Abstract
Objectives
Mice harboring c-Myb hypomorphic mutations display enhanced thrombopoiesis because of increased numbers of megakaryocytes and their progenitors. Thrombopoietin induces these same effects, which lead us to hypothesize that the hormone acts through modulation of c-Myb expression, as c-Myb levels falls during thrombopoietin-induced MK maturation. Micro RNAs (miRs) down-regulate gene expression by binding to the 3′ untranslated region (UTR) of specific mRNAs; we noted that the 3′UTR of c-Myb contains four miR-150 binding sites.
Methods
We used quantitative RT-PCR, western blotting and reporter gene analyses to assess the response of c-Myb to thrombopoietin stimulation and to gain of- and loss of-miR-150 expression.
Results
We found that thrombopoietin reduced c-Myb mRNA and protein levels within 7 hr in megakaryocytes and UT7/TPO cells. Using a reporter gene containing the c-Myb 3′UTR region, including its 4 miR150 binding sites, we found that expression of miR150 reduced luciferase expression to 50% of baseline at 24 hr and to 25% at 48 hr in UT7/TPO cells. Quantitative-PCR and western blotting also revealed that miR-150 reduced endogenous c-Myb mRNA and protein to 50% in UT7/TPO cells, and to 65% in mature megakaryocytes. Converse experiments utilizing anti-miR150 increased luciferase activity 2-fold over control anti-miR. Finally, thrombopoietin increased miR150 expression 1.8-fold within 24 hr and 3.4-fold within 48 hr.
Conclusions
These findings establish that miR150 down-modulates c-Myb levels, and since thrombopoietin affects miR150 expression, our results indicate that in addition to affecting MK progenitor cell growth, thrombopoietin down-modulates c-Myb expression through the induction of miR-150.
Introduction
Thrombopoiesis, the process by which hematopoietic stem cells ultimately differentiate into platelet producing megakaryocytes, is characterized by a series of developmental decisions dependent on the expression of several lineage specific transcription factors, hematopoietic growth factors and supportive marrow stromal and endothelial cells (1). One of the transcription factors that influences megakaryocyte development is c-Myb, the gene for which was discovered as the cellular homologue of the transforming gene of two avian retroviruses (2). c-Myb recognizes the core sequence PyAACT/GG (3) and is expressed in immature hematopoietic cells, where it plays an important role; fetal c-myb null mice die at E15 because of anemia (4). Several targets of c-Myb have been identified, including those in G1 and G2 phases of the cell cycle (5,6). Recently, 3 different mutations of c-Myb (MybD152V, MybD384V, MybM303V) that reduce its function to 20–60% of normal were identified and shown to cause anemia and thrombocytosis (7,8). The mice bearing such mutations display greatly reduced numbers of erythroid progenitors and 5–10 fold expanded numbers of megakaryocytic progenitors (6,9). Based on these studies and others, a model has developed which places c-Myb as a switch that influences the lineage fate decision of bi-potent megakaryocyte-erythroid progenitors, with high c-Myb levels favoring erythroid development, and low c-Myb levels resulting in a megakaryocytic fate (10). However, while several intracellular processes have been found to influence the transcriptional activity of c-Myb, the regulation of c-Myb expression has not yet been fully explored.
Thrombopoietin, the primary humoral regulator of thrombopoiesis acts by binding to the cellular proto-oncogene c-Mpl, a homodimeric member of the type I hematopoietic cytokine receptor family (11). Upon thrombopoietin binding c-Mpl undergoes a conformational change allowing cross-phosphorylation and activation of JAK2, a kinase constitutively tethered to the box1 region of the two receptor cytoplasmic domains. The phosphorylation of JAK2 activates its kinase, which in turn phosphorylates tyrosine residues within the cytoplasmic domain of the receptor, and other inducibly tethered secondary signaling mediators. Many thrombopoietin-dependent signaling pathways influence cellular events, including signal transducers and activator of transcription (STAT)3, STAT5, phosphoinositol-3-kinase (PI3K) and mitogen activated protein kinase (MAPK), which contribute to hematopoietic cell survival and proliferation (reviewed in 12). For example, STAT5 enhances the expression of the anti-apoptotic molecule BclXL (13), and PI3K reduces expression of the cell cycle inhibitor p27 (14) and enhances expression of the G1 cell cycle regulator cyclin D1 (15). Since both thrombopoietin and reduced levels of c-Myb enhance thrombopoiesis, we determined whether the hormone reduces expression of the transcription factor, thereby also testing the more general hypothesis that hematopoietic growth factors influence the expression of lineage specific transcription factors.
Recently, small non-coding micro(mi) RNAs have been described that regulate gene expression by targeting mRNA for degradation, if matched perfectly, or that cause translational repression, if slightly mismatched. miRNAs are transcribed as part of long precursor RNAs that are further processed (16). Literally hundreds of miRNAs have been identified in the mammalian genome and there is now ample evidence that they play an important role in human gene expression, including in normal and neoplastic hematopoiesis (17–19). More specifically, several miRNAs have been found to affect hematopoietic differentiation, including miR150 on B and T lymphocyte development (20), miR221 and 222 on erythroid development, (21) and miR223 and miR142 on T cell production (17). Using a variety of predictive algorithms we found several potential miR target sequences in the 3′ untranslated region of the c-Myb gene. As miRNAs bind cooperatively to targeted 3′ untranslated regions (UTR) of the genes they regulate (22), the possibility arose that c-Myb expression was regulated by miRNA. Based upon the results of our current study, we have established that thrombopoietin-induced megakaryopoiesis is characterized by down-regulation of c-Myb expression, mediated at least in part by enhanced expression of miR150.
Materials and Methods
Culture of UT7/TPO cells
UT7/TPO cells, a primitive hematopoietic cell line, were obtained from Dr. Norio Komatsu and cultured in 10% FBS in Iscove’s Modified Dulbecco’s Medium (IMDM) supplemented with 10ng/ml recombinant human thrombopoietin. For stimulation, the thrombopoietin concentration was adjusted to 100 ng/ml, and for starvation, the cells were washed 3 times with PBS and grown in 0.1% FBS/IMDM for 24h.
