Abstract
The link between Ras transformation and enhanced cell migration due to altered integrin signaling is well established in tumorigenesis, however there remain gaps in our understanding of its mechanism. The Ras suppressor, Rsu-1, has recently been linked to the IPP (integrin-linked kinase {ILK}, PINCH-1/LIMS1, parvin) focal adhesion complex based on its interaction with the LIM 5 domain of PINCH1. Defining the role of the Rsu1-PINCH1-ILK-parvin complex in tumorigenesis is important because both ILK and PINCH1 are elevated in certain tumors while ectopic expression of Rsu-1 blocks tumorigenesis. Our studies previously identified an alternatively-spliced isoform of Rsu-1 in high-grade gliomas. We report here the detection of a truncated (p29) Rsu-1 protein, which correlates with the presence of the alternatively spliced Rsu-1 RNA. This RNA and the respective protein were detected in human tumor cell lines that contain high levels of activated Ras, and inhibitor studies demonstrate that the Mek-ERK pathway regulates expression of this truncated Rsu-1 product. We also show that Rsu-1 colocalizes with ILK at focal contacts and co-immunoprecipitates with the ILK-PINCH1 complex in non-transformed cells, but following Ras transformation the association of Rsu-1 with the PINCH1-ILK complex is greatly reduced. Using a human breast cancer cell line, our in vitro studies demonstrate that the depletion of Rsu-1 full-length protein enhances cell migration coincident with an increase in Rac-GTP while the depletion of the p29 Rsu-1 truncated protein inhibits migration. These findings indicate that Rsu-1 may inhibit cell migration by stabilizing the IPP adhesion complex and that Ras activation perturbs this inhibitory function by modulating both Rsu-1 splicing and association of full-length Rsu-1 with IPP. Hence, our findings demonstrate that Rsu-1 links the Ras pathway with the IPP complex and the perturbations of cell attachment-dependent signaling that occur in the malignant process.
Keywords: Rsu-1, PINCH1, Integrin-linked kinase, Adhesion, Ras, Focal adhesion, Migration
Introduction
The loss of integrin engagement and detachment of cells from the extracellular matrix can lead to apoptosis in a process referred to as anoikis (Frisch and Francis, 1994). Many tumor cells are insensitive to detachment-induced cell death and this insensitivity contributes to their motility and metastatic potential as well as the capacity to grow in an anchorage-independent manner. Oncogene activation enhances the resistance to anoikis observed in tumor cells, and the ectopic expression of tumor suppressor genes can restore sensitivity to detachment in some circumstances (Davies et al., 1998; Khwaja et al., 1997; Koul et al., 2001; Lu et al., 1999; Rosen et al., 2000). Hence, the regulation of cell attachment signaling is critical for control of tumor growth.
Studies in our laboratory have focused on characterization of the Rsu-1 protein, which was originally isolated in an expression cloning assay based on its ability to suppress transformation by the Ras oncogene (Cutler et al., 1992). Rsu-1 is a highly conserved, ubiquitously expressed single-copy gene that encodes an LRR (leucine-rich repeat) protein (Cutler et al., 1992; Tsuda and Cutler, 1993). Ectopic expression of Rsu-1 prevented Ras oncogene-induced phenotypic transformation, inhibited anchorage-independent growth of rodent and human tumor cell lines and blocked tumor formation in a nude mouse xenograft model (Cutler et al., 1992; Tsuda et al., 1995; Vasaturo et al., 2000). The human Rsu-1 locus maps to10p13, a region that is deleted in high-grade gliomas, and an alternatively spliced Rsu-1 mRNA that encodes a truncated and unstable protein product occurs in 30% of high-grade gliomas (Chunduru et al., 2002). Ectopic Rsu-1 expression altered actin cytoskeleton organization and blocked the activation of Jun kinase and ROCK, but not ERK, by growth factor stimulation (Masuelli and Cutler, 1996; Vasaturo et al., 2000).
Recently we and others reported that Rsu-1 binds to the LIM-domain protein PINCH1 (Dougherty et al., 2005; Kadrmas and Beckerle, 2004). PINCH1 contains five LIM domains (LIM 1-5) and functions as a scaffolding protein. The LIM 1 domain of PINCH1 binds to the aminoterminal ankyrin repeat domain of the integrin-linked kinase (ILK) and can modulate ILK activity (Tu et al., 1999). The LIM 4 domain of PINCH1 binds to the SH2-SH3 protein Nck2 (Tu et al., 1998). Our data revealed that the LIM 5 domain of PINCH1 binds to Rsu-1, and that Rsu-1 colocalized with PINCH1 in focal adhesions (Dougherty et al., 2005). PINCH1, in association with ILK and α-parvin, mediates cell matrix-adhesion functions in part by linking focal adhesion contacts to the actin cytoskeleton (Herreros et al., 2000; Nikolopoulos and Turner, 2000, 2001; Tu et al., 1999; Zhang et al., 2002a,b). ILK binds to the cytoplasmic domain of the β-integrins (Hannigan et al., 1996) through the respective carboxyterminal domain. The aminoterminal ankyrin repeat region of ILK interacts with the LIM-domain proteins PINCH1 and 2 (Tu et al., 1999, 2001) and paxillin (Nikolopoulos and Turner, 2001). Parvin, which also binds to ILK, can bind F-actin as well as paxillin, providing another mechanism to localize ILK to focal adhesions (Legate et al., 2006; Nikolopoulos and Turner, 2002). Studies demonstrated that both PINCH1 and ILK are required for the localization of the complex to focal adhesions (Tu et al., 2001), and the inhibition of PINCH-ILK interaction in mammalian cells inhibited spreading and reduced motility, suggesting that PINCH is required for ILK activity (Zhang et al., 2002a, b). In this study we examined the relationship of Rsu-1 to the IPP complex in the context of Ras activation. We demonstrate that expression of a truncated form, p29 Rsu-1, initially observed in high-grade gliomas, correlates with Ras activation in tumor cell lines. We also demonstrate that the p29 Rsu-1 does not bind to PINCH1. The association of full-length Rsu-1 with the IPP complex is reduced by Ras transformation, and the Ras-dependent effects on Rsu-1 can be partially restored by blocking the Mek-Erk pathway. Additionally, MDA-MB-468 breast cancer cells depleted of p33 Rsu-1 showed enhanced cell migration and Rac activation. We conclude that Rsu-1 promotes adhesion and inhibition of migration as a component of the IPP complex. This function is actively altered by Ras signaling and these data lend insight to the tumor suppressor effects of Rsu-1.
Materials and methods
Cell lines
The human breast cancer cell lines used in the study (MCF7, T47D, MDA-MB-231, MDA-MB-468) were obtained from the American Type Culture Collection. The immortalized human astrocytes (E6/E7/hTERT) and the Ras-transformed version (E6/E7/hTERT/Ras) were provided by Dr. Russell Pieper and the cells were propagated as described (Sonoda et al., 2001a,b). The A7r5 (rat vascular smooth muscle) cell line was cultured as previously described (Burgstaller and Gimona, 2004). Cos-1 cells were propagated as described (Dougherty et al., 2005).
Western blotting
The preparation of the cell lysates in RIPA buffer and the Western blotting procedures were performed as described previously (Dougherty et al., 2005; Galbaugh et al., 2006). The anti-Rsu-1 carboxyterminal antibody has been described (Dougherty et al., 2005). The anti-aminoterminal Rsu-1 antibody was prepared in rabbits by immunization with an aminoterminal peptide and followed by affinity purification. The antibodies used for Western blotting include mouse anti-PINCH1 clone 49 (BD Biosciences, San Diego, CA, USA), mouse anti-ILK clone 65.1.9 (Upstate Biotechnology, Charlottesville, VA, USA), rabbit anti-ILK (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-Rac1 clone 102 (BD Biosciences), and anti-β-actin clone AC15 (Sigma, St. Louis, MO, USA). Immunoprecipitations were performed following the lysis of the cells with buffer consisting of 1% NP40, 10% glycerol, 50 mM NaCl, 25 mM Tris (pH 7.5), and protease and phosphatase inhibitors, and clearing by centrifugation at 12,000g for 5 min. Immunoprecipitates were collected with 1 µg of primary antibody and protein A agarose for 2 h on a rocker platform at 4°C. The precipitates were washed 4 times with lysis buffer prior to addition of gel loading buffer and boiling for 5 min.
RT-PCR
The RT-PCR of total RNA from human tumor cell lines was performed as described previously (Chunduru et al., 2002). Primers TT11 (forward 5’-GCTACCTTCCGTGACCATGT) and TT15 (reverse 5’-CCCTTCCTTATC TTTCTTGG) were used. The RT-PCR products were separated by agarose gel electrophoresis and analyzed by Southern blot following transfer to nylon membranes and hybridization to a probe for Rsu-1. In addition, the RT-PCR products were TA-cloned and sequenced to confirm their identity as the Rsu-1 altered splice product (Chunduru et al., 2002).
siRNA
Depletion of Rsu-1 and truncated Rsu-1 was accomplished using siRNA in a reverse transfection protocol with RNAiMax lipofection reagent (Invitrogen, Carlsbad CA, USA). The siRNAs were used at 75 nM concentration in cell culture. The control siRNA was a documented negative control siRNA (Qiagen, Valencia, CA, USA). The sequences of the sense strands of the siRNAs (Invitrogen) are: Rsu-1: 5’UCAACGGCCUCUUUACCUUdTdT, Truncated Rsu-1 (RsuJ): 5’AGAACUAGCCUCUACGGCAUU.
