proliferation cluster.'
The p53-dependent repression of miRNAs from the ‘cell-cycle-associated co-cluster' is mediated through E2F1
To experimentally test whether the cell-cycle-associated miRs and their host mRNAs are transcriptionally activated by E2F1, we infected primary WI-38 cells with E1A, a viral oncoprotein that disrupts pRb-E2F complexes and leads to an upregulation of the endogenous E2F activity (Fattaey et al, 1993). As expected, stable overexpression of E1A resulted in elevated levels of candidate miRNAs and host mRNAs, which were part of the ‘cell-cycle-associated co-cluster,' that together represent all three paralogous polycistrons (Figure 4B). Specifically, the MCM7 gene and its resident miRNAs miRs-106b/93/25; the non-coding RNA c13orf25 and its resident miR-17-5p; and miR-106a, which represents the miR-106a-92 polycistron, were all upregulated following E2F activation. We note that the level of miR-155, which belongs to the ‘immune response co-cluster', was not upregulated by E2F (Supplementary Figure S3E).
To investigate the kinetics of the miRNA's transcriptional activation by E2F1, we infected WI-38 cells with a retrovirus encoding for an E2F1 protein fused to a modified estrogen receptor (ER) ligand-binding domain. Treatment of ER-E2F1-expressing cells with 4-hydroxytamoxifen (4-OHT) permits ER-E2F1 translocation to the nucleus, thereby inducing its transactivation activity. As depicted in Figure 4C, as early as 4 h after ER-E2F1 activation by 4-OHT, significant upregulation of the candidate miRNAs and host transcripts was already evident. The miRNAs and their hosts peaked after 8–10 h of 4-OHT treatment, similarly to Cyclin E, a known E2F1 target. It is noted that upregulation of MCM7 and its resident miRNAs following 4-OHT treatment was also observed in ER-E2F1 expressing lung carcinoma cells (H1299) and osteosarcoma cells (U2OS) (Supplementary Figure S3B and C). Finally, to strengthen the notion that E2F1 directly transactivates the miRNAs, we treated ER-E2F1 expressing WI-38 cells with 4-OHT in the presence or absence of cycloheximide, which inhibits protein biosynthesis and should attenuate the induction of the miRNAs if translation of a secondary mediator is required. As depicted in Supplementary Figure S3D, the induction of the miRNAs was not inhibited by cycloheximide. Altogether, these results indicate that E2F1 can directly bind its cognate sites upstream of the polycistronic miRNAs and activate their transcription.
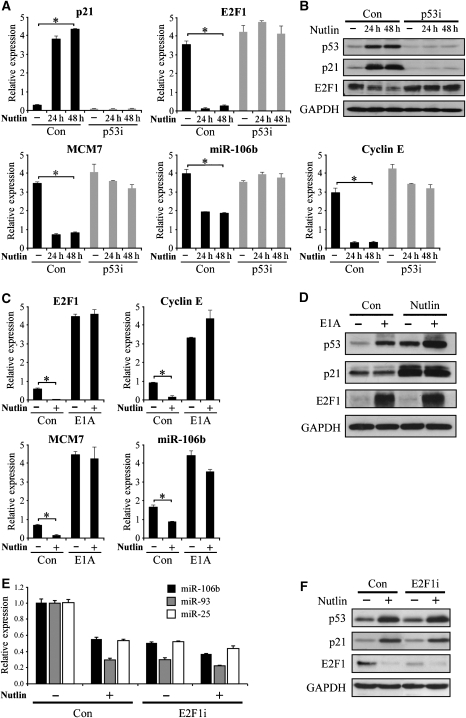
Having shown that representative miRs are activated by E2F1 in our system, we set to test whether their p53-dependent repression is mediated through modulation of E2F1 activity. To that end, we infected WI-38 cells with a retrovirus encoding for either an shRNA targeting p53 (p53i) or a control shRNA (con) and treated them with Nutlin-3, a small molecule that stabilizes the p53 protein by inhibiting its Mdm2-dependent ubiquitylation and degradation (Vassilev et al, 2004), thereby inducing p53 activation in a non-genotoxic manner. Nutlin treatment resulted in a robust p53 protein accumulation, accompanied by p21 mRNA and protein induction (Figure 5A and B), which was completely abrogated in the p53i cells. Remarkably, E2F1 mRNA and protein levels were downregulated upon Nutlin treatment in a p53-dependent manner. Cyclin E showed a similar pattern, supporting the notion that E2F1 downregulation was accompanied by a reduction in E2F activity. Accordingly, both MCM7 and its resident miR-106b were significantly downregulated in a p53-dependent manner (Figure 5A) along with other miRs from the ‘cell-cycle-associated co-cluster' (data not shown). Thus, treatment with Nutlin, a non-genotoxic p53 activator, resulted in a p53-dependent transcriptional repression of mRNAs and miRNAs with associated cell-cycle functions. To substantiate the causal relationship between the p53-dependent reduction of E2F1 activity and the repression of the miRs and their hosts, we treated control and E1A-expressing WI-38 cells with Nutlin. As depicted in Figure 5C and D, E1A induced the expression and prevented the Nutlin-dependent repression of E2F1 as well as of its target Cyclin E. Most significantly, E1A abolished the downregulation of MCM7 and miR-106b upon Nutlin treatment. A similar pattern was observed for miR-17-5p and its host c13orf25 (data not shown). Finally, we stably knocked down E2F1 using retroviral-encoded shRNA in WI-38 cells in combination with Nutlin treatment, and measured the levels of miRs-106b/25/93 (Figure 5E), as well as the protein levels of p53, p21 and E2F1 (Figure 5F). Indeed, the knock down of E2F1 resulted in reduced levels of the miRNAs. Supporting the notion that repression of the miR-106b/93/25 polycistron by p53 is mediated through E2F1 inhibition, Nutlin treatment of the E2F1-knockdown cells had very little effect as compared with the control cells.