Isolation of murine megakaryocytes
Primary murine bone marrow cells were isolated (~2–3×109 cells) and treated with ammonium chloride to lyse red cells and grown for 1–3 days in serum-free Optimem medium (Stem Cell Technologies, Vancouver, Canada) supplemented with 5% conditioned medium from thrombopoietin-producing BHK cells; immature megakaryocytes were isolated using CD41-PE labeled antibodies and magnetic cell sorting with PE-beads (MACS) (Miltenyi Biotech, Germany) while mature megakaryocytes were purified by BSA gradient.
Quantitative PCR of c-Myb and other gene transcript levels
Cells were starved of thrombopoietin for 24 hr and then stimulated with the hormone for 1 to 48hr; total RNA was extracted from cell lines and megakaryocytes using the RNEasy Kit (Qiagen, Chatsworth, CA) and reversed transcribed using Superscript III supermix (InVitrogen, Carlsbad, CA) for the SYBR Greener two-step Q-RT-PCR kit for the iCycler (BioRad, Hercules, CA). Q-PCR cycling conditions were 50°C for 2 min, 95°C for 8 min 30sec, and 45 cycles of 95°C for 15s followed by 60°C for 1 min. Melting curve analysis immediately followed Q-PCR with 95°C for 1 min, 55°C for 1 min, and 80 cycles of 55°C + 0.5°C/cycle for 10s. c-Myb and RUNX1 values were then normalized against human β-actin or murine glyceraldehyde dehydrogenase (GAPDH). Primers for Q-PCR were: human c-Myb (forward) 5′-gtcacaaattgactgttacaacaccat-3′, (reverse) 5′-ttctactagatgagagggtgtctgagg-3′; murine c-Myb (forward) 5′-ccaagtgacgctttccagatt-3′, (reverse) 5′-ccccgacacagcatctacg-3′; murine RUNX1 (forward) 5′-tacctgggatccatcacctc-3′, (reverse) 5′-gagatggacggcagagtagg-3′; murine C/EBPβ forward) 5′-caagctgagcgacgagtaca-3′, (reverse) 5′-agctgctccaccttcttctg-3′; murine GATA1 (forward) 5′-ttgtgaggccagagagtgtg-3′, (reverse) 5′-tccgccagagtgttgtagtg-3′; murine FOG-1 (forward) 5′-agcggtgtctgtcacaactg-3′, (reverse) 5′-ggctgtctggaggaagtttg-3′; murine Fli-1 (forward) 5′-caaccagccagtgagagtca-3′, (reverse) 5′-gccgttcttctcatccatgt-3′; human β-actin (forward) 5′-ccaaccgcgagaagatga-3′, (reverse) 5′-ccagaggcgtacagggatag-3′; murine GAPDH (forward) 5′-ggtgaaggtcggtgtgaacggat-3′, (reverse) 5′-cctggaagatggtgatgggcttc-3′. Each Q-PCR was performed in triplicate. The reported data represent the mean results from at least three independent experiments.
Western blot analyses
UT7/TPO cell cultures or purified megakaryocytes were lysed in 1%NP40/0.1%NaDodSO4 buffer containing complete protease inhibitor and PhosStop (Roche Applied Science, Indianapolis, IN). 20 μg these cell lysates were loaded onto 4–% NaDodSO4 polyacrylamide gels (BioRad), size fractionated by electrophoresis, western blotted onto PVDF membranes (BioRad), probed with anti-c-Myb (05–75, Upstate, Millipore, Billerica, MA), and stripped and re-probed with anti-β-actin (A2228, Sigma-Aldrich, St Louis, MO). The relative intensity of c-Myb protein signals was determined by HP Scanjet and ImageJ software; the signal intensity was calculated by comparing each c-Myb signal to the corresponding β-actin signal and then compared to the ratios for unstimulated cells (set at 100%). Densitometric results were then averaged and p-values calculated, and a representative western blot displayed in the corresponding figure.
Q-miRNA PCR using Taqman miRNA assays
Specific miRNA-specific reverse transcriptase (RT) primer and PCR primers were obtained for miRs that were modulated by thrombopoietin in UT7/TPO cells and had target sites in the c-Myb 3′ UTR, including miR-150 and miR-195 (ABI, Foster City, CA). RNA for miR quantitation was obtained using the miRvana Isolation Kit (Ambion/ABI) for small RNA species. Further purification and enrichment of small RNA from total RNA was achieved by differential ethanol concentrations and sequential binding onto the glass fiber filter according to the manufacturer’s recommendations. The QmiRNA assays consist of a two-step RT-PCR for quantification of small amounts of miRNA (from 1–10 ng of total RNA); the first step is RT of miRNA using miRNA-specific looped RT primers to generate cDNA, and the second step is PCR using miRNA-specific forward and reverse primers and a miRNA-specific Taqman MGB probe. The assay detects only mature, active miRNA and not the inactive precursor miRNA, with single-base discrimination. Q-RT-PCR for RNU-6B served as a control for miR quantitation.
Construction of the c-Myb 3′ UTR luciferase reporter vectors
miRNA binding sites in the 3′UTR of c-Myb were determined using the TargetScan 4.0 program (Whitehead Bioinformatics and Research Computing; http://www.targetscan.org/). The 3′ UTR of human c-Myb (NM_005375.2 nt 2151–3285) was cloned by PCR amplification from UT7 cDNA using a Mlu I-tailed forward primer (5′-ccgacgcgtcagaacacttcaagttgacttgg-3′) and a Hind III-tailed reverse primer (5′-cccaagcttgctacaaggcagtaagtacaccgtc-3′). The 1.2 kb PCR product was digested with Hind III and MluI and cloned into Hind III/Mlu I digested pMIR-Report-luc vector (Ambion/ABI), which drives luciferase expression from the CMV promoter. The fidelity of the resulting construct, pCMV-luc-3′UTRcMyb, was verified by restriction analysis and DNA sequencing with luc forward primer (5′-gaggtagatgagatgtga-3′) and a polyA reverse primer (5′-aggcgattaagttgggta-3′). pMIR-Report-luc (pCMV-luc) served as a control plasmid.
Transfection of UT7 cells
UT7/TPO cells (4 × 106) were transduced by nucleofection with 1 μg of luciferase reporter plasmid together with 2 μg of miR-150, miR-195 or miR Negative Control #1 (Ambion), and anti-miR-150, anti-miR-195 or anti-miR Negative Control #1 (Ambion) and 250 ng of pMXGFP using Nucleofector Reagent R and program T-024 (Amaxa, Gaithersburg, MD). Cells were cultured in 10% FBS in IMDM for 24h. Transfection was monitored by immunofluorescence microscopy and FACS for GFP. After 24 hr, the cells were washed with 10% FBS in IMDM and split into subcultures for RNA extraction or cultured further for up to 48 hr in 0–100 ng/ml thrombopoietin.