Immunofluorescence microscopy
The following cell lines were used to localize endogenous Rsu-1: A7r5, Cos-7, and E6/E7/hTERT (E6/7) and E6/E7/hTERT/Ras (E6/7/Ras) human astrocytes. All cells were plated on glass coverslips coated with human plasma fibronectin at 10 µg/ml (Roche, Indianapolis, IN, USA), cultured overnight at 37°C, and assayed for immunofluorescence the next day in the following protocol: cells were rinsed briefly in phosphate-buffered saline (PBS), then fixed on ice with cold methanol for 5 min. Cells were washed three times in PBS and incubated in 0.25 % Triton X-100 for 10 min. After three additional washes in PBS, the cells were blocked in 2% bovine serum albumin (BSA) for 1 h. Samples were incubated with primary antibodies diluted in 2% BSA for 1 h followed by three washes in PBS. Alexa Fluor anti-rabbit and anti-mouse secondary antibodies (Invitrogen) were diluted in 2% BSA, with or without ToPro-3 iodide nuclear stain (Molecular Probes/Invitrogen) and incubated for 30 min followed by three washes in PBS. Coverslips were mounted on glass slides with Prolong Antifade solution (Invitrogen) and dried overnight. High magnification fluorescence images were obtained with a Zeiss Axiovert 100 M confocal microscope equipped with a 100× 1.4 N.A. objective. Image acquisition and post-acquisition analysis were performed with Zeiss LSM software and Adobe Photoshop 7.0 software. Anti-aminoterminal Rsu-1 rabbit polyclonal, monoclonal anti-ILK antibody clone 65.1.9 (BD Biosciences), monoclonal anti-focal adhesion kinase (FAK) antibody clone 77 (BD Biosciences), and monoclonal anti-phosphotyrosine (pTyr) antibody clone PY99 (Santa Cruz) were used for immunofluorescence analysis.
Cell migration assays
Cell migration and invasion assays on human tumor cell lines were performed at 72 h post transfection of siRNA. The cells were seeded in serum-free medium at 2 × 104 cells per chamber into the top part of a Boyden chamber (BD Biosciences) and the chamber was inserted into a well containing complete medium with 10% fetal bovine serum. The cells were seeded in 8-µm pore chambers with or without matrigel coating. At 20 h post seeding the cells were removed from the top chamber by swabbing and the cells on the bottom of the membrane were fixed and stained (Albini et al., 1987). The membranes were mounted on slides and photographed, and the cells were enumerated microscopically. The percentage of migrating cells and the percentage of invading cells were calculated from quadruplicate wells. Migration was determined as the number of membrane invading cells/total number of cells seeded × 100. The invasion results were calculated by normalizing the number of cells migrating through matrigel to those migrating through the uncoated membrane.
Detection of GTP-Ras and GTP-Rac
Breast cancer cell lines were serum starved for 24 h then stimulated with EGF for 7.5 min or left untreated. The level of Ras-GTP was determined as described (Taylor and Shalloway, 1996). Cells were lysed in buffer containing 25 mM HEPES (pH 7.5), 150 mM NaCl, 1 % NP-40, 0.25 % Na deoxycholate, 10 % glycerol, 25 mM NaF, 10 mM MgCl2, 1 mM EDTA, 1 mM Na vanadate, 10 mg/ml leupeptin, and 10 mg/ml aprotinin, and centrifuged at 12,000g for 5 min. Equal amounts of lysate containing 1 mg protein were used for affinity binding to glutathione– Sepharose-GST-RBD, which contains amino acids 1–149 of cRaf-1 fused to GST, for 30 min at 4°C with agitation. As controls lysates were incubated with or without 1 mM GDP or 0.5 mM GTPγS for 10 min prior to binding. Beads were washed three times with lysis buffer, resuspended in SDS–PAGE sample buffer, and boiled for 5 min. The bound proteins were resolved on 12 % gels and subjected to Western blotting; 5% of the total lysate was subjected to SDS–PAGE for detection of “total Ras”. Blots were probed with anti-pan Ras clone OP22 (Calbiochem, San Diego, CA, USA).
The levels of Rac-GTP were determined by a similar method. Lysates of stimulated cells were prepared as described above and bound to GST-p21 binding domain of Pak1 (GST-PBD) (BD Biosciences) (Usui et al., 2003). The bound and unbound proteins were detected by Western blotting with mouse anti-Rac1 clone 102 (BD Biosciences).
Results
A truncated p29 Rsu-1 protein is specifically detected in Ras-activated tumor cell lines
Rsu-1 was isolated based on its ability to suppress transformation by activated Ras. The main Rsu-1-interacting protein, PINCH1, is an adaptor protein that binds to ILK. Since both PINCH1 and ILK exhibit increased expression in tumors and in tumor stroma (Ahmed et al., 2003; Dai et al., 2003; Gao et al., 2004; Graff et al., 2001; Ito et al., 2003; Marotta et al., 2001, 2003; Wang-Rodriguez et al., 2002), we examined both the level of Rsu-1 expression in tumor cell lines and its relationship to that of PINCH1 and ILK, in the context of tumor cells that exhibit Ras activation.
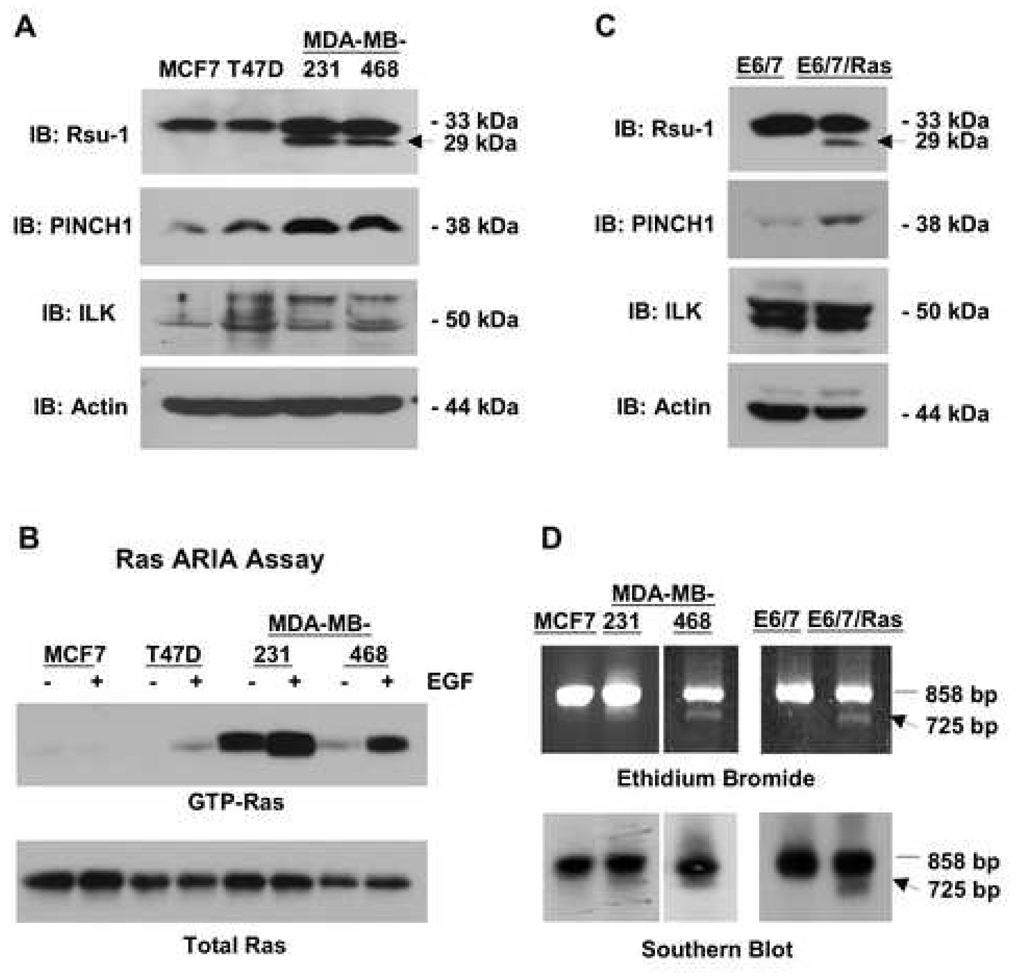
We examined the level of Rsu-1, PINCH1 and ILK in a series of human breast cancer cell lines including estrogen receptor-positive (ER+) MCF7 and T47D and estrogen receptor-negative (ER−) MDA-MD-231 and MDA-MB-468 cell lines. The cell lines chosen include the less malignant (i.e. estrogen-dependent) MCF7 and T47D cells that exhibit limited tumorigenicity in mice and grow poorly in anchorage-independent conditions, as well as more malignant MDA-MB-231 and 468 cells that readily proliferate without the benefit of hormone, grow in an anchorage-independent manner and rapidly give rise to metastatic tumors in mice (Kenny et al., 2007). The results in Figure 1A indicate that in human breast tumor cell lines the levels of PINCH1 protein tend to increase as the cells become more malignant, as there appears to be elevated PINCH1 in both MDA-MB-231 fibroblastic breast cancer cell line, as well as in the less adherent MDA-MB-468 cells. However, the levels of p50 ILK (Fig. 1A) and parvin (data not shown) remain constant. The appearance of a higher-molecular-weight protein reacting with anti-ILK is detected in cell lines except for MCF7 and this may represent ILK-2 or possibly a phosphorylated version of ILK (Li et al., 1999; Tan et al., 2002). The other notable finding from this analysis is the appearance of a 29-kDa protein that is detected by anti-amino-terminal Rsu-1 antibody in the more malignant ER− breast tumor cell lines; both the p29 Rsu-1 and p33 Rsu-1, the full-length Rsu-1 protein, are expressed in the ER− cells. Our previous studies identified an alternatively-spliced form of Rsu-1 RNA that occurs in approximately 30% of high-grade gliomas (Chunduru et al., 2002). Due to exon skipping there is a shift in the Rsu-1 reading frame near the end of the LRR region in this RNA that introduces a premature termination codon. The RNA sequence predicts a protein product of approximately 29 kDa and expression of this Rsu-1 protein revealed that it is highly unstable and lacking the carboxyterminal domain, therefore it was not recognized by the anti-carboxyterminal Rsu-1 antibody (Chunduru et al., 2002).