Figure 5.
MCM7 and miR-106b are repressed by Nutlin-activated p53 in an E2F-dependent manner. (A, B) WI-38 were infected with a retrovirus encoding for either a small hairpin RNA targeting p53 (p53i) or a control shRNA (Con) and treated with 10 μM Nutlin-3 for 24 or 48 h. QRT–PCR (A) and western blotting (B) analyses demonstrated p53 stabilization by Nutlin, which resulted in activation of p21 and repression of E2F1 mRNA and protein levels. MCM7 and its resident miR-106b were repressed in a p53-dependent manner upon Nutin-3 treatment. (C, D) WI-38 cells were infected with E1A or an empty vector control (Con) and treated with 10 μM Nutlin-3 for 24 h. E1A elevated E2F transactivation activity, resulting in the induction of Cyclin E and E2F1 itself as well as of MCM7 and miR-106b. Nutlin treatment of the control cells repressed transcription of E2F1 and its targets. E1A abolished this repression, indicating that the repression of E2F1 by p53 is necessary for the p53-dependent downregulation of MCM7 and miR-106b. In (A, C), statistically significant difference in expression (t-test; P-value<0.01) between the non-treated samples and the Nutlin-treated samples (at both 24 and 48 h) is marked by asterisks. (E, F) WI-38 cells were infected with a retrovirus encoding for either a small hairpin RNA targeting E2F1 (E2F1i) or a control shRNA (Con) and treated with 10 μM Nutlin-3 for 48 h. QRT–PCR (E) and western blot (F) analyses demonstrated repression of E2F1 and its targets miRs-106b/93/25, as well as activation of p53 and p21 upon Nutlin treatment. E2F1 knockdown mimicked the effect of Nutlin treatment in repressing the miR-106b/93/25 polycistron. Nutlin treatment in the presence of E2F1 shRNA had little effect on the miRs, indicating that E2F1 inhibition mediates the repression of the miRs by Nutlin-activated p53. GAPDH protein levels serve as loading controls in (B, D, F). QRT–PCR data are represented as mean±s.d.
We therefore conclude that E2F1 inhibition by p53 is necessary for the downregulation of MCM7 and its resident miRNAs. The same mechanism may underlie the p53-dependent downregulation of additional miRs from the ‘cell-cycle co-cluster' and, more specifically, the three paralogous polycistrons.
Cell-cycle-associated miRNAs target key cell-cycle regulators and affect pivotal characteristics of proliferation
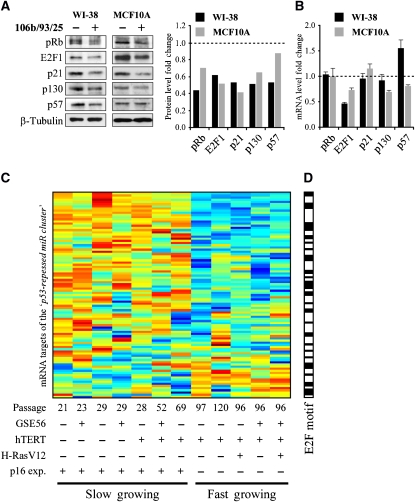
Next, we set out to identify the functions of the p53-repressed miRs. We focused on the miR-106b/93/25 polycistron as a representative member of the large family of miRs that includes also the miR-17-92 and miR-106a-92 polycistrons. We overexpressed the genomic region encoding miRs-106b/93/25, which corresponds to an intron of the MCM7 gene in young WI-38 cells and in MCF10A mammary epithelial cells, both characterized by low basal expression of these miRs. Following our previous computational prediction of E2F and miR-106b/93/25 involvement in a FFL, in which they both target a mutual set of genes (Shalgi et al, 2007), we compiled a list of their mutual predicted targets (Supplementary Table S5). Interestingly, many of these predicted targets participate in cell-cycle regulation (P-value for ‘cell-cycle' annotation enrichment=1.4 × 10−10). Another key cell-cycle regulator, p21, was recently reported as a target for miR-106b (Ivanovska et al, 2008), and is a known target of both E2F1 (Gartel et al, 1998) and p53. We then measured the protein levels of selected predicted targets in the miR-106b/93/25-overexpressing cells (Figure 6). We observed downregulation of p21, as well as of pRB and p130, which were suggested earlier, based on reporter assays, as potential targets of miR-106a and the miR-17-92 cluster, respectively (Volinia et al, 2006; Wang et al, 2008). Interestingly, E2F1, which was shown to be a target of miR-17-5p and miR-20a (O'Donnell et al, 2005), and is predicted by PicTar (Krek et al, 2005) to be a target for both miR-106b and miR-93, was significantly downregulated as well. We also observed downregulation of p57 in WI-38 cells, in agreement with PicTar predictions (Figure 6A; Supplementary Figure S4). Notably, these proteins have defined functions in the regulation of the cell cycle, most of them being negative regulators of proliferation. As the mRNA levels of p57, p21, pRb and p130 did not decrease in WI-38 cells and only marginally in MCF10A cells (Figure 6B), the reduction in their protein levels most likely stems from translational inhibition and not mRNA degradation. Considering the above, it is unlikely that the reduction in the targets' protein levels stems from reduced E2F transcriptional activity. In contrast, E2F1 mRNA levels were reduced in both cell lines that express the miR-106b/93/25 polycistron, in agreement with Petrocca et al (2008).
Figure 6.