Reporter gene expression assays
UT7/TPO cell cultures or purified megakaryocytes were lysed with 1x Lysis Buffer (Roche) and assayed for luciferase activity using Steady-Glo (Promega, Madison, WI) reagents and read in a luminometer (Wallac 1420 Victor plate reader, Perkin Elmer, Waltham, MA, USA).
Results
Thrombopoietin regulates c-Myb mRNA expression and protein function in UT7/TPO cells and primary murine megakaryocytes
Based on the inverse relationship between the level of c-Myb expression and that of megakaryopoiesis (7,8), and the profound effects of thrombopoietin on megakaryocyte and platelet development (11), we hypothesized that the hormone regulates expression of c-Myb. Using Q-RT-PCR we found that thrombopoietin reduces c-Myb expression in UT7/TPO cells which closely represent the megakaryocytic lineage. A detailed study of c-Myb expression in these cells revealed that thrombopoietin treatment reduced c-Myb mRNA expression two-fold within 3 hr (p≤0.03) (Figure 1A). To verify these changes occur in primary cells, we treated freshly isolated murine megakaryocytes with thrombopoietin and found similar results; following 2 d treatment with thrombopoietin, megakaryocyte c-Myb levels fell by ~15-fold, compared to a 2-fold increase in RUNX1, no change in GATA1, and a 4-fold increases in FOG1 and Fli1 (Figure 1B and 1C). To determine if these changes are reflective of direct effects of thrombopoietin, we performed a shorter time course analysis, and used western blotting to confirm the changes also occur at the level of protein expression. We found that thrombopoietin treatment reduced c-Myb protein expression to 51% of baseline within 7 hr in UT7/TPO and to 65% of baseline in murine megakaryocytes (Figures 2A/B). Densitometric analysis of the c-Myb to β-actin ratio revealed that reduction of c-Myb protein levels were significant from 5h onward in both UT7/TPO cells (p≤0.01) (n=3) and murine megakaryocytes (p≤0.01). To test if these changes in c-Myb mRNA and protein levels resulted in a corresponding reduction in c-Myb function we introduced a c-Myb reporter gene into UT7/TPO cells, in which the three c-Myb binding sites in the c-mim1 promoter drives luciferase activity (cMim-luc; kindly provided by Scott Ness [23]). We found that thrombopoietin reduced reporter gene activity to 62% of baseline at 5 hr (p=0.038) (Figure 2C). In contrast, when the mim1 promoter orientation was reversed, thrombopoietin did not change luciferase expression (data not shown). While these changes in c-Myb protein expression and function seem modest, it did not escape our attention that a ~50% reduction in c-Myb expression results in an ~10-fold increase in the number of mature progenitors committed to the megakaryocyte lineage (7,8).
Figure 1. Expression of c-Myb and other megakaryocytic transcription factors in megakaryocytic cell lines and mature megakaryocytes.
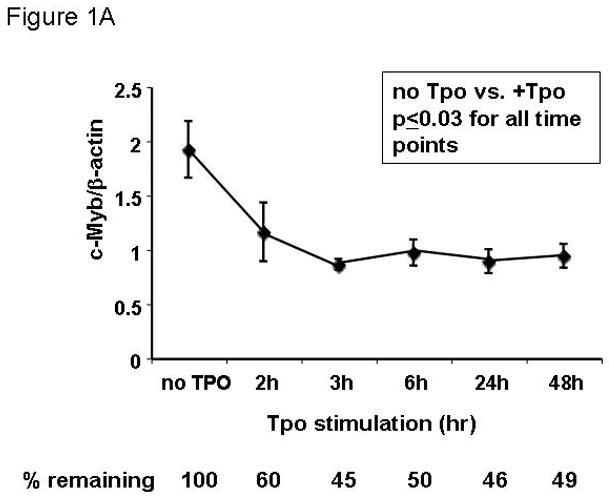
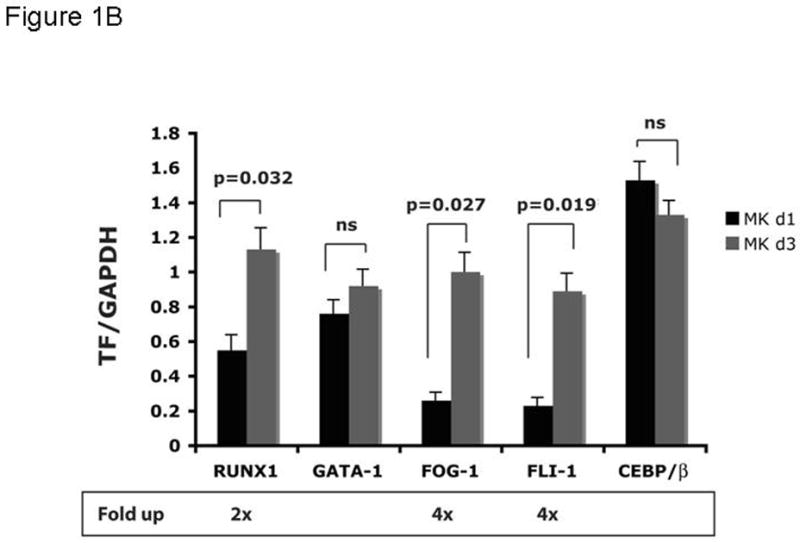
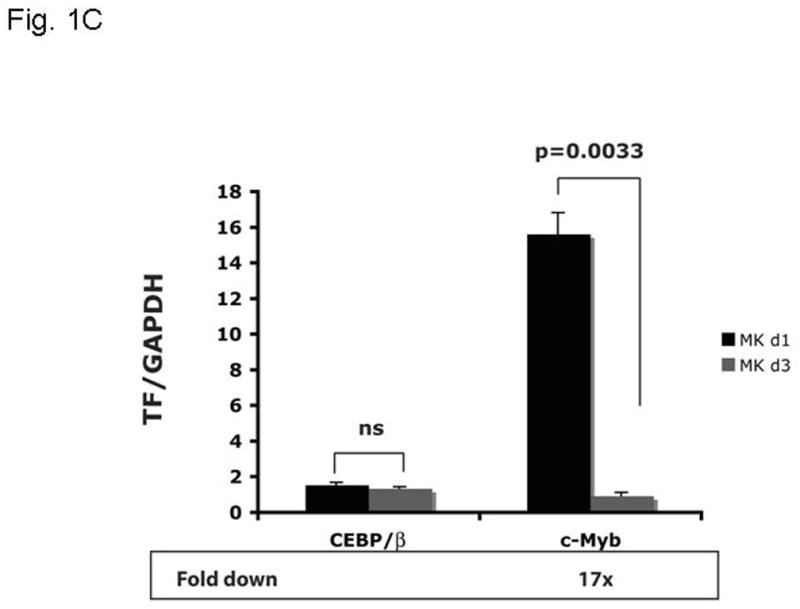
UT7/TPO cells (A) were starved for 24h and then stimulated with 100 ng/ml thrombopoietin for up to 48 hr. RNA was extracted and c-Myb levels were assessed by Q-RT-PCR, with β-actin as internal control. Each assay was performed in triplicate. c-Myb levels significantly declined (p≤0.03) for all time points with Tpo stimulation (n=3). Immature murine megakaryocytes (B–C) were induced to differentiate over 2 days with thrombopoietin. RNA was harvested from immature and mature megakaryocytes, and the levels of RUNX1, GATA1, FOG1, Fli1, C/EBPβ and c-Myb were assessed by quantitative real-time PCR with glyceraldehyde dehydrogenase (GAPDH) as an internal control. The results shown are the relative values of signal intensity for the various transcription factors compared to the internal controls. Each experiment was performed in triplicate and the results compared using the t-test for paired values. The results represented in figures 1B and 1C were performed together, but are shown on separate figures due to the range of scales.