Fig. 1. Ras-activated human tumor cell lines and immortalized human astrocytes express a truncated 29-kDa Rsu-1 protein.
(A) Lysates (100 µg in RIPA buffer) of human breast tumor cell lines were examined for expression of Rsu-1, PINCH1, ILK, and β-actin by Western blotting. Detection of actin was used as a loading control. Ras activation correlates with expression of a truncated 29-kDa Rsu-1 protein (arrow). (B) Cell lines expressing the truncated form of Rsu-1 contain high levels of Ras-GTP. Breast cancer cell lines were serum starved for 24 h and then stimulated with EGF for 7.5 min or left untreated. The level of Ras-GTP was determined by binding 1 mg lysate proteins to GST-Raf-RBD beads. The Ras-GTP bound to the beads and the total Ras protein in the lysates were detected by Western blotting using anti-pan Ras antibody. (C) Lysates (100 µg in RIPA buffer) of immortalized human astrocytes and their Ras-transformed counterparts were examined for expression of Rsu-1, PINCH1, ILK, and β-actin by Western blotting. Ras activation correlates with expression of a truncated 29-kDa Rsu-1 protein (arrow). (D) An alternatively-spliced transcript of Rsu-1 is detected in human breast cancer cell lines and Ras-transformed astrocytes; 1 µg RNA was used for RT-PCR to amplify the Rsu-1 open reading frame-specific sequence. RT-PCR products were separated on 1% agarose gels, stained with ethidium bromide and transferred to filters for Southern blotting with an Rsu-1 open reading frame-specific probe. A 725-bp product that encodes the p29 Rsu-1 protein is specifically detected in cell lines with Ras activation but not in matched controls.
The highly transformed (ER-) breast cancer cell lines in which p29 Rsu-1 was detected contain either a mutant form of Ki-Ras (MDA-MB-231) or activation of high-level ErbB signaling (MDA-MB-468) leading to a high level of Ras activation. To confirm the level of Ras activation in the breast cancer cell lines, we examined the levels of Ras-GTP in these cells (Fig. 1B). The Ras ARIA assay, which uses binding of Ras-GTP but not Ras-GDP to a GST fusion protein containing the Ras-binding domain of Raf (GST-Raf-RBD), was performed on lysates of cells with or without EGF stimulation. Cells with Ki-Ras mutation (MDA-MB-231) and cells with elevated ErbB1 signaling (MDA-MB-468) exhibit constitutively elevated Ras-GTP and higher levels upon EGF stimulation. Increased Ras activation correlated with the appearance of the 29-kDa Rsu-1 product in breast cancer cell lines.
To extend the investigation to a tumor cell line model in which the contribution of Ras activation could be more directly evaluated, we examined a non-transformed human astrocytic cell line (E6/7) and its Ha-Ras (V12)-transformed counterpart (E6/7/Ras) (Sonoda et al., 2001a,b). In the E6/7 cells only p33 Rsu-1 was detected, but p29 Rsu-1 was also detected as a consequence of expression of a constitutively activated mutant of Ha-Ras in the E6/7-Ras cells (Fig. 1C). Similar to the results observed in breast cancer cell lines, the level of PINCH1 protein is elevated in Ras-transformed astrocytes but the levels of ILK and parvin (data not shown) are unchanged.
Since expression of mutant Ras or high levels of GTP-Ras resulted in the appearance of the 29-kDa truncated Rsu-1 protein product we searched for the alternatively spliced Rsu-1 RNA in the breast cancer and astrocytic cell lines. Reverse transcription and amplification with primers from the 5’ and 3’ ends of the Rsu-1 open reading frame results in production of an 858-bp fragment. If exon 8 is not included in the mature Rsu-1 mRNA, then a 725-bp fragment is the resulting PCR product. The RT-PCR products derived from MDA-MB-231 and MDA-MB-468 cells, but not the MCF7 cells, contain this 725-bp fragment (Fig. 1D). The size and the sequence of the RT-PCR products in these cells is identical to Rsu-1 alternatively-spliced products previously observed in human gliomas or from human cell lines that express mutant forms of Ras (i.e. HT1080 and LS174) (Chunduru et al., 2002). The RT-PCR product resulting from the exon 8-deleted RNA was also detected in the E6/7/Ras cell line but not the immortalized E6/7 astrocytic cell line (Fig. 1D). Together these results indicate that the presence of activated Ras correlates with the presence of exon-deleted Rsu-1 RNA that encodes a truncated 29-kDa Rsu-1 protein.
Ras transformation regulates alternative splicing of the adhesion molecule CD44 via the splicing co-activator SRM160 as well as the RNA-binding protein SAM68 (Cheng and Sharp, 2006; Cheng et al., 2006). Our studies indicated that SAM68 levels did not change in response to Ras activation in breast cancer cell lines (data not shown), and it is unclear whether Ras activation regulates Rsu-1 splicing by a SRM160-dependent mechanism similar to that for CD44.
The 29-kDa Rsu-1 does not co-immunoprecipitate with endogenous PINCH1
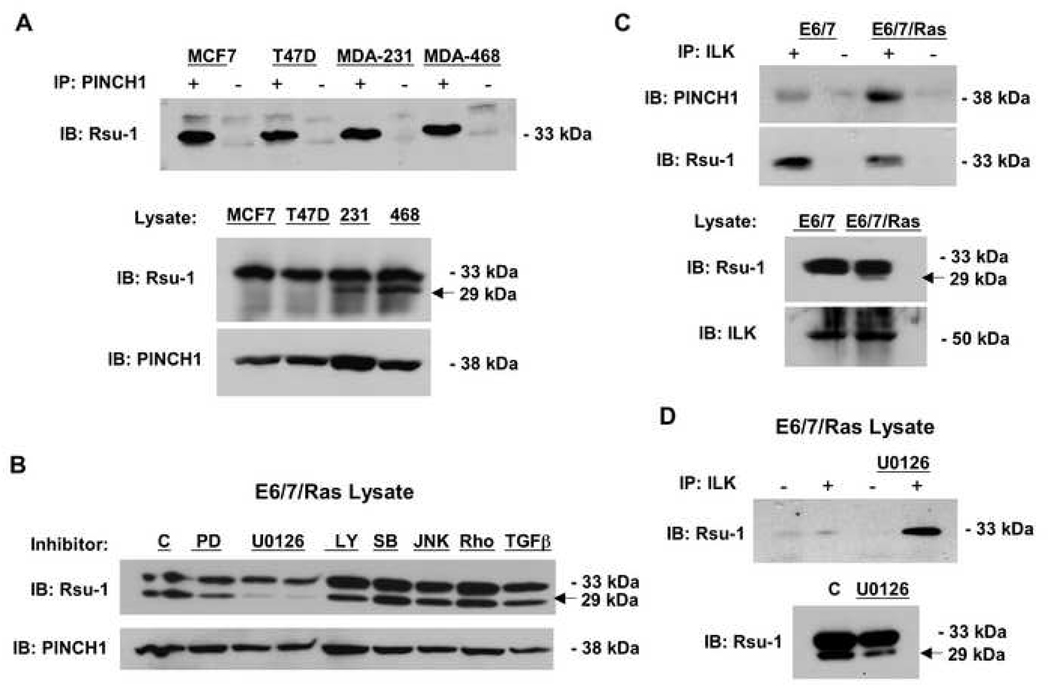
Our previous studies demonstrated that Rsu-1 binds to the adaptor protein PINCH1 and that Rsu-1 co-immunoprecipitated with PINCH1 in mammalian cells in culture (Dougherty et al., 2005). We therefore tested whether PINCH1 could also immunoprecipitate the 29-kDa truncated Rsu-1 protein in ER− breast cancer cell lines. As shown in Figure 2A, the full-length p33 Rsu-1 co-precipitated with PINCH1 in all the breast cancer cell lines; however, the p29 Rsu-1 was not detected in PINCH1 precipitates from ER− human breast cancer cell lines that expressed the p29 Rsu-1. PINCH1 immunoprecipitates from E6/7/Ras human astrocytes also did not contain p29 Rsu-1 (data not shown). We conclude that the p29 Rsu-1 isoform binds poorly or not at all to PINCH1, and likely does not compete directly with full-length Rsu-1 for PINCH1 binding.
Fig. 2. The association of Rsu-1 with the PINCH1-ILK complex is altered in tumor cell lines.
(A) Lysates of human breast cancer cell lines were immunoprecipitated with anti-PINCH1. The immunoprecipitates were analyzed by Western blotting for co-immunoprecipitation of endogenous Rsu-1. Only p33 Rsu-1 is detected in PINCH1 immunoprecipitates. (B) The Ras-transformed immortalized human astrocytes were treated with inhibitors at the indicated concentrations for 36 h and the effect on p33 and p29 Rsu-1 protein expression was determined by Western blotting. Inhibitors: 10 µM PD98059, 10 and 20 µM U0126 (left and right lanes, respectively), 10 µM LY29402, 500 nM SB20350, 100 nM JNKII (SP600125), 100 nM RhoK inhibitor (Y27632), 5 ng/ml TGFβ. (C) Lysates of immortalized human astrocytes and the Ras-transformed astrocyte cell line were immunoprecipitated with anti-ILK. The immunoprecipitates were analyzed by Western blotting for co-immunoprecipitation of endogenous Rsu-1 and PINCH1. (D) The ILK-immunoprecipitates of Ras-transformed astrocytes were analyzed by Western blotting for co-immunoprecipitation of endogenous Rsu-1 with and without pre-treatment of the cells with U0126.