Overexpression of miR-106b/93/25 polycistron results in silencing of cell-cycle-related genes. WI-38 primary fibroblasts and MCF10A mammary cells were infected with a retrovirus encoding for either the genomic region that contains miRs-106b/93/25 or an empty vector control. (A) Western blot analysis of cell-cycle-regulating targets of the overexpressed miRs. Overexpression of miRs-106b/93/25 reduced the protein levels of E2F1, pRb, p130, E2F1 and p21 in both cell types and of p57 in WI-38 cells. β-Tubulin levels serve as a loading control. The scanned blots were analyzed using the ImageJ software. Values represent the fold change of each protein relative to the empty vector-infected cells, and were normalized to the levels of β-tubulin. (B) QRT–PCR analysis of the mRNA levels of the genes presented in (A). Values represent the fold change of each mRNA relative to the empty vector-infected cells. Data are represented as mean±s.d. (C) Expression pattern of predicted targets of at least five miRs from the ‘p53-repressed miR cluster.' mRNA expression levels were derived from WI-38 cells that underwent immortalization and gradual in vitro transformation (the cell status is indicated below) as described by Milyavsky et al (2003). This expression pattern was found to be significantly coherent (EC score=0.14, EC P-value=5 × 10−3). See Supplementary dataset S4 for the expression values. (D) Promoter analysis performed on the genes from (C) (using AMADEUS) revealed enrichment for E2F motif (P-value=2.2 × 10−13). Genes with E2F motif in their promoter are indicated in black in the bar on the right (and in Supplementary dataset S4).
To gain independent support for the role of the ‘p53-repressed miR cluster,' we investigated the expression pattern of genes that are targeted by these miRs in the above-mentioned transformation system, where primary WI-38 cells were gradually transformed into tumorigenic cells (Milyavsky et al, 2005). Interestingly, targets harboring predicted sites for multiple miRs from the ‘p53-repressed miR cluster' within their 3′-UTR had significantly coherent expression patterns (Pilpel et al, 2001) during the transformation process (Figure 6C). This observation means that genes that are targeted by multiple miRs from the ‘p53-repressed miR cluster' are significantly co-expressed during the transformation process as compared with random sets of genes. Furthermore, not only are these targets expressed similarly to one another but also the actual expression pattern of many of them is consistent with the pro-proliferative role of the miRs that regulate them, i.e. the expression of the majority of the genes in this target set was decreased when cells gained the accelerated proliferation phenotype (designated as ‘fast growing'). Furthermore, promoter analysis, using the AMADEUS algorithm (Linhart et al, 2008), of these predicted target genes, revealed the E2F-binding site as one of the most highly enriched motifs (P-value=2.2 × 10−13). This supports our general notion that E2F cooperates with the ‘p53-repressed miR cluster' in regulating shared targets at the transcriptional and post-transcriptional levels. This suggests that the FFL consisting of E2F and miRs-106b/93/25 may be deregulated during cancer progression. We note that a fifth of the predicted targets of the miR cluster show a very different expression pattern (Figure 6C, bottom part), which may indicate more complex regulatory interactions.
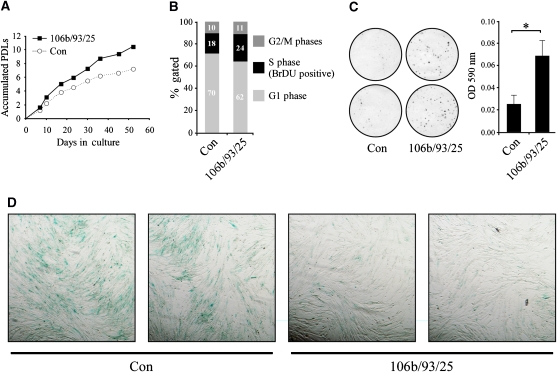
Having shown the molecular effects of the overexpression of the miR-106b/93/25 polycistron, we tested whether proliferation-related parameters such as growth rate, colony formation efficiency (CFE) and replicative senescence are affected by these miRs. As these miRs are significantly repressed by p53 during senescence, and considering the fact that they target several antiproliferation regulators, we predicted that their overexpression, similarly to p53 inactivation, would accelerate cellular growth rate and delay senescence. Indeed, as depicted in Figure 7, the miR-106b/93/25-overexpressing WI-38 cells demonstrated a moderate acceleration in proliferation rate and an increased fraction of S-phase cells (24% compared to 18%). Strikingly, these cells displayed a pronounced increase in the efficiency of young cells to form colonies when seeded at low density and reduced senescence-associated beta-galactosidase (SA-β-Gal) staining at late passages, indicating a delay of replicative senescence. Additionally, we evaluated the effect of miR-106b/93/25 overexpression in WI-38 cells on their CFE in combination with p53 inactivation. As depicted in Supplementary Figure S5, miR-106b/93/25 enhanced the CFE of both active and inactive p53-expressing cells. However, the effect of the overexpressed miRs was much more pronounced in the active p53 cells, augmenting their CFE by 20-fold as compared with only 2.6-fold increase in CFE in the inactive p53 cells. In fact, the effect of overexpression of the miRs on the CFE of the control cells was comparable to that of p53 inactivation. These observed phenotypes suggest that the transcriptional repression of miR-106b/93/25 and their paralogs mediates part of the antiproliferative effects of p53.
Figure 7.
Overexpression of miR-106b/93/25 polycistron in WI-38 cells promotes proliferation. Overexpression of miR-106b/93/25 polycistron in WI-38 cells promotes proliferation. (A) Growth curves for control (empty vector) and miR-106b/93/25-overexpressing cells. PDLs, population doublings. The difference between the growth curves was analyzed by paired t-test of the number of PDLs in each passage, and was found to be statistically significant (P-value=1.2 × 10−4) (B) Cell-cycle analysis of BrDU-labeled cells using fluorescence cytometer. miR-106b/93/25-expressing cells demonstrated increased proportion of S phase (BrDU positive) cells. (C) Colony formation assay. Cells were plated at low density and grown for 2 weeks. Plates were stained with crystal violet (left). The crystal violet was extracted with acetic acid and quantified with a spectrophotometer using a 590 nm filter (right). The difference was statistically significant (P-value<0.05). (D) Senescence-associated β-galactosidase staining depicting decreased level of senescence in miR-106b/93/25-expressing cells as compared with their empty vector control counterparts (Con).