Figure 2. Thrombopoietin reduces c-Myb protein and function in UT7/TPO cells and in murine megakaryocytes.
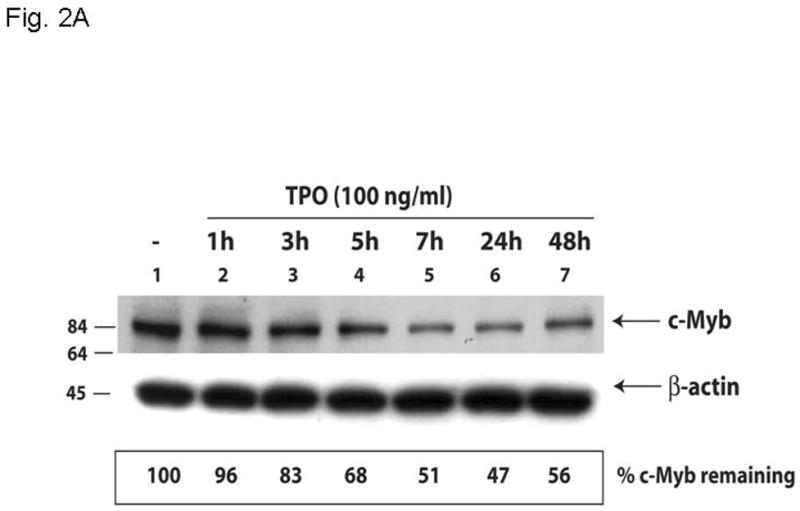
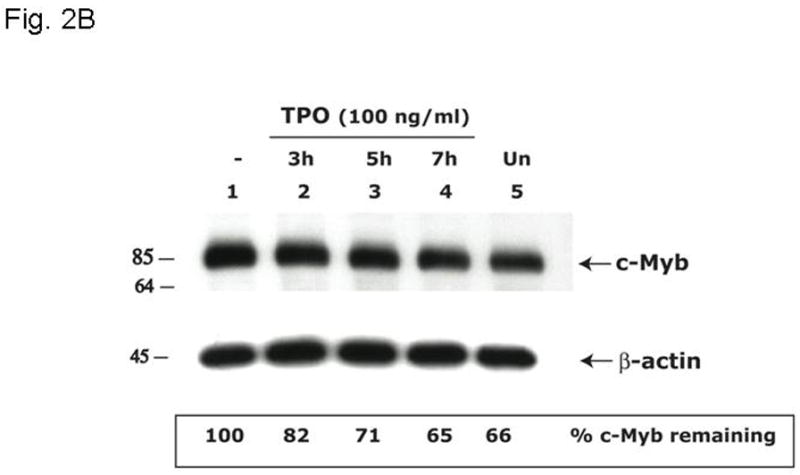
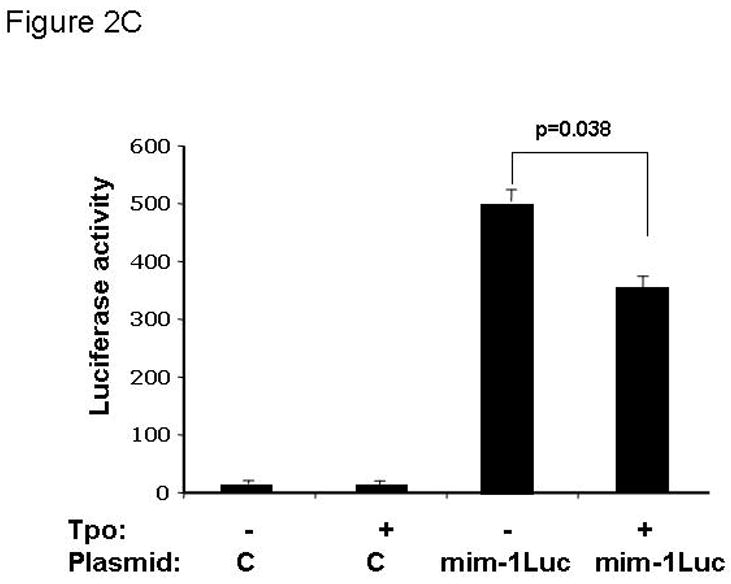
A. UT7/TPO cells were starved overnight and then stimulated with 100 ng/ml thrombopoietin for 0–48 hr, cell lysates were prepared, size fractionated, transferred to PVDF membranes and probed for c-Myb. Densitometric analysis revealed a significant decrease in c-Myb protein levels at 5h–48h (p≤0.01 for all time points). The fraction of c-Myb remaining is shown, along with a representative western blot. B. Mature murine megakaryocytes were produced by marrow cell culture in thrombopoietin and the cells purified by BSA gradient and starved for 20 hr in StemPro serum-free medium. The cells were then cultured in 100 ng/ml thrombopoietin for up to 7 hr, cell lysates prepared and subjected to western blotting for c-Myb and β-actin,. Densitometric analyses (n=3) showed c-Myb protein levels were significantly decreased at 5–7h (p≤0.01). The fraction of c-Myb remaining is shown, along with a representative western blot. C. The c-Myb responsive reporter construct c-mim-luc (mim1Luc), which contains three c-Myb response elements was introduced into UT7/TPO cells and following overnight starvation was cultured an additional 5 hr with thrombopoietin or control culture medium. Sham transfected cells served as a control (C). The data represent the mean of three experiments. The difference between thrombopoietin and no thrombopoietin in c-mim1-luc transduced cultures achieved statistical significance (p=0.038).