The expression of truncated Rsu-1 requires Mek-ERK activation
Based on our observation that Ras activation correlated with expression of the p29 Rsu-1 protein, we tested specific chemical inhibitors of Ras-dependent signal transduction to determine if p29 Rsu-1 expression could be blocked, and if so, to identify which Ras effector pathways regulated expression. The E6/7/Ras-transformed human astrocytes that expressed p29 Rsu-1 were treated with various inhibitors of Ras-dependent signaling and the effect on the expression of the p29 Rsu-1 truncated protein product was determined by Western blotting. The results shown in Figure 2B demonstrate that the MEK inhibitor U0126 strongly reduced expression of the p29 Rsu-1 protein. The Erk inhibitor, PD98059, partially inhibited expression of p29 Rsu-1 but the constitutive inhibition of Mek activation was more effective. A higher concentration of PD98059 (20 µM) also resulted in additional inhibition of p29 Rsu-1 expression (data not shown). In contrast, inhibitors of Rho kinase (Y27632), Jun kinase (JNK) II (SP600125), and p38 kinase (SB202190) had little or no effect on p29 Rsu-1 expression at any concentration tested. In addition, inhibition of PKC and stimulation with, or neutralization of, TGFβ did not affect the level of p29 Rsu-1 expression. This indicated that a major Ras-dependent signal transduction pathway (Mek-Erk) was responsible for expression of p29 Rsu-1.
Ras transformation alters the association of p33 Rsu-1 with the IPP complex
We next tested whether Ras activation altered the association of p33 Rsu-1 with ILK and PINCH1. Immunoprecipitation of ILK from the immortalized human astrocytes or their Ras-transformed counterparts was followed by Western blotting. We identified both PINCH1 and Rsu-1 in the immunoprecipitates from both the control and Ras-transformed cells. However, the ILK immunoprecipitates from E6/7/Ras astrocytes contained less p33 Rsu-1 than the E6/7 counterparts (Fig. 2C) whereas the association of PINCH1 with ILK appeared to increase with Ras transformation. These results demonstrate that Ras transformation reduces the association of p33 Rsu-1 with the IPP complex.
We also tested whether inhibition of Mek altered the association of p33 Rsu-1 with ILK (Fig. 2D). Exposure of E6/7/Ras cells to U0126 for 24 h prior to lysis and immunoprecipitation of ILK increased the amount of p33 Rsu-1 in the ILK-PINCH1 complex relative to untreated cells. As previously observed, pretreatment of E6/7/Ras astrocytes with U0126 reduced the amount of p29 Rsu-1 protein. This data suggests that inhibition of Mek activation can restore the association of p33 Rsu-1 with the IPP complex in the E6/7 astrocytes despite the presence of activated Ras.
Rsu-1 localization at focal adhesions is altered by Ras transformation
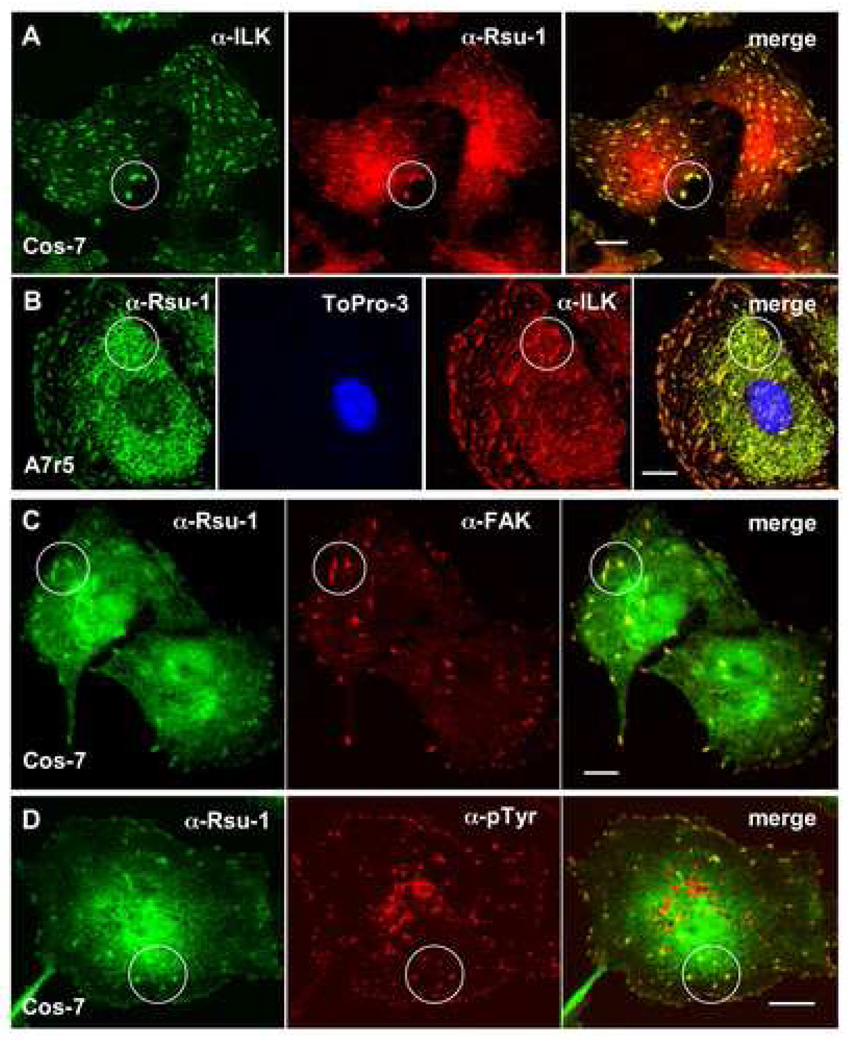
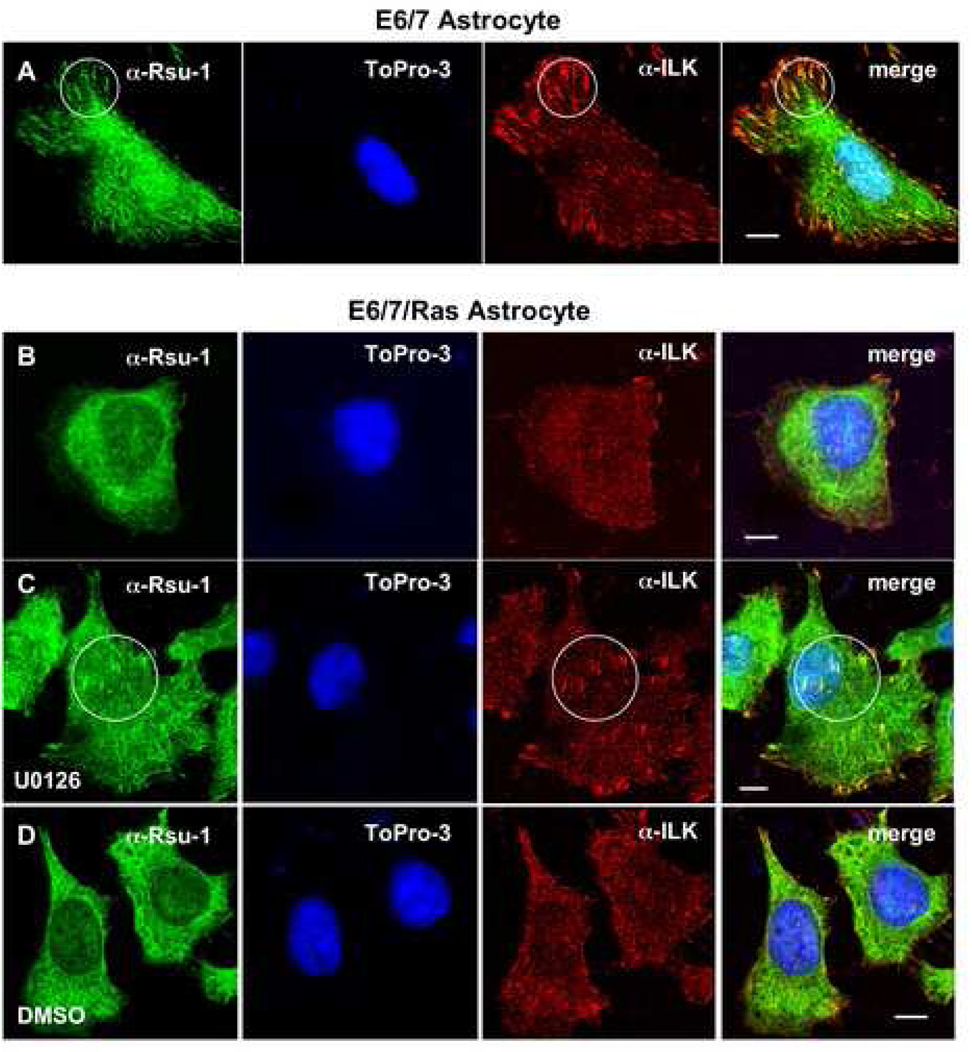
We previously noted colocalization of epitope- and fluorescently-tagged forms of Rsu-1 and PINCH1 at focal adhesions (Dougherty et al., 2005). To test whether endogenous Rsu-1 also localized at focal adhesions, we examined several cell lines for colocalization with ILK, a marker for both focal and fibrillar adhesions (Dedhar, 2000). Using anti-amino-terminal Rsu-1 antibody, we detected colocalization of Rsu-1 and ILK in Cos-7 (Fig. 3A) and A7r5 (Fig. 3B) cell lines. We also detected colocalization of Rsu-1 with other markers of focal adhesions, including focal adhesion kinase (FAK) and phosphotyrosine (Fig. 3C, D). This data demonstrates that endogenous Rsu-1 is a component of focal adhesions. To examine the localization of Rsu-1 in the context of Ras transformation, we also tested E6/7 and E6/7/Ras astrocytes. In E6/7 astrocytes, Rsu-1 localization appears similar to the localization pattern observed in Cos-7 and A7r5 cells, and we confirmed colocalization of Rsu-1 with ILK at focal adhesions in this cell line (Fig. 4A). Expression of activated H-Ras in cell lines has been demonstrated to suppress integrin activation (Hughes et al., 1997) and significantly affect cell morphology, including a decrease in the number and size of focal adhesions (Nobes and Hall, 1999; Furuhjelm and Peranen, 2003). We note that Rsu-1 localization at focal adhesions appears reduced in E6/7/Ras astrocytes, and we confirmed that Rsu-1 colocalizes poorly if at all, with ILK at focal adhesions (Fig. 4B). The effect of Ras on focal adhesion turnover has been shown to require ERK activation but not JNK activation (Hughes et al., 1997), and the inhibitor of ERK activation, U0126, has been shown to inhibit focal adhesion disassembly (Orr et al., 2002; Webb et al., 2004). We examined E6/7/Ras astrocytes under conditions where p29 Rsu-1 expression was blocked to specifically determine the subcellular location of p33 Rsu-1. After treatment of E6/7/Ras astrocytes with U0126 (24 h, 20 µM) or the control treatment with DMSO, the cells were assayed by immunofluorescence. In contrast to the control DMSO treatment, we observed partial restoration of Rsu-1 colocalization with ILK at focal adhesions in Ras-transformed astrocytes treated with U0126 (Fig. 4C, D). Interestingly, in E6/7 astrocytes treated with U0126, we observed enlarged cells with focal adhesions of increased size and number (data not shown). We conclude that Rsu-1 colocalization with ILK at focal adhesions is Ras and ERK dependent. This data also suggests that p29 Rsu-1 expression correlates with conditions of increased focal adhesion turnover. Additionally, localization of p33 Rsu-1 to focal adhesions correlates with non-transformed cell lines (Cos-7, A7r5) or in transformed cell lines in which focal adhesion disassembly is inhibited (E6/7/Ras treated with U0126). This also agrees with our observation that Ras signaling reduces p33 Rsu-1 in ILK immunoprecipitates in E6/7/Ras astrocytes.