Discussion
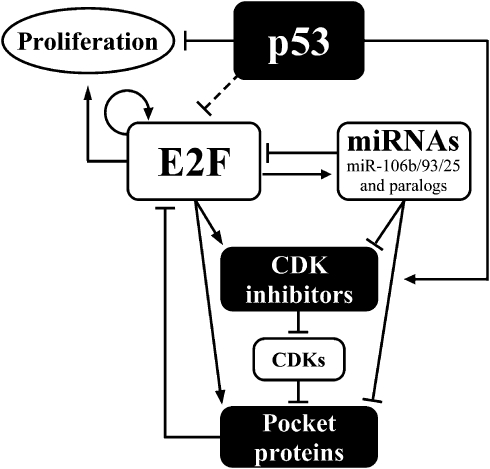
In the present study, we elucidate a complex regulatory network involving a group of cancer-related miRs. In this network, E2F1 transcriptionally controls the miR-106b/93/25 polycistron and its paralogs, and together they regulate a mutual set of target genes. In concordance with the growth acceleration that resulted from the overexpression of these miRs, many of their targets are considered antiproliferative cell-cycle regulators. Importantly, this intricate FFL is repressed by p53 through inhibition of E2F1. A schematic model for the proposed network is presented in Figure 8.
Figure 8.
A schematic model for the cell-cycle regulatory network comprising E2F, p53, miRs and other cell-cycle regulators. Arrows correspond to direct transcriptional activation, whereas bar-headed lines represent direct or indirect inhibition mediated by the following mechanisms: post-transcription gene silencing (miRNAs and their targets), protein binding and inactivation (pocket proteins and E2Fs; as well as CDK inhibitors and CDKs, that in turn inhibit pocket proteins by phosphorylation). The circular arrow represents E2F self-activating ability. Possible mechanisms underlying the repression of E2F by p53 are detailed in the discussion.
Employing three independent experiments, we identified a novel miR signature that is transcriptionally repressed by p53 in human primary cells and in breast cancers. Consistent with p53 function, many signature members, including the three paralogous polycistrons and miR-155, are considered oncogenic miRs and are overexpressed in diverse types of tumors (Eis et al, 2005; He et al, 2005; Volinia et al, 2006; Yanaihara et al, 2006). miR-15b and miR-16, which are considered to be tumor suppressor miRs (Cimmino et al, 2005), are exceptions in this regard.
Upon diverse stress stimuli, p53 is known to regulate different subsets of genes, resulting in alternative cellular outcomes (Oren, 2003). Consistently, the repression of the miRs was restricted to non-genotoxic contexts, namely, replicative senescence and Nutlin-induced Mdm2 inhibition, as doxorubicin treatment did not result in transcriptional repression of the miR cluster despite p53 activation. Our earlier studies have shown that Nutlin treatment induces p53-dependent senescence accompanied by upregulation of miR-34 (Kumamoto et al, 2008). Therefore, the repression of the miRs might be specific to p53-induced senescence triggered in this study by cellular aging or Nutlin treatment. As p53 is capable of inducing senescence in vivo (Xue et al, 2007), the observed miRNA repression in the wild-type p53-harboring breast tumors may be associated with increased senescence in these samples.
Co-clustering of coding mRNAs and microRNAs from the breast cancer study separated the p53-repressed miRs into two functional and regulatory categories, namely ‘cell cycle' and ‘immune response' (Figure 3). The expression-based division perfectly mirrored earlier reported functions of these miRs, including members of the three paralogous polycistrons that were co-clustered with cell-cycle-associated mRNAs, and were shown here and in additional reports (Hossain et al, 2006; Lu et al, 2007; Ivanovska et al, 2008) to promote cell proliferation. Another p53-repressed polycistron, miR-15b/16, was clustered with the cell-cycle genes. Indeed, members of this polycistron have been implicated in the regulation of cell-cycle progression (Linsley et al, 2007). The ‘immune response-associated co-cluster' included miR-155, which is commonly overexpressed in lymphomas (Eis et al, 2005), participates in the germinal center response (Thai et al, 2007), and is upregulated in chronic gastritis (Petrocca et al, 2008). The remaining members of this co-cluster were also implicated in immune responses, including miR-150 and miR-146 (Lodish et al, 2008) as well as miR-142 (Wu et al, 2007). Overexpression of miR-155 in primary cells did not affect the rate of proliferation (Supplementary Figure S3F), suggesting distinct functions for the members of the ‘immune response-associated co-cluster.' Interestingly, in addition to many cell-cycle-related coding genes reported to be repressed by p53, recent evidence indicates p53-mediated repression of immune response-related genes (e.g. interleukin-1β, interleukin-6 and Cxcl1 (Buganim et al, submitted) and SDF-1 (Moskovits et al, 2006)).
For the first time, we demonstrate that the three paralogous polycistronic miRNAs are coordinately activated by E2F1. Importantly, we establish E2F1 as the mediator of the p53-dependent repression of miRs-106b/93/25 and suggest that this mechanism underlies the repression of the two additional paralogous polycistrons. Upon Nutlin treatment, E2F1 protein levels were dramatically downregulated in a p53-dependent manner. A similar phenomenon was described earlier, and was attributed to enhanced ubiquitylation of E2F1 by an unknown ligase, resulting in proteasome-mediated degradation (Ambrosini et al, 2007). However, we demonstrate that Nutlin treatment also results in a robust p53-dependent E2F1 mRNA repression, in agreement with an earlier observation that overexpression of p53, as well as p21, results in downregulation of E2F1 mRNA (Ookawa et al, 2001). It is plausible that p53-mediated reduction in E2F1 protein inhibits E2F1 transcription as this gene contains an E2F motif in its promoter and is itself an E2F target gene (Johnson et al, 1994). Thus, even a slight reduction in E2F1 protein level might trigger a feedback loop that will result in significant reduction of both protein and mRNA levels. Considering this feedback loop, inhibition of E2F1 activity could also explain the observed repression of its mRNA and protein levels. Such inactivation may be indirectly mediated by the p53 target gene p21 through the inhibition of CDKs that inactivate the pocket proteins, which in turn inhibit E2F activity. Another mechanism for E2F1 inactivation by p53 was recently suggested by identifying BTG3 as a p53 target gene that directly binds E2F1 and inhibits its activity (Ou et al, 2007). Yet another possible mediator for p53-dependent E2F1 suppression is miR-34a, a direct transcriptional target of p53, which was recently suggested to induce senescence and to repress the E2F pathway (Tazawa et al, 2007).