c-Myb RNA and protein levels are regulated by miRNA species
To begin to explore the molecular mechanism(s) by which thrombopoietin affects c-Myb mRNA expression, we hypothesized that the hormone might act to alter the levels of miRNAs that target the 3′ UTR of c-Myb. Previous work by others using microarrays had demonstrated several miRNA changes in thrombopoietin-stimulated hematopoietic cells (24), including several for which both high and modest affinity miRNA binding sites in the murine and human c-Myb genes were found. Findings in our microRNA microarray comparing thrombopoietin-stimulated megakaryocytes vs. thrombopoietin-starved megakaryocytes revealed several miRNAs against c-Myb were upregulated, including miR-150 and miR-195 (data not shown). In this study we focused on miR-150 and miR-195, as our in silico analysis revealed that the c-Myb 3′ UTR displays two high and two low affinity binding sites for miR-150 and one high affinity binding site for miR-195 (Figure 3A). To directly test if these miR species regulate c-Myb expression, we created a reporter gene, linking the CMV promoter driven luciferase gene and the 1.2 kb 3′UTR of c-Myb (pCMV-luc-3′UTRcMyb; Figure 3A) and introduced the construct into UT7/TPO cells together with either miR-150, miR-195, anti-miR-150, anti-miR-195 or the corresponding controls. We found that miR-150 significantly down-regulated luciferase activity to 40% of baseline 24 hr following co-transfection with pCMV-luc-3′UTRcMyb (Figure 3B) compared to a miR negative control (p≤0.002). miR195 exerted a significant, but quantitatively lesser effect. Luciferase activity in cells transfected with a control luc plasmid lacking the 3′UTR of c-Myb was not modulated by introduction of miR-150 (Figure 3B, column 1 vs. column 4). Since UT7/TPO cells express miR-150 at baseline, we performed converse experiments utilizing anti-miRs, which inhibit expression of endogenous miRs. We found that when co-transfected with pCMV-luc-3′UTRcMyb, anti-miR-150 significantly up-modulated luciferase activity to 180% of baseline compared to an anti-miR-negative control (p=0.0001) (Figure 3C, column 1 vs. column 3), or to a significantly lesser extent in an otherwise identical reporter construct that lacks the 3′UTR of c-Myb (p=0.016) (Figure 3C, column 1 vs. column 4). However, in contrast to our results with miR195, anti-miR-195 had little effect on luciferase activity (Figure 3C, column 2 vs. column 3). Both miR negative control and anti-miR negative control did not display non-specific effects, as transfection of pCMV-luc-3′UTRcMyb alone or in combination with either miR negative control or anti-miR negative control resulted in similar luciferase activities (data not shown).