Fig. 3. Rsu-1 colocalizes with focal adhesion components.
Cos-7 (A, C, D) or A7r5 (B) cells were plated on fibronectin-coated coverslips and assayed by immunofluorescence using anti-amino-terminal Rsu-1 (A–D), anti-ILK (A, B), anti-FAK (C), and anti-pTyr antibodies (D). Colocalizations are highlighted in circles. Nuclei were counterstained with Topro-3 in (B). Bars: 10 µm (A–D).
Fig. 4. Ras transformation reduces Rsu-1 colocalization with ILK at focal adhesions.
E6/7 astrocytes (A) and E6/7 astrocytes transformed with activated Ras (E6/7/Ras) (B–D) were plated on fibronectin-coated coverslips and assayed by immunofluorescence using anti-amino-terminal Rsu-1 and anti-ILK antibodies. Nuclei were counterstained with Topro-3 (to exclude cells with aberrant or multiple nuclei.). Colocalizations are highlighted in circles. Colocalization at focal adhesions is reduced in E6/7/Ras (B). Inhibition of ERK activation (20 µM U0126 for 24 h prior to plating) partially restores Rsu-1 and ILK colocalization at focal adhesions (C). (D) Solvent control (DMSO). Bars: 10 µm (A–D).
p33 Rsu-1 and p29 Rsu-1 depletion have opposing effects on tumor cell migration
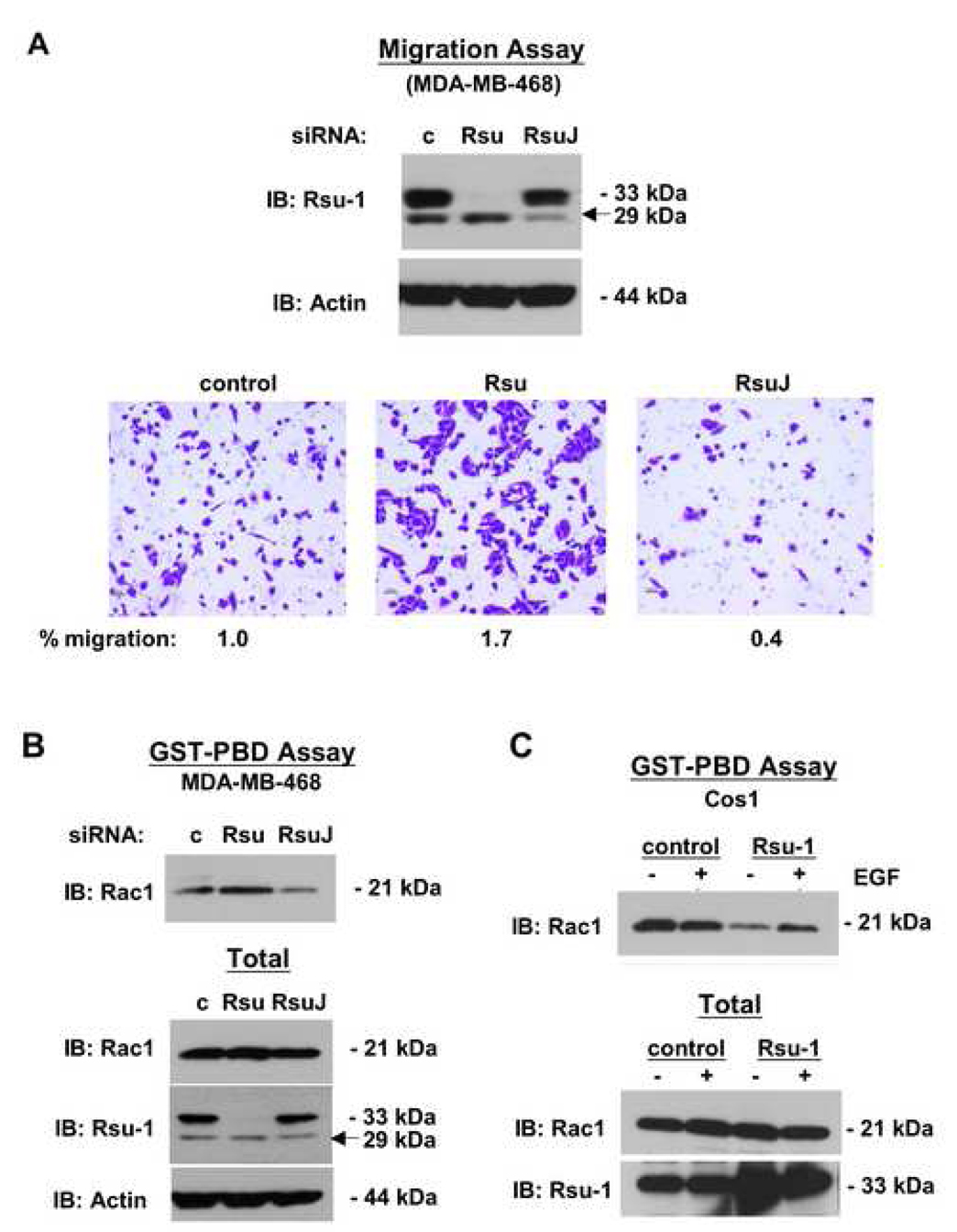
Our previous studies demonstrated that RNAi-directed reduction of Rsu-1 inhibited cell adhesion (Dougherty et al., 2005) whereas elevated expression of Rsu-1 enhanced cell spreading (Masuelli and Cutler, 1996). Hence, we tested the influence of Rsu-1 depletion on tumor cell line migration or invasion. MDA-MB-468 cells were transfected with control, p33 Rsu-1- or p29 Rsu-1-specific RNAi. For p29 Rsu-1 depletion, an siRNA (RsuJ) directed against the aberrant exon-exon junction generated in the alternative splicing of the Rsu-1 RNA was synthesized. At 72 h post transfection of siRNA the cells were analyzed for migration through uncoated membranes or invasion through matrigel-coated membranes in Boyden chamber assays. The results shown in Figure 5A demonstrated that the siRNAs efficiently depleted the respective proteins. Compared to control siRNA, the depletion of p33 Rsu-1 enhanced transit of cells across the membrane regardless of the presence of matrigel coating. However, the depletion of p29 Rsu-1 inhibited migration of MDA-MB-468 cells, again regardless of the presence of matrigel. The normalized data, where migration of control siRNA-transfected cells was set as 1.0, indicated a significant increase when p33 Rsu-1 was depleted (1.7) and a significant decrease when p29 Rsu-1 was depleted (0.4). Thus, we conclude that p33 Rsu-1 depletion enhanced cell migration. In addition, because depletion of the p29 Rsu-1 inhibited migration, we conclude that its presence in cells may contribute to the migratory phenotype.
Fig. 5. Effect of Rsu-1 expression on cell migration and levels of Rac-GTP.