Earlier studies have reported the effect of a single miR on a single target gene, such as miR-106b effect on p21 (Ivanovska et al, 2008). Others have described E2F-dependent activation of a single polycistron, miR-17-92 (Sylvestre et al, 2007; Woods et al, 2007), and miRs-106b/93/25 (Petrocca et al, 2008). We suggest that the three polycistrons, consisting of a family of 15 different miRs, are in fact transcriptionally co-regulated directly by E2F and indirectly by p53. In addition, many miRs in this family share highly similar seed sequences (Tanzer and Stadler, 2004). Thus, when an entire miR family is coordinately activated, its combinatorial and cumulative effects on mRNA targets may be profound. To recapitulate natural conditions, we combinatorially expressed three of the family miRs (miRs-106b/93/25), which are naturally co-transcribed, and demonstrated their effect on a set of target proteins. Future technologies allowing combinatorial knock down of an entire miR family may further establish their effects on other target genes. We show that miRs-106b/93/25 silence key members of the E2F pathway, including negative regulators of proliferation such as the pocket proteins pRb and p130 and the CDK inhibitors p21 and p57 (Figure 6). These and many other cell-cycle regulators are known E2F targets and are predicted to be silenced by miRs-106b/93/25 (Supplementary Table S5). Thus, we provide experimental evidence for our recent in silico predicted FFL motif (Shalgi et al, 2007).
We demonstrate here (Figure 6) a cancer-related manifestation of the concept of miR target avoidance (Farh et al, 2005; Stark et al, 2005). These studies introduced the concept of ‘spatio-temporal avoidance,' showing that miRs and their targets tend to avoid being expressed in the same tissue or at the same developmental time, thereby assisting to determine differentiation boundaries and transitions. This avoidance could reflect a direct negative regulatory effect of the miR on its target. Alternatively, miR target avoidance may be mediated through a common transcriptional program controlling both the miRs and their targets.
The overexpression of miRs-106b/93/25 phenotypically mimicked p53 inactivation in WI-38 cells, as evident from an elevated rate of proliferation, increased CFE and delay of senescence. Importantly, induction of senescence, which we suggest to be partially mediated by the repression of the polycistronic miRs described above, is considered one of the main mechanisms by which p53 suppresses tumor formation (Xue et al, 2007).
In summary, we present here another arm of p53's tight control of cell proliferation, senescence and tumor suppression. This involves an elaborate network encompassing miRs and their targets, which modulate cell fate both during normal growth and in cellular senescence.
Materials and methods
Cell culture
WI-38, MRC5, IMR90 (obtained from the ATCC) and PFCA179 (provided by Dr H Klocker) cells were cultured in MEM with 10% FCS (fetal calf serum), 1 mM sodium pyruvate, 2 mM L-glutamine and antibiotics. U2OS and H1299 cell lines were cultured in DMEM and RPMI, respectively, with 10% FCS and antibiotics. MCF10A cells were maintained in DMEM F12 supplemented with 5% horse serum, 0.5 μg/mlhydrocortisone, 0.1 mg/ml insulin, 0.1 μg/ml cholera toxin and 10 ng/ml EGF. All cells were maintained in a humidified incubator at 37°C and 5% CO2. Primary fibroblasts were passaged every 5–6 days. PDLs were calculated using the formula: PDLs=log(cell output/cell input)/log2. For colony formation assays, cells were plated at low density (0.1–0.2 cells/mm2), grown for 10–14 days and stained with crystal violet.
Plasmids and retroviral infections
The retrovirus encoding for GSE56 was described by Milyavsky et al (2005). shRNAs targeting p53 (p53i), or mouse NOXA (control shRNA) were stably expressed using pRetroSuper and were described by Berkovich and Ginsberg (2003). pRetroSuper-E2F1 was described by Korotayev et al (2008). ER-E2F1 was described by Vigo et al (1999). E1A was expressed from pBabe-puro-E1A12S (a gift from K Helin). For expression of miRs-106b/93/25, a 1-kb human genomic fragment was cloned with the primers 5′-ggatcctatcctgcgcctttcc-3′ and 5′-cacatggccacagaagac-3′ into miR-Vec (Voorhoeve et al, 2006). For expression of miR-155, a 238-bp human genomic fragment was cloned with the primers 5′-gtggcacaaaccaggaag-3′ and 5′-tatccagcagggtgactc-3′ into miR-Vec. Retrovirus infection procedures were described by Milyavsky et al (2003).