Figure 3. miR150 affects a c-Myb 3′ UTR reporter gene.
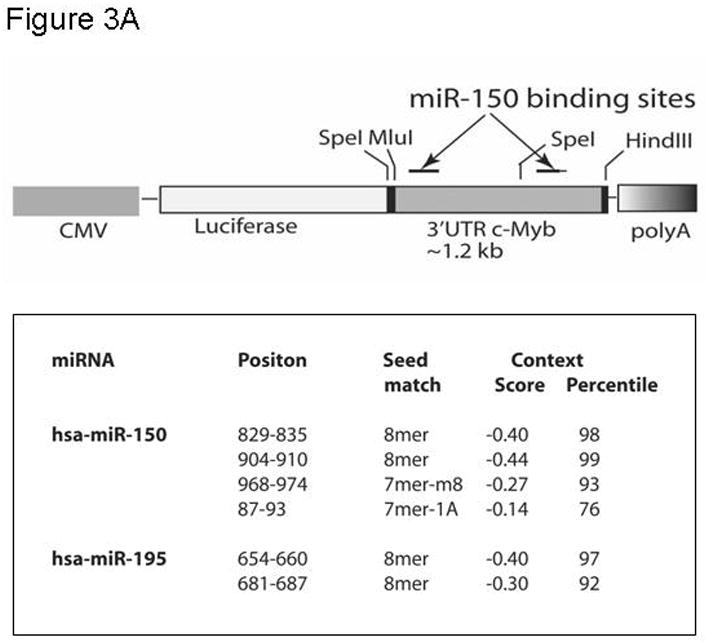
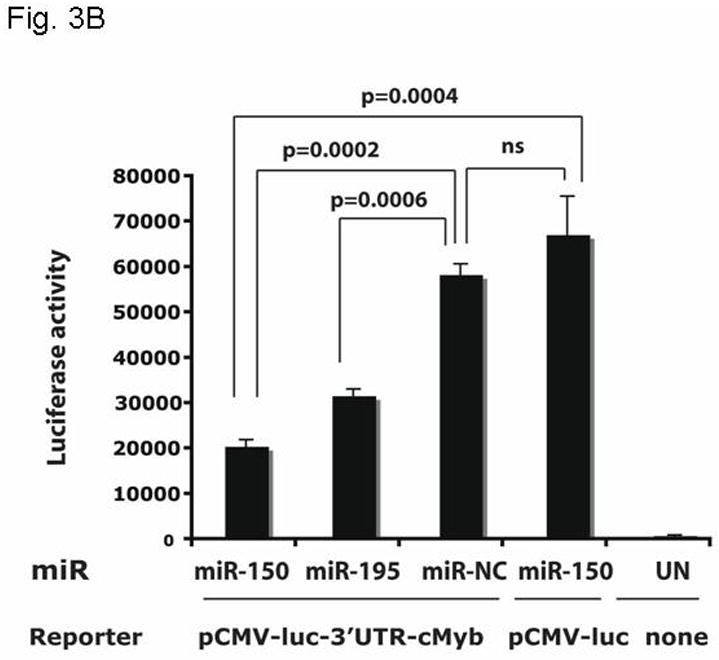
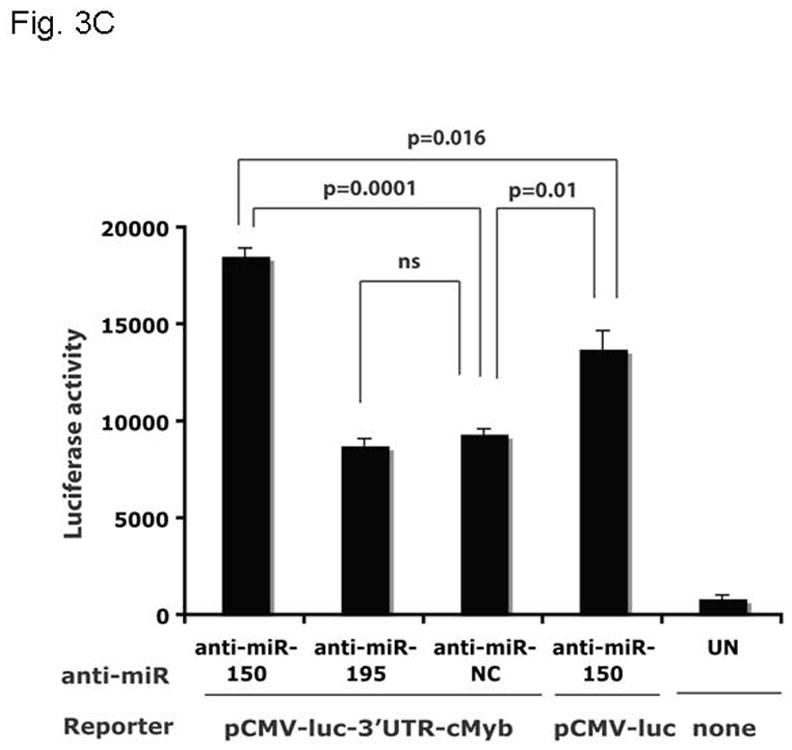
A. The pCMV-luc-3′UTRc-Myb reporter gene is illustrated [with the list of miR binding sites]. The construct was generated by introducing the 1.2 kb 3′UTR of c-Myb into pMIR-Report-luc. B. pCMV-luc-3′UTRc-Myb was transfected into UT7/TPO cells with miR150, miR195 or a negative miR control. After 24 hr, luciferase activity was assessed in duplicate cultures. Values reported are the mean ± S.E.M. of three separate experiments and reported as relative luciferase activity between the various conditions. The same vector, but without the 3′UTR of c-myb (pCMV-luc), were also utilized as controls, and results are shown. C. pCMV-luc-3′UTRc-Myb was transfected into UT7/TPO cells with anti-miR150, anti-miR195 or a negative anti-miR control. After 24 hr luciferase activity was assessed in duplicate cultures and is reported as the mean ± S.E.M. of three separate experiments. The same controls as in figure 3B were performed. pCMV-luc-3′UTRc-Myb was also transfected into the cells with miR negative control and anti-miR negative control, and these gave the same values as the vector without the 3′UTRcMyb (data not shown).
The introduction of miRs and anti-miRs into UT7/TPO cells also provided us the opportunity to monitor their effects on endogenous c-Myb mRNA and protein levels. Using Q-PCR, we found that endogenous c-Myb mRNA was significantly down-regulated to 55% of baseline upon transfection of miR-150 compared to the miR negative control (p=0.008) (Figure 4A), with a lesser effect of miR195, while the essential megakaryocytic transcription factor, RUNX1, remained unaltered (Figure 4B). Western blotting of these cell lysates revealed that c-Myb protein expression was down-regulated to 30% of baseline following transduction with miR-150 but not with the miR negative control or miR195 (Figure 4C).
Figure 4. miR150 affects c-Myb expression.
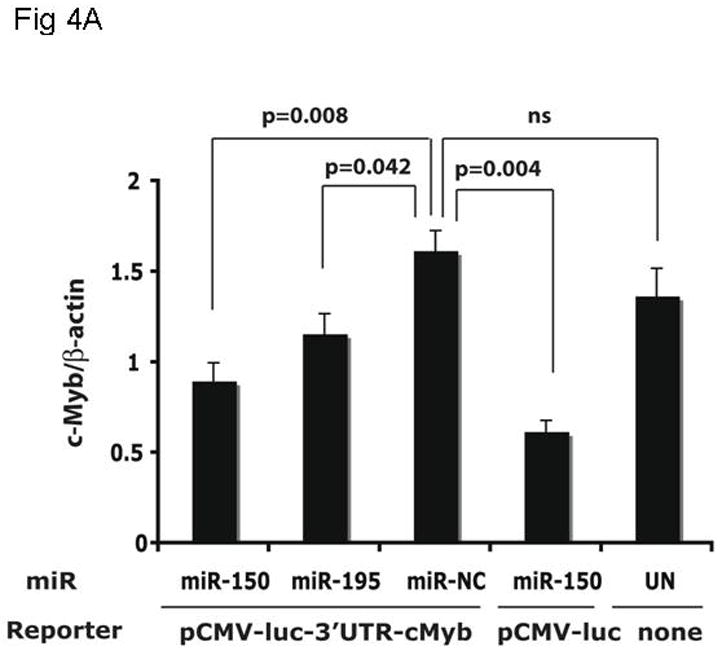
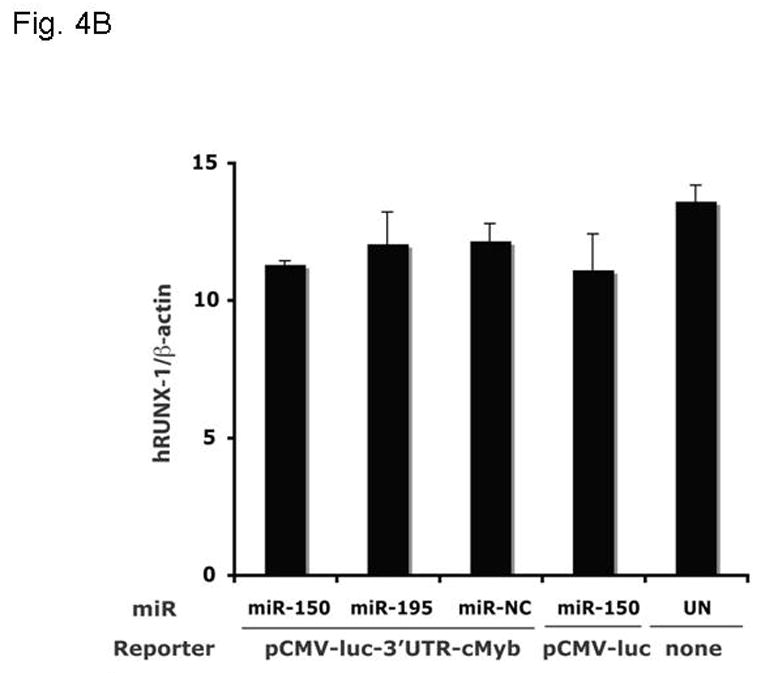
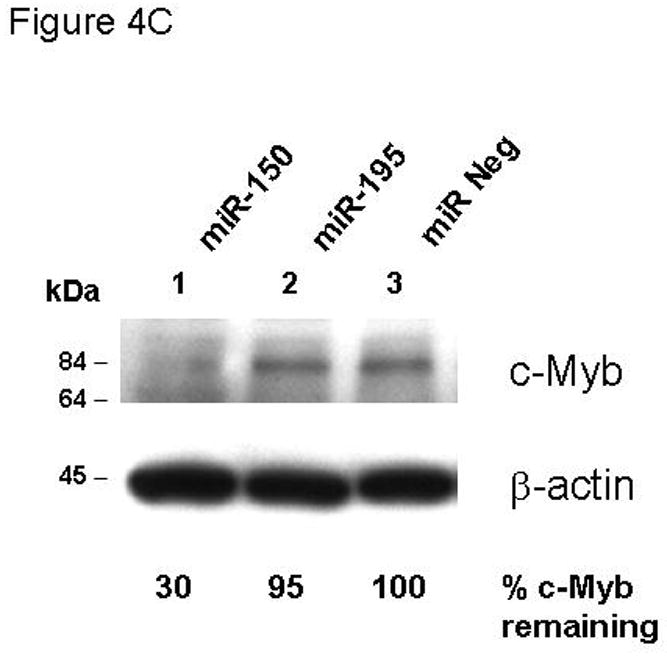