(A, B) MDA-MB-468 cells were depleted of p33 Rsu-1 (siRNA Rsu) or p29 Rsu-1 (siRNA RsuJ) for 72 h. (A) siRNA-treated cells (5 × 104) were seeded on matrigel-coated membranes in Boyden chambers. At 20 h post seeding the cells migrating through the membrane were fixed, stained and enumerated (lower panels). The percentage of cells migrating was normalized to that of control siRNA-treated cells (siRNA c) which was set at 1.0. Upper panels: Verification of reduced Rsu-1 expression after siRNA treatment. (B) siRNA-treated cells were harvested and the level of Rac-GTP was determined by binding to the GST-Rac-binding domain of Pak1 (GST-PBD) (upper panel). (C) Cos-1 cells were transfected with an empty vector or a vector encoding p33 Rsu-1. At 72 h post transfection the cells lysates were prepared from cells with or without EGF stimulation (100 ng/ml, 7 min), and the level of Rac-GTP was determined (upper panel). Five percent of the total cell lysate was included as control for cellular levels of Rac (B, C; lower panels).
Rsu-1 levels contribute to the control of Rac activation
Rac1 is a small GTPase that when activated (GTP-bound) promotes cellular migration through localized polymerization of actin at the leading edge of cells (Raftopoulou and Hall, 2004). Depletion of Rac1 significantly decreases lamella formation and multi-directional cell migration, thus activated Rac1 is a useful marker for this activity (Pankov et al., 2005). Members of the IPP complex have been reported to affect Rac activation. Ectopic ILK expression activated Rac1 and Cdc42 in fibroblasts (Qian et al., 2005), and inhibition or depletion of ILK inhibited Rac activation in HeLa and other cells (Filipenko et al., 2005; Lu et al., 2006; Zhang et al., 2004). Depletion of PINCH1 inhibited Rac activation whereas depletion of α-parvin, but not β-parvin, enhanced Rac activation (Zhang et al., 2004). Hence, we examined Rac1 activation in MDA-MB-468 breast cancer cell lines depleted of Rsu-1.
The GST-PBD assay, which uses binding of Rac-GTP to a GST fusion protein containing the p21-binding domain of Pak1 (GST-PBD), was performed on lysates of MDA-MB-468 cells depleted of p29 or p33 Rsu-1. Our data indicate that depletion of p33 Rsu-1 enhanced Rac1 activation, as might be expected in cells with increased migratory capacity (Fig. 5B). Conversely, depletion of p29 Rsu-1 in this cell line decreased Rac1 activation compared to control siRNA treatment. We next examined whether ectopic transient expression of p33 Rsu-1 could influence Rac-GTP levels in Cos-1 cells (Fig. 5C). Cos-1 cells were transfected with empty vector or vector encoding p33 Rsu-1 and tested for Rac-GTP levels with or without EGF stimulation. In the vector control cells EGF stimulation resulted in elevated Rac-GTP levels. The expression of p33 Rsu-1 resulted in a decrease in Rac-GTP levels compared to the control cells, regardless of EGF stimulation. Taken together these data demonstrate that loss of p33 Rsu-1 expression correlates with increased cell migration and activation of Rac1, whereas a decrease in p29 Rsu-1 correlates with decreased migration and decreased Rac-GTP. Additionally, these data indicate that p33 Rsu-1, in contrast to PINCH1 or ILK, has an inhibitory effect on cell migration.
Discussion
Previous work demonstrated that Rsu-1 binds to PINCH1 and is required for cell adhesion (Dougherty et al., 2005). The results reported here indicate that Rsu-1 can also function to inhibit cell migration. It appears that the association of p33 Rsu-1 with PINCH1 is required for inhibition of migration, as the reduction of p33 Rsu-1 association with the IPP complex correlates with Rac1 activation and increased migration. This is further supported by our observation that Rsu-1 localizes to focal adhesions in non-transformed cells but is significantly reduced at focal adhesions in cells expressing activated Ras. The IPP complex is linked to actin cytoskeletal activities via parvin as well as proteins bound to PINCH1 and ILK (Legate et al., 2006). Nck2, which binds the LIM 4 domain of PINCH1 (Tu et al., 2001), also binds a number of effectors involved in regulating cytoskeletal organization such as the WASP/Scar family of proteins (Buday et al., 2002; Li et al., 2001). The Nck small adaptor proteins have been implicated in linking the PDGF receptor (Li and She, 2000) and other receptor tyrosine kinases via Rho effectors (e.g. Pak) to cytoskeleton rearrangement (Braverman and Quilliam, 1999). More recently, a role for Nck1 and Nck2 in directed cell migration and lamellapodia formation has been demonstrated (Bladt et al., 2003; Rivera et al., 2006). Ectopic expression of Rsu-1 in NIH3T3 cells also resulted in increased cell spreading and abnormal actin staining, suggesting a link between Rsu-1 and the actin cytoskeleton (Masuelli and Cutler, 1996). Rsu-1 expression blocked activity of RasGAP (Masuelli and Cutler, 1996), a protein that has been reported in a complex with Nck1 and Dok (Becker et al., 2000; Jones and Dumont, 1998), and Rsu-1 also blocked ROCK activation (Masuelli and Cutler, 1996; Vasaturo et al., 2000). Thus, binding of Rsu-1 to PINCH1 may stabilize the IPP complex, and the activities of Nck2 or similar effectors of PINCH1 may be modulated by p33 Rsu-1 association.
An alternative model whereby p33 Rsu-1 may promote cell adhesion and inhibit migration, is by reducing either direct or indirect activation of JNK and Rac1. Some recent studies suggest that JNK activation regulates cell migration (Huang et al., 2003). Activated JNK localizes to focal adhesions (Almeida et al., 2000), and activation of JNK is correlated with an increase in migration (Abassi and Vuori, 2002; Hauck et al., 2001; Huynh-Do et al., 2002). Additionally, the activation of the integrin-ILK-PINCH1 pathway in Drosophila activates JNK signaling (Clark et al., 2003; Harden et al., 1996). Previous work from our laboratory demonstrated that ectopic expression of Rsu-1 inhibited JNK activity (Masuelli and Cutler, 1996; Vasaturo et al., 2000), and our recent demonstration that the knockdown of Rsu-1 was accompanied by elevation of JNK and p38 kinase activity supports this finding (Dougherty et al., 2005). Moreover, the disruption of Rsu-1 and PINCH1 expression in Drosophila interfered with JNK activity during dorsal closure in embryogenesis (Kadrmas and Beckerle, 2004). Thus, p33 Rsu-1 may regulate cell migration in part by blocking JNK activation. This may involve effectors linked to the IPP complex, for example the guanine nucleotide exchange factor (GEF) α-PIX. Evidence suggests that activation of Rac1 by ILK may occur via binding of β-parvin to α-PIX (Feng et al., 2002; Filipenko et al., 2005; Mishima et al., 2004). α-PIX, in association with an additional GEF, GIT1, as well as p65 PAK, regulates focal adhesion turnover (Manabe et al., 2002; Obermeier et al., 1998). These components are directly controlled by Rac1 and required for efficient cell migration (Raftopoulou and Hall, 2004). Thus, p33 Rsu-1 may also affect Rac exchange activity through its association with PINCH1 and components that influence α-PIX activity.
Increasing evidence suggests that Rsu-1 acts as a tumor-suppressor gene by enhancing adhesion and inhibiting cell migration, in part through its association with PINCH1 of the IPP complex. While Rsu-1 can suppress the transformed phenotype elicited by the Ras oncogene, elevation of Ras activation can modulate the function of Rsu-1 by regulating expression of an isoform of Rsu-1 which does not inhibit, but rather may promote migration. Future studies will aim to clarify the mechanism whereby these Rsu-1 isoforms regulate focal adhesion stability, cell adhesion and cell migration.