RNA preparation and quantitative real-time PCR
RNA was extracted with TRI reagent (Molecular Research Center Inc.). For mRNA quantification, a 2 μg aliquot of total RNA was reverse transcribed using Bio-RT (Biolab) and random hexamers. Quantitative real-time PCR (QRT–PCR) was performed using Platinum SYBR Green qPCR SuperMix (Invitrogen). mRNA levels were normalized to the level of GAPDH in the same sample. Primer sequences are listed in Supplementary Table S6A. For miRNA quantification, TaqMan miRNA assays (Applied Biosystems) were used according to the manufacturer's protocol. Levels were normalized to the U6 control gene. All QRT–PCR reactions were performed on ABI7300 machine. Results are presented as mean and standard deviation for two or three duplicate runs.
miRNA microarrays, data analysis and clustering
The miRNA profiling presented in Figure 1A was performed as follows: RNA was extracted from WI-38 cells using TRI reagent as described above, labeled with Hy5 and hybridized on Exiqon's miRCURY™ LNA Array (v.8.1) with a common reference Hy3-labeled RNA pool. Data are provided as Supplementary dataset S1. Two biological replicates were performed for each sample type. Hy5/Hy3 ratios were log2 transformed and filtered such that miRs that were undetected in 11 or 12 samples were discarded. Duplicates were averaged, such that each miR was represented by six values, corresponding to the six different samples. For each miR, a credibility value was calculated as one minus the average of the six standard deviations (s.d.) between the duplicates. A duplicate that had one missing value was set as the detected value and was assigned with high s.d. The 5% most non-credible miRs were discarded. Data were clustered using hierarchical clustering (average linkage), with 20 clusters. The miRNA profiling presented in Figure 1B was performed as follows: RNA was extracted from young and old WI-38 and MRC5 cells using TRIZOL (Invitrogen) according to the manufacturer's protocol and was used for biotin labeling and hybridization on the version 4.0 miR arrays (Ohio State University) as described by Liu et al (2004). Data were normalized and log2-transformed. Data are provided as Supplementary dataset S2. Data were clustered using hierarchical clustering (average linkage), with 10 clusters. The miRNA profiling presented in Figure 1C was performed as follows: RNA was extracted as described by Sorlie et al (2006). Samples were hybridized on Agilent's miRNA arrays (beta version of V1) at the Agilent's facilities in St Clara, US by HJ. Data are provided as Supplementary dataset S3. Two outlier samples (both belonging to the mutant p53 set) were discarded (see Supplementary Figure S6). Data were clustered using hierarchical clustering (average linkage), with 25 clusters.
Microarray data can be downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). Accession numbers are as follows: GSE12450 (the WI-38 p53-dependent senescence study), GSE12821 (the WI-38 and MRC5 senescence study) and GSE12848 (The Breast Cancer Study).
Co-clustering of miRNA and mRNA expression data from human breast cancer samples
mRNA array data were filtered and normalized as in Sorlie et al (2006) (GEO accession number GSE3155) and only samples that were included in the miRNA analysis were used. Data were log2-transformed, and replicate mRNA probes were averaged. Only variable mRNAs that differ by at least 1.5-fold from their median expression in at least 40% of the samples were considered. miRNA data were centered such that the intensity values for each miR were divided by their mean, and log2 transformed. The combined mRNA and miRNA data were then clustered using hierarchical clustering (average linkage) with 40 clusters. miRs from the ‘breast cancer p53-repressed miR cluster' were mapped to the resulting co-clusters, and were found to reside in two clusters, that were then analyzed for functional enrichment using DAVID (Dennis et al, 2003), and for enrichment of sequence motifs in their corresponding ENSMBL gene promoters using AMADEUS (Linhart et al, 2008).
Analysis of miRNA targets expression coherence
The entire set of miRNA expression profiles was clustered into 20 miR clusters based on the above expression data (WI-38 young versus senescent, along with p53 inactivation). Then, we compiled a set of predicted targets for the miRs from each cluster using PicTar (Krek et al, 2005). Specifically, for each of the 20 miR clusters, a series of potential sets of targets were created. The first set consisted of mRNAs predicted to be targeted by at least one miR from the miR cluster. The second set consisted of mRNAs predicted to be targeted by at least two miRs from the miR cluster, and so on. The expression coherence (EC) score, a measure of expression similarity (Pilpel et al, 2001), was then computed for each set of targets according to their expression described by Milyavsky et al (2005) (expression data are also available as Supplementary dataset S5). The most significantly coherent expression pattern belonged to the set of genes that had target sites for at least five miRs from the ‘p53-repressed miR cluster' (EC P-value=5 × 10−3). The expression values for this gene set in the data from Milyavsky et al (2005) and prediction of E2F sites are found in Supplementary Figure S4.
Immunoblot analysis
Western blots were performed as described by Milyavsky et al (2005). The following antibodies were used: α-p53 pAb H-47 (produced in our laboratory), α-p21 sc-377 (Santa Cruz), α-E2F1 sc-193 (Santa Cruz), α-GAPDH MAB374 (Chemicon), α-p130 sc-317 (Santa Cruz), α-p57 sc-8298 (Santa Cruz), α-pRb 554136 (Pharmingen), and α–β-tubulin T7816 (Sigma).
Cell-cycle analysis
Cells were labeled for 30 min with 10 μM BrdU (Sigma), fixed with 70% EtOH/HBSS (2 h, −20°C), treated with 2 M HCl/0.5% Triton, washed and treated with 0.1 M Na2B4O7 pH 8.5, and stained with FITC-conjugated anti-BrdU (Becton Dickinson) and 10 μg/ml propidium iodide. Samples were analyzed using a FACSort machine (Becton Dickinson). At least 1 × 104 events were recorded per sample.
SA-β-Gal activity assay
Cells were fixed with 3% formaldehyde/PBS for 5 min, washed with PBS and incubated for 16 h at 37°C with a solution containing 1 mg/ml X-gal, 40 mM citric acid, sodium phosphate, pH 6.0, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 150 mM NaCl and 2 mM MgCl2.
Chromatin immunoprecipitation
DNA–protein complexes were immunoprecipitated from U2OS cells using the ChIP assay kit (Upstate Biotechnology), according to the manufacturer's protocol with the following polyclonal antibodies: α-E2F1 sc-193 (Santa Cruz) and α-HA sc-805 (Santa Cruz); the latter served as a control for nonspecific DNA binding. The precipitated DNA was subjected to QRT–PCR analysis using specific primers corresponding to each predicted E2F site, as well as primers for normalization (β-tubulin) and negative control for E2F1 binding (β-actin coding region). Primer sequences are listed in Supplementary Table S6B.