The same cultures assessed for reporter gene activity in Figure 3B were also assayed for expression of the endogenous c-Myb (A,C) and RUNX-1 (B) genes using Q-RT-PCR (A,B) and quantitative western blotting (C).
Thrombopoietin regulates miR150 expression
The previous results establish that miR-150 can significantly alter c-Myb expression. To test whether miR-150 responds to thrombopoietin stimulation, we used a miR-specific Q-PCR assay and tested the effects of the hormone on its expression. We found that thrombopoietin increased miR-150 expression to 144% of baseline at 3 hr, 183% at 24 hr and 340% at 48 hr in UT7/TPO cells (Figure 5). In contrast, erythropoietin did not affect miR-150 levels in UT7/EPO cells (data not shown).
Figure 5. Thrombopoietin affects miR-150 expression.
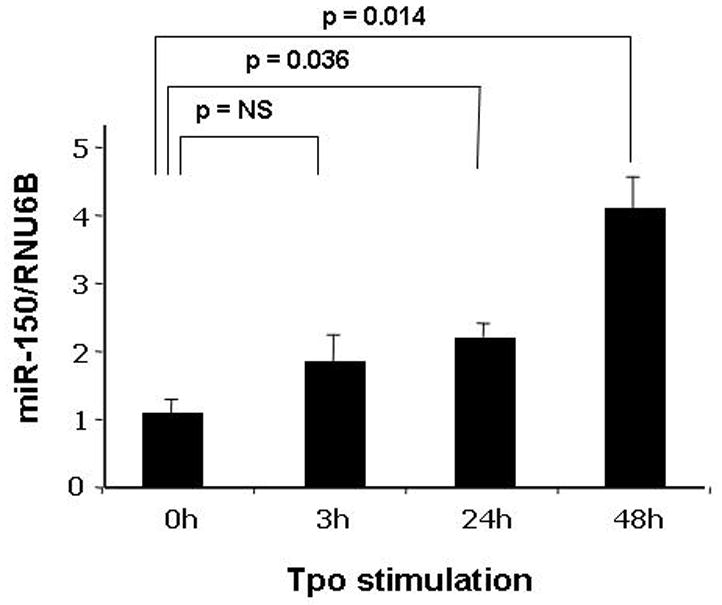
A Q-RT-PCR assay for miR-150 using the RNU6B control microRNA as an internal control was developed and used to assess the miR-150 cellular response to stimulation with thrombopoietin. We found that UT-7/TPO cells stimulated for 3–48 hr with the hormone up-regulated miR-150 expression substantially and significantly. The results represent the mean ± S.E.M. of three independent experiments with assays performed in triplicate.
Discussion
Several features of the biology of Tpo and c-Myb are consistent with the hypothesis that the two genes lie on the same genetic pathway and that thrombopoietin reduces c-Myb expression. For example, the 10-fold increase in hematopoietic stem cells found in the hypomorphic c-mybM303V animals matches closely the ~8-fold reduction in these cells seen in c-mpl −/− mice (25) or the ~15-fold decrease in stem cell expansion seen when normal marrow cells are transplanted into tpo−/− mice (26). Moreover, the level of c-Myb mRNA in thrombopoietin-stimulated CD41+ cord blood derived cells is 7.4-fold lower than seen in BFU-E or CD34+ cells (27). In the present study we sought to mechanistically link thrombopoietin and c-Myb expression. We found thrombopoietin suppresses c-Myb transcription and protein levels to 40–65% of resting levels in UT7/TPO cells and mature megakaryocytes within a few hours of its addition to thrombopoietin depleted cells; while the direct effect of thrombopoietin on c-Myb may seem modest, this level is physiologically relevant since the c-MybM303V mice, which display anemia and thrombocytosis, are characterized by a ~50% reduction in c-Myb function.