Acknowledgements
The studies were supported by grants from the NIH (R01CA90908) and the USUHS to M.L. Cutler, and from the European Union (Marie Curie Excellence grant 002573) to M. Gimona.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abassi YA, Vuori K. Tyrosine 221 in Crk regulates adhesion-dependent membrane localization of Crk and Rac and activation of Rac signaling. EMBO J. 2002;21:4571–4582. doi: 10.1093/emboj/cdf446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed N, Riley C, Oliva K, Stutt E, Rice GE, Quinn MA. Integrin-linked kinase expression increases with ovarian tumour grade and is sustained by peritoneal tumour fluid. J. Pathol. 2003;201:229–237. doi: 10.1002/path.1441. [DOI] [PubMed] [Google Scholar]
- Albini A, Iwamoto Y, Kleinman HK, Martin GR, Aaronson SA, Kozlowski JM, McEwan RN. A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res. 1987;47:3239–3245. [PubMed] [Google Scholar]
- Almeida EA, Ilic D, Han Q, Hauck CR, Jin F, Kawakatsu H, Schlaepfer DD, Damsky CH. Matrix survival signaling: from fibronectin via focal adhesion kinase to c-Jun NH(2)-terminal kinase. J. Cell Biol. 2000;149:741–754. doi: 10.1083/jcb.149.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker E, Huynh-Do U, Holland S, Pawson T, Daniel TO, Skolnik EY. Nck-interacting Ste20 kinase couples Eph receptors to c-Jun N-terminal kinase and integrin activation. Mol. Cell. Biol. 2000;20:1537–1545. doi: 10.1128/mcb.20.5.1537-1545.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bladt F, Aippersbach E, Gelkop S, Strasser GA, Nash P, Tafuri A, Gertler FB, Pawson T. The murine Nck SH2/SH3 adaptors are important for the development of mesoderm-derived embryonic structures and for regulating the cellular actin network. Mol. Cell. Biol. 2003;23:4586–4597. doi: 10.1128/MCB.23.13.4586-4597.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman L, Quilliam L. Identification of Grb4/Nckb, a src homology 2 and 3 domain-containing adapter protein having similar binding and biological properties to Nck. J. Biol. Chem. 1999;274:5542–5549. doi: 10.1074/jbc.274.9.5542. [DOI] [PubMed] [Google Scholar]
- Buday L, Wunderlich L, Tamas P. The Nck family of adapter proteins: regulators of actin cytoskeleton. Cell Signal. 2002;14:723–731. doi: 10.1016/s0898-6568(02)00027-x. [DOI] [PubMed] [Google Scholar]
- Burgstaller G, Gimona M. Actin cytoskeleton remodelling via local inhibition of contractility at discrete microdomains. J. Cell Sci. 2004;117:223–231. doi: 10.1242/jcs.00839. [DOI] [PubMed] [Google Scholar]
- Cheng C, Sharp PA. Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol. Cell. Biol. 2006;26:362–370. doi: 10.1128/MCB.26.1.362-370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Yaffe MB, Sharp PA. A positive feedback loop couples Ras activation and CD44 alternative splicing. Genes Dev. 2006;20:1715–1720. doi: 10.1101/gad.1430906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chunduru S, Kawami H, Gullick R, Monacci WJ, Dougherty G, Cutler ML. Identification of an alternatively spliced RNA for the Ras suppressor RSU-1 in human gliomas. J. Neurooncol. 2002;60:201–211. doi: 10.1023/a:1021130620178. [DOI] [PubMed] [Google Scholar]
- Clark KA, McGrail M, Beckerle MC. Analysis of PINCH function in Drosophila demonstrates its requirement in integrin-dependent cellular processes. Development. 2003;130:2611–2621. doi: 10.1242/dev.00492. [DOI] [PubMed] [Google Scholar]
- Cutler M, Bassin R, Zanoni L, Talbot N. Isolation of rsp-1, a novel cDNA capable of suppressing v-ras. Mol. Cell. Biol. 1992;12:3750–3756. doi: 10.1128/mcb.12.9.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai DL, Makretsov N, Campos EI, Huang C, Zhou Y, Huntsman D, Martinka M, Li G. Increased expression of integrin-linked kinase is correlated with melanoma progression and poor patient survival. Clin. Cancer Res. 2003;9:4409–4414. [PubMed] [Google Scholar]
- Davies MA, Lu Y, Sano T, Fang X, Tang P, LaPushin R, Koul D, Bookstein R, Stokoe D, Yung WK, Mills GB, Steck PA. Adenoviral transgene expression of MMAC/PTEN in human glioma cells inhibits Akt activation and induces anoikis. Cancer Res. 1998;58:5285–5290. [PubMed] [Google Scholar]
- Dedhar S. Cell-substrate interactions and signaling through ILK. Curr. Opin. Cell Biol. 2000;12:250–256. doi: 10.1016/s0955-0674(99)00083-6. [DOI] [PubMed] [Google Scholar]
- Dougherty GW, Chopp T, Qi SM, Cutler ML. The Ras suppressor Rsu-1 binds to the LIM 5 domain of the adaptor protein PINCH1 and participates in adhesion-related functions. Exp. Cell Res. 2005;306:168–179. doi: 10.1016/j.yexcr.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Feng Q, Albeck JG, Cerione RA, Yang W. Regulation of the Cool/Pix proteins: key binding partners of the Cdc42/Rac targets, the p21-activated kinases. J. Biol. Chem. 2002;277:5644–5650. doi: 10.1074/jbc.M107704200. [DOI] [PubMed] [Google Scholar]
- Filipenko NR, Attwell S, Roskelley C, Dedhar S. Integrin-linked kinase activity regulates Rac- and Cdc42-mediated actin cytoskeleton reorganization via alpha-PIX. Oncogene. 2005;24:5837–5849. doi: 10.1038/sj.onc.1208737. [DOI] [PubMed] [Google Scholar]
- Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhjelm J, Peranen J. The C-terminal end of R-Ras contains a focal adhesion targeting signal. J. Cell Sci. 2003;116:3729–3738. doi: 10.1242/jcs.00689. [DOI] [PubMed] [Google Scholar]
- Galbaugh T, Cerrito MG, Jose CC, Cutler ML. EGF-induced activation of Akt results in mTOR-dependent p70S6 kinase phosphorylation and inhibition of HC11 cell lactogenic differentiation. BMC Cell Biol. 2006;7:34. doi: 10.1186/1471-2121-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Arbman G, Rearden A, Sun XF. Stromal staining for PINCH is an independent prognostic indicator in colorectal cancer. Neoplasia. 2004;6:796–801. doi: 10.1593/neo.04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff JR, Deddens JA, Konicek BW, Colligan BM, Hurst BM, Carter HW, Carter JH. Integrin-linked kinase expression increases with prostate tumor grade. Clin. Cancer Res. 2001;7:1987–1991. [PubMed] [Google Scholar]
- Hannigan G, Leung-Hagesteijn C, Fitz-gibbon L, Coppolino M, Radeva G, Filmus J, Bell J, Dedhar S. Regulation of cell adhesion and anchorage dependent growth by a novel B1 integrin linked kinase. Nature. 1996;379:91–96. doi: 10.1038/379091a0. [DOI] [PubMed] [Google Scholar]
- Harden NJL, Loh H, Ong Y, Tan I, Leung T, Manser E, Lim L. A Drosophila homolog of Rac- and cdc42-activated serine/threonine kinase Pak is a potential focal adhesion and focal complex protein that co-localizes with dynamic actin structures. Mol. Cell. Biol. 1996;16:1896–1908. doi: 10.1128/mcb.16.5.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck CR, Sieg DJ, Hsia DA, Loftus JC, Gaarde WA, Monia BP, Schlaepfer DD. Inhibition of focal adhesion kinase expression or activity disrupts epidermal growth factor-stimulated signaling promoting the migration of invasive human carcinoma cells. Cancer Res. 2001;61:7079–7090. [PubMed] [Google Scholar]
- Herreros L, Rodriguez-Fernandez JL, Brown MC, Alonso-Lebrero JL, Cabanas C, Sanchez-Madrid F, Longo N, Turner CE, Sanchez-Mateos P. Paxillin localizes to the lymphocyte microtubule organizing center and associates with the microtubule cytoskeleton. J. Biol. Chem. 2000;275:26436–26440. doi: 10.1074/jbc.M003970200. [DOI] [PubMed] [Google Scholar]
- Huang C, Rajfur Z, Borchers C, Schaller MD, Jacobson K. JNK phosphorylates paxillin and regulates cell migration. Nature. 2003;424:219–223. doi: 10.1038/nature01745. [DOI] [PubMed] [Google Scholar]
- Hughes PE, Renshaw MW, Pfaff M, Forsyth J, Keivens VM, Schwartz MA, Ginsberg MH. Suppression of integrin activation: a novel function of a Ras/Raf-initiated MAP kinase pathway. Cell. 1997;88:521–530. doi: 10.1016/s0092-8674(00)81892-9. [DOI] [PubMed] [Google Scholar]
- Huynh-Do U, Vindis C, Liu H, Cerretti DP, McGrew JT, Enriquez M, Chen J, Daniel TO. Ephrin-B1 transduces signals to activate integrin-mediated migration, attachment and angiogenesis. J. Cell Sci. 2002;115:3073–3081. doi: 10.1242/jcs.115.15.3073. [DOI] [PubMed] [Google Scholar]
- Ito R, Oue N, Zhu X, Yoshida K, Nakayama H, Yokozaki H, Yasui W. Expression of integrin-linked kinase is closely correlated with invasion and metastasis of gastric carcinoma. Virchows Arch. 2003;442:118–123. doi: 10.1007/s00428-002-0718-6. [DOI] [PubMed] [Google Scholar]
- Jones N, Dumont DJ. The Tek/Tie2 receptor signals through a novel Dok-related docking protein, Dok-R. Oncogene. 1998;17:1097–1108. doi: 10.1038/sj.onc.1202115. [DOI] [PubMed] [Google Scholar]
- Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Kenny P, Lee G, Myers C, Neve R, Semeiks J, Spellman P, Lorenz K, Lee E, Barcellos-Hoff M, Peterson O, Gray J, Bissell M. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol. Oncol. 2007;1:84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khwaja A, Rodriguez-Viciana P, Wennstrom S, Warne PH, Downward J. Matrix adhesion and Ras transformation both activate a phosphoinositide 3-OH kinase and protein kinase B/Akt cellular survival pathway. EMBO J. 1997;16:2783–2793. doi: 10.1093/emboj/16.10.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul D, Parthasarathy R, Shen R, Davies MA, Jasser SA, Chintala SK, Rao JS, Sun Y, Benvenisite EN, Liu TJ, Yung WK. Suppression of matrix metalloproteinase-2 gene expression and invasion in human glioma cells by MMAC/PTEN. Oncogene. 2001;20:6669–6678. doi: 10.1038/sj.onc.1204799. [DOI] [PubMed] [Google Scholar]