Supplementary Material
Supplementary Information
Supplementary Dataset 1
Supplementary Dataset 2
Supplementary Dataset 3
Supplementary Dataset 4
Supplementary Dataset 5
Acknowledgments
This study was supported by a Center of Excellence grant from the Flight Attendant Medical Research Institute; EC FP6 funding (contract no. 502983); FP7 funding (ONCOMIRS, agreement 201102); the Ben May Charitable Trust; the Israel Science Foundation and the Yad Abraham Center for Cancer Diagnosis and Therapy. This research was supported in part by the Intramural Research Program, NIH, NCI and CCR. VR is the incumbent of the Norman and Helen Asher Professorial Chair Cancer Research at the Weizmann institute. YP is an incumbent of the Rothstein Career Development Chair in Genetic Diseases. RS is a fellow of the Horowitz Foundation for Complexity Sciences. GHN was supported by the Howard Hughes Medical Institute Grant for Graduate Medical Education. This publication reflects only authors' views. The European Commission is not liable for any use that may be made of the information herein. We thank Agilent Technologies for access to the early version of their miRNA arrays and for their support.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ambrosini G, Sambol EB, Carvajal D, Vassilev LT, Singer S, Schwartz GK (2007) Mouse double minute antagonist Nutlin-3a enhances chemotherapy-induced apoptosis in cancer cells with mutant p53 by activating E2F1. Oncogene 26: 3473–3481 [DOI] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Berkovich E, Ginsberg D (2003) ATM is a target for positive regulation by E2F-1. Oncogene 22: 161–167 [DOI] [PubMed] [Google Scholar]
- Campisi J, d'Adda di Fagagna F (2007) Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol 8: 729–740 [DOI] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT (2007) Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 26: 745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM (2005) miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA 102: 13944–13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G. Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4: P3. [PubMed] [Google Scholar]
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE (2005) Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA 102: 3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ (2006) Oncomirs––microRNAs with a role in cancer. Nat Rev Cancer 6: 259–269 [DOI] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP (2005) The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science 310: 1817–1821 [DOI] [PubMed] [Google Scholar]
- Fattaey AR, Harlow E, Helin K (1993) Independent regions of adenovirus E1A are required for binding to and dissociation of E2F–protein complexes. Mol Cell Biol 13: 7267–7277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartel AL, Goufman E, Tevosian SG, Shih H, Yee AS, Tyner AL (1998) Activation and repression of p21(WAF1/CIP1) transcription by RB binding proteins. Oncogene 17: 3463–3469 [DOI] [PubMed] [Google Scholar]
- Ginsberg D, Mechta F, Yaniv M, Oren M (1991) Wild-type p53 can down-modulate the activity of various promoters. Proc Natl Acad Sci USA 88: 9979–9983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ (2007) A microRNA component of the p53 tumour suppressor network. Nature 447: 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM (2005) A microRNA polycistron as a potential human oncogene. Nature 435: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J, Benchimol S (2003) Transcriptional repression mediated by the p53 tumour suppressor. Cell Death Differ 10: 404–408 [DOI] [PubMed] [Google Scholar]
- Hossain A, Kuo MT, Saunders GF (2006) Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol 26: 8191–8201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain SP, Harris CC (1999) p53 mutation spectrum and load: the generation of hypotheses linking the exposure of endogenous or exogenous carcinogens to human cancer. Mutat Res 428: 23–32 [DOI] [PubMed] [Google Scholar]
- Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson AL, Linsley PS, Cleary MA (2008) MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol 28: 2167–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DG, Ohtani K, Nevins JR (1994) Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev 8: 1514–1525 [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ (2005) RAS is regulated by the let-7 microRNA family. Cell 120: 635–647 [DOI] [PubMed] [Google Scholar]
- Kern SE, Kinzler KW, Baker SJ, Nigro JM, Rotter V, Levine AJ, Friedman P, Prives C, Vogelstein B (1991) Mutant p53 proteins bind DNA abnormally in vitro. Oncogene 6: 131–136 [PubMed] [Google Scholar]
- Korotayev K, Chaussepied M, Ginsberg D (2008) ERK activation is regulated by E2F1 and is essential for E2F1-induced S phase entry. Cell Signal 20: 1221–1226 [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N (2005) Combinatorial microRNA target predictions. Nat Genet 37: 495–500 [DOI] [PubMed] [Google Scholar]
- Kumamoto K, Spillare ES, Fujita K, Horikawa I, Yamashita T, Appella E, Nagashima M, Takenoshita S, Yokota J, Harris CC (2008) Nutlin-3a activates the p53 tumor suppressor to both down-regulate ING2 and up-regulate mir-34a, b and c expression and induce senescence. Cancer Res 68: 3193–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhart C, Halperin Y, Shamir R (2008) Transcription factor and microRNA motif discovery: the Amadeus platform and a compendium of metazoan target sets. Genome Res 18: 1180–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, Chau N, Cleary M, Jackson AL, Carleton M, Lim L (2007) Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol 27: 2240–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, Alder H, Bullrich F, Negrini M, Croce CM (2004) An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA 101: 9740–9744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish HF, Zhou B, Liu G, Chen CZ (2008) Micromanagement of the immune system by microRNAs. Nature Rev 8: 120–130 [DOI] [PubMed] [Google Scholar]
- Lu Y, Thomson JM, Wang HY, Hammond SM, Hogan BL (2007) Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol 310: 442–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milyavsky M, Shats I, Erez N, Tang X, Senderovich S, Meerson A, Tabach Y, Goldfinger N, Ginsberg D, Harris CC, Rotter V (2003) Prolonged culture of telomerase-immortalized human fibroblasts leads to a premalignant phenotype. Cancer Res 63: 7147–7157 [PubMed] [Google Scholar]