Several lines of evidence suggest that c-Myb enhances erythropoiesis and is a negative regulator of megakaryopoiesis (28). Based on our finding that thrombopoietin affects c-myb mRNA levels, we next determined if this is due to miRNA induced modulation of c-myb mRNA stability and/or translation efficiency. To test the effect of various miRNA species, we generated and introduced into UT7/TPO cells a reporter construct that reflects the post-transcriptional modulation of c-Myb; we found that luciferase activity was significantly reduced by miR-150, and that endogenous miR-150 regulates c-Myb as an anti-miR150 increased reporter gene activity. Next we assessed the effects of miR-150 on c-Myb mRNA and protein levels in UT7/TPO cells; we found that miR-150 reduced c-Myb mRNA levels to 55% of control levels, and c-Myb protein to 30% of control levels. These results were found in multiple experiments and strongly suggest that miR-150 affects both c-Myb mRNA stability and translation efficiency.
Regulation of c-Myb transcription is complex, dependent on stage of cell differentiation and cell cycle, and levels of [Ca++]i, c-Myb, Ets1, GATA1, NF-κB and E2F (29–33). Cell context is critical; c-Myb itself upregulates c-Myb expression in fibroblasts but down-modulates itself in T cells (30). In addition, once transcription initiates, it can stall within intron 1, a step that is controlled during B cell development (34). Once transcribed and translated into protein, c-Myb is subject to further regulation, including lysine acetylation (35) and SUMOylation (36), which increase and decrease its transcriptional function, respectively. Thus, our results add another dimension to the regulation of c-Myb expression, modulation by a microRNA, miR-150.
Our results with miR-150 during megakaryopoiesis differ from that of Garzon and colleagues (24), who found using a miR gene array that miR-150 was down-modulated 5.3-fold during thrombopoietin-supported development of human megakaryocytes from primitive CD34+ bone marrow hematopoietic progenitors. While these results appear opposite in direction to our own, levels of the miR were not monitored throughout the developmental period. Thus it is possible that expression of miR-150 is biphasic, high in primitive hematopoietic cells, dropping down as cells begin to differentiate, and then rising again in response to lineage specific signals. Alternately, the differences in the two studies might relate to the use of a gene array in one, and specific Q-RT-PCR analyses in our current work.
Taken together, our results, based on both gain-of-function and loss-of-function strategies, have identified a direct mechanistic link between thrombopoietin-induced miR-150 expression and c-Myb down-modulation during megakaryopoiesis. This novel molecular pathway could thus account for some of the favorable effects of thrombopoietin on platelet production. These studies also establish that hematopoietic growth factors can influence transcription factor expression through modulation of microRNA species.
Acknowledgments
This work was supported by NIH grant R01 DK49855 and R01 CA 31615 to KK, and 2006 and 2007 Training Grants from the American Society of Hematology to HP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kaushansky K. The molecular mechanisms that control thrombopoiesis. J Clin Invest. 2005;115:3339–3347. doi: 10.1172/JCI26674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramsey RG. c-Myb a stem-progenitor cell regulator in multiple tissue compartments. Growth Factors. 2005;23:253–261. doi: 10.1080/08977190500233730. [DOI] [PubMed] [Google Scholar]
- 3.Ganter B, Chao ST, Lipsick JS. Transcriptional activation by the Myb proteins requires a specific local promoter structure. FEBS Lett. 1999;460:401–410. doi: 10.1016/s0014-5793(99)01373-3. [DOI] [PubMed] [Google Scholar]
- 4.Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J. 2003;22:4478–4488. doi: 10.1093/emboj/cdg434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman AD. Runx1, c-Myb, and C/EBPalpha couple differentiation to proliferation or growth arrest during hematopoiesis. J Cell Biochem. 2002;86:624–629. doi: 10.1002/jcb.10271. [DOI] [PubMed] [Google Scholar]
- 6.Nakata Y, Shetzline S, Sakashita C, Kalota A, Rallapalli R, Rudnick SI, Zhang Y, Emerson SG, Gewirtz AM. c-Myb Contributes to G2/M Cell Cycle Transition in Human Hematopoietic Cells by Direct Regulation of Cyclin B1 Expression. Mol Cell Biol. 2007;27:2048–2058. doi: 10.1128/MCB.01100-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpinelli MR, Hilton DJ, Metcalf D, Antonchuk JL, Hyland CD, Mifsud SL, Di Rago L, Hilton AA, Willson TA, Roberts AW, Ramsay RG, Nicola NA, Alexander WS. Suppressor screen in Mpl−/− mice: c-Myb mutation causes supraphysiological production of platelets in the absence of thrombopoietin signaling. Proc Natl Acad Sci U S A. 2004;101:6553–6558. doi: 10.1073/pnas.0401496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandberg ML, Sutton SE, Pletcher MT, Wiltshire T, Tarantino LM, Hogenesch JB, Cooke MP. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev Cell. 2005;8:153–166. doi: 10.1016/j.devcel.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 9.Metcalf D, Carpinelli MR, Hyland C, Mifsud S, Dirago L, Nicola NA, Hilton DJ, Alexander WS. Anomalous megakaryocytopoiesis in mice with mutations in the c-Myb gene. Blood. 2005;105:3480–3487. doi: 10.1182/blood-2004-12-4806. [DOI] [PubMed] [Google Scholar]
- 10.Mukai HY, Motohashi H, Ohneda O, Suzuki N, Nagano M, Yamamoto M. Transgene insertion in proximity to the c-myb gene disrupts erythroid-megakaryocytic lineage bifurcation. Mol Cell Biol. 2006;26:7953–7965. doi: 10.1128/MCB.00718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaushansky K. Thrombopoietin: The primary regulator of platelet production. Blood. 1995;86:419–431. [PubMed] [Google Scholar]
- 12.Kaushansky K, Drachman JG. The molecular and cellular biology of thrombopoietin: the primary regulator of platelet production. Oncogene. 2002;21:3359–3367. doi: 10.1038/sj.onc.1205323. [DOI] [PubMed] [Google Scholar]
- 13.Socolovsky M, Fallon AE, Wang S, Brugnara C, Lodish HF. Fetal anemia and apoptosis of red cell progenitors in Stat5a−/−5b−/− mice: a direct role for Stat5 in Bcl-X(L) induction. Cell. 1999;98:181–191. doi: 10.1016/s0092-8674(00)81013-2. [DOI] [PubMed] [