- Legate KR, Montanez E, Kudlacek O, Fassler R. ILK, PINCH and parvin: the tIPP of integrin signalling. Nat. Rev. Mol. Cell Biol. 2006;7:20–31. doi: 10.1038/nrm1789. [DOI] [PubMed] [Google Scholar]
- Li F, Zhang Y, Wu C. Integrin-linked kinase is localized to cell-matrix focal adhesions but not cell-cell adhesion sites and the focal adhesion localization of integrin-linked kinase is regulated by the PINCH-binding ANK repeats. J. Cell Sci. 1999;112:4589–4599. doi: 10.1242/jcs.112.24.4589. [DOI] [PubMed] [Google Scholar]
- Li W, She H. The SH2 and SH3 adapter Nck: a two-gene family and a linker between tyrosine kinases and multiple signaling networks. Histol. Histopathol. 2000;15:947–955. doi: 10.14670/HH-15.947. [DOI] [PubMed] [Google Scholar]
- Li W, Fan J, Woodley DT. Nck/Dock: an adapter between cell surface receptors and the actin cytoskeleton. Oncogene. 2001;20:6403–6417. doi: 10.1038/sj.onc.1204782. [DOI] [PubMed] [Google Scholar]
- Lu H, Fedak PW, Dai X, Du C, Zhou YQ, Henkelman M, Mongroo PS, Lau A, Yamabi H, Hinek A, Husain M, Hannigan G, Coles JG. Integrin-linked kinase expression is elevated in human cardiac hypertrophy and induces hypertrophy in transgenic mice. Circulation. 2006;114:2271–2279. doi: 10.1161/CIRCULATIONAHA.106.642330. [DOI] [PubMed] [Google Scholar]
- Lu Y, Lin YZ, LaPushin R, Cuevas B, Fang X, Yu SX, Davies MA, Khan H, Furui T, Mao M, Zinner R, Hung MC, Steck P, Siminovitch K, Mills GB. The PTEN/MMAC1/TEP tumor suppressor gene decreases cell growth and induces apoptosis and anoikis in breast cancer cells. Oncogene. 1999;18:7034–7045. doi: 10.1038/sj.onc.1203183. [DOI] [PubMed] [Google Scholar]
- Manabe R, Kovalenko M, Webb DJ, Horwitz AR. GIT1 functions in a motile, multimolecular signaling complex that regulates protrusive activity and cell migration. J. Cell Sci. 2002;115:1497–1510. doi: 10.1242/jcs.115.7.1497. [DOI] [PubMed] [Google Scholar]
- Marotta A, Tan C, Gray V, Malik S, Gallinger S, Sanghera J, Dupuis B, Owen D, Dedhar S, Salh B. Dysregulation of integrin-linked kinase (ILK) signaling in colonic polyposis. Oncogene. 2001;20:6250–6257. doi: 10.1038/sj.onc.1204791. [DOI] [PubMed] [Google Scholar]
- Marotta A, Parhar K, Owen D, Dedhar S, Salh B. Characterisation of integrin-linked kinase signalling in sporadic human colon cancer. Br. J. Cancer. 2003;88:1755–1762. doi: 10.1038/sj.bjc.6600939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuelli L, Cutler M. Increased expression of the Ras suppressor, Rsu-1, enhances Erk-2 activation and inhibits Jun kinase activation. Mol. Cell. Biol. 1996;16:5466–5476. doi: 10.1128/mcb.16.10.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima W, Suzuki A, Yamaji S, Yoshimi R, Ueda A, Kaneko T, Tanaka J, Miwa Y, Ohno S, Ishigatsubo Y. The first CH domain of affixin activates Cdc42 and Rac1 through alphaPIX, a Cdc42/Rac1-specific guanine nucleotide exchanging factor. Genes Cells. 2004;9:193–204. doi: 10.1111/j.1356-9597.2004.00717.x. [DOI] [PubMed] [Google Scholar]
- Nikolopoulos SN, Turner CE. Actopaxin, a new focal adhesion protein that binds paxillin LD motifs and actin and regulates cell adhesion. J. Cell Biol. 2000;151:1435–1448. doi: 10.1083/jcb.151.7.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolopoulos SN, Turner CE. Integrin-linked kinase (ILK) binding to paxillin LD1 motif regulates ILK localization to focal adhesions. J. Biol. Chem. 2001;276:23499–23505. doi: 10.1074/jbc.M102163200. [DOI] [PubMed] [Google Scholar]
- Nikolopoulos S, Turner C. Molecular dissection of actopaxin-integrin-linked kinase-paxillin interactions and their role in sub-cellular localization. J. Biol. Chem. 2002;277:1568–1575. doi: 10.1074/jbc.M108612200. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol. 1999;144:1235–1244. doi: 10.1083/jcb.144.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermeier A, Ahmed S, Manser E, Yen SC, Hall C, Lim L. PAK promotes morphological changes by acting upstream of Rac. EMBO J. 1998;17:4328–4339. doi: 10.1093/emboj/17.15.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr AW, Pallero MA, Murphy-Ullrich JE. Thrombospondin stimulates focal adhesion disassembly through Gi- and phosphoinositide 3-kinase-dependent ERK activation. J. Biol. Chem. 2002;277:20453–20460. doi: 10.1074/jbc.M112091200. [DOI] [PubMed] [Google Scholar]
- Pankov R, Endo Y, Even-Ram S, Araki M, Clark K, Cukierman E, Matsumoto K, Yamada KM. A Rac switch regulates random versus directionally persistent cell migration. J. Cell Biol. 2005;170:793–802. doi: 10.1083/jcb.200503152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Zhong X, Flynn DC, Zheng JZ, Qiao M, Wu C, Dedhar S, Shi X, Jiang BH. ILK mediates actin filament rearrangements and cell migration and invasion through PI3K/Akt/Rac1 signaling. Oncogene. 2005;24:3154–3165. doi: 10.1038/sj.onc.1208525. [DOI] [PubMed] [Google Scholar]
- Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev. Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- Rivera GM, Antoku S, Gelkop S, Shin NY, Hanks SK, Pawson T, Mayer BJ. Requirement of Nck adaptors for actin dynamics and cell migration stimulated by platelet-derived growth factor B. Proc. Natl. Acad. Sci. USA. 2006;103:9536–9541. doi: 10.1073/pnas.0603786103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen K, Rak J, Leung T, Dean N, Kerbel R, Filmus J. Activated Ras prevents down regulation of Bcl-X(L) triggered by detachment from the extracellular matrix. J. Cell Biol. 2000;149:447–456. doi: 10.1083/jcb.149.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda Y, Ozawa T, Aldape KD, Deen DF, Berger MS, Pieper RO. Akt pathway activation converts anaplastic astrocytoma to glioblastoma multiforme in a human astrocyte model of glioma. Cancer Res. 2001a;61:6674–6678. [PubMed] [Google Scholar]
- Sonoda Y, Ozawa T, Hirose Y, Aldape KD, McMahon M, Berger MS, Pieper RO. Formation of intracranial tumors by genetically modified human astrocytes defines four pathways critical in the development of human anaplastic astrocytoma. Cancer Res. 2001b;61:4956–4960. [PubMed] [Google Scholar]
- Tan C, Mui A, Dedhar S. Integrin-linked kinase regulates inducible nitric oxide synthase and cyclooxygenase-2 expression in an NF-kappa B-dependent manner. J. Biol. Chem. 2002;277:3109–3116. doi: 10.1074/jbc.M108673200. [DOI] [PubMed] [Google Scholar]
- Taylor S, Shalloway D. Cell cycle-dependent activation of Ras. Curr. Biol. 1996;6:1621–1627. doi: 10.1016/s0960-9822(02)70785-9. [DOI] [PubMed] [Google Scholar]
- Tsuda T, Cutler M. Human RSU-1 is highly homologous to mouse Rsu-1 and localizes to human chromosome 10. Genomics. 1993;18:461–462. doi: 10.1006/geno.1993.1503. [DOI] [PubMed] [Google Scholar]
- Tsuda T, Marinetti M, Masuelli L, Cutler M. The Ras suppressor RSU-1 localizes to 10p13 and its expression in the U251 glioblastoma cell line correlates with a decrease in growth rate and tumorigenic potential. Oncogene. 1995;11:397–403. [PubMed] [Google Scholar]
- Tu Y, Li F, Wu C. Nck-2, a novel Src homology2/3-containing adaptor protein that interacts with the LIM-only protein PINCH and components of growth factor receptor kinase-signaling pathways. Mol. Biol. Cell. 1998;9:3367–3382. doi: 10.1091/mbc.9.12.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y, Li F, Goicoechea S, Wu C. The LIM-only protein PINCH directly interacts with integrin-linked kinase and is recruited to integrin-rich sites in spreading cells. Mol. Cell. Biol. 1999;19:2425–2434. doi: 10.1128/mcb.19.3.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y, Huang Y, Zhang Y, Hua Y, Wu C. A new focal adhesion protein that interacts with integrin-linked kinase and regulates cell adhesion and spreading. J. Cell Biol. 2001;153:585–598. doi: 10.1083/jcb.153.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui I, Imamura T, Huang J, Satoh H, Olefsky JM. Cdc42 is a Rho GTPase family member that can mediate insulin signaling to glucose transport in 3T3-L1 adipocytes. J. Biol. Chem. 2003;278:13765–13774. doi: 10.1074/jbc.M208904200. [DOI] [PubMed] [Google Scholar]
- Vasaturo F, Dougherty GW, Cutler ML. Ectopic expression of Rsu-1 results in elevation of p21CIP and inhibits anchorage-independent growth of MCF7 breast cancer cells. Breast Cancer Res. Treat. 2000;61:69–78. doi: 10.1023/a:1006462323260. [DOI] [PubMed] [Google Scholar]
- Wang-Rodriguez J, Dreilinger A, Alsharabi G, Rearden A. The signaling adaptor protein PINCH is up-regulated in the stroma of common cancers, at the invasive edges. Cancer. 2002;95:1387–1395. doi: 10.1002/cncr.10878. [DOI] [PubMed] [Google Scholar]
- Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, Horwitz AF. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Tu Y, Vaelyvis A, Yang Y, Qin J, Wu C. Assembly of the PINCH-ILK-CH-ILKBP complex precedes and is essential for localization of each component to cell-matrix adhesion sites. J. Cell Sci. 2002a;115:4777–4786. doi: 10.1242/jcs.00166. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Guo L, Chen K, Wu C. A critical role of the PINCH-integrin-linked kinase interaction in the regulation of cell shape change and migration. J. Biol. Chem. 2002b;277:318–326. doi: 10.1074/jbc.M108257200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Tu Y, Wu C. Distinct roles of two structurally closely related focal adhesion proteins, alpha-parvins and beta-parvins, in regulation of cell morphology and survival. J. Biol. Chem. 2004;279:41695–41705. doi: 10.1074/jbc.M401563200. [DOI] [PubMed] [Google Scholar]