- Milyavsky M, Tabach Y, Shats I, Erez N, Cohen Y, Tang X, Kalis M, Kogan I, Buganim Y, Goldfinger N, Ginsberg D, Harris CC, Domany E, Rotter V (2005) Transcriptional programs following genetic alterations in p53, INK4A, and H-Ras genes along defined stages of malignant transformation. Cancer Res 65: 4530–4543 [DOI] [PubMed] [Google Scholar]
- Moskovits N, Kalinkovich A, Bar J, Lapidot T, Oren M (2006) p53 attenuates cancer cell migration and invasion through repression of SDF-1/CXCL12 expression in stromal fibroblasts. Cancer Res 66: 10671–10676 [DOI] [PubMed] [Google Scholar]
- Naume B, Zhao X, Synnestvedt M, Borgen E, Russnes EG, Lingjærde OC, Strømberg M, Wiedswang G, Kvalheim G, Kåresen R, Nesland JM, Børresen-Dale AL, Sørlie T (2007) Presence of bone marrow micrometastasis is associated with different recurrence risk within molecular subtypes of breast cancer. Mol Oncol 1: 160–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT (2005) c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435: 839–843 [DOI] [PubMed] [Google Scholar]
- Ookawa K, Tsuchida S, Kohno T, Yokota J (2001) Alterations in expression of E2F-1 and E2F-responsive genes by RB, p53 and p21(Sdi1/WAF1/Cip1) expression. FEBS Lett 500: 25–30 [DOI] [PubMed] [Google Scholar]
- Oren M (2003) Decision making by p53: life, death and cancer. Cell Death Differ 10: 431–442 [DOI] [PubMed] [Google Scholar]
- Ossovskaya VS, Mazo IA, Chernov MV, Chernova OB, Strezoska Z, Kondratov R, Stark GR, Chumakov PM, Gudkov AV (1996) Use of genetic suppressor elements to dissect distinct biological effects of separate p53 domains. Proc Natl Acad Sci USA 93: 10309–10314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou YH, Chung PH, Hsu FF, Sun TP, Chang WY, Shieh SY (2007) The candidate tumor suppressor BTG3 is a transcriptional target of p53 that inhibits E2F1. EMBO J 26: 3968–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, Cavazzini L, Volinia S, Alder H, Ruco LP, Baldassarre G, Croce CM, Vecchione A (2008) E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 13: 272–286 [DOI] [PubMed] [Google Scholar]
- Pilpel Y, Sudarsanam P, Church GM (2001) Identifying regulatory networks by combinatorial analysis of promoter elements. Nat Genet 29: 153–159 [DOI] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M (2007) Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 26: 731–743 [DOI] [PubMed] [Google Scholar]
- Ren B, Yu G, Tseng GC, Cieply K, Gavel T, Nelson J, Michalopoulos G, Yu YP, Luo JH (2006) MCM7 amplification and overexpression are associated with prostate cancer progression. Oncogene 25: 1090–1098 [DOI] [PubMed] [Google Scholar]
- Ryan KM, Phillips AC, Vousden KH (2001) Regulation and function of the p53 tumor suppressor protein. Curr Opin Cell Biol 13: 332–337 [DOI] [PubMed] [Google Scholar]
- Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC (2008) MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 299: 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R, Lieber D, Oren M, Pilpel Y (2007) Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput Biol 3: e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Wang Y, Xiao C, Johnsen H, Naume B, Samaha RR, Borresen-Dale AL (2006) Distinct molecular mechanisms underlying clinically relevant subtypes of breast cancer: gene expression analyses across three different platforms. BMC Genomics 7: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM (2005) Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 123: 1133–1146 [DOI] [PubMed] [Google Scholar]
- Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P (2007) An E2F/miR-20a autoregulatory feedback loop. J Biol Chem 282: 2135–2143 [DOI] [PubMed] [Google Scholar]
- Tabach Y, Milyavsky M, Shats I, Brosh R, Zuk O, Yitzhaky A, Mantovani R, Domany E, Rotter V, Pilpel Y (2005) The promoters of human cell cycle genes integrate signals from two tumor suppressive pathways during cellular transformation. Mol Syst Biol 1: 2005.0022 10.1038/msb4100030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer A, Stadler PF (2004) Molecular evolution of a microRNA cluster. J Mol Biol 339: 327–335 [DOI] [PubMed] [Google Scholar]
- Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H (2007) Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G(1)-arrest. Cell Cycle 6: 1586–1593 [DOI] [PubMed] [Google Scholar]
- Tazawa H, Tsuchiya N, Izumiya M, Nakagama H (2007) Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA 104: 15472–15477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K (2007) Regulation of the germinal center response by microRNA-155. Science 316: 604–608 [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA (2004) In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303: 844–848 [DOI] [PubMed] [Google Scholar]
- Vigo E, Muller H, Prosperini E, Hateboer G, Cartwright P, Moroni MC, Helin K (1999) CDC25A phosphatase is a target of E2F and is required for efficient E2F-induced S phase. Mol Cell Biol 19: 6379–6395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103: 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH, Agami R (2006) A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124: 1169–1181 [DOI] [PubMed] [Google Scholar]
- Wang Q, Li YC, Wang J, Kong J, Qi Y, Quigg RJ, Li X (2008) miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Natl Acad Sci USA 105: 2889–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ML, George LK, Grant GD, Perou CM (2006) Common markers of proliferation. Nat Rev Cancer 6: 99–106 [DOI] [PubMed] [Google Scholar]
- Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, Botstein D (2002) Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell 13: 1977–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods K, Thomson JM, Hammond SM (2007) Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem 282: 2130–2134 [DOI] [PubMed] [Google Scholar]
- Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N (2007) miRNA profiling of naive, effector and memory CD8 T Cells. PLoS ONE 2: e1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Shalgi R, Fodstad O, Pilpel Y, Ju J (2006) Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin Cancer Res 12: 2014–2024 [DOI] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW (2007) Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445: 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC (2006) Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9: 189–198 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Dataset 1
Supplementary Dataset 2
Supplementary Dataset 3
Supplementary Dataset 4
Supplementary Dataset 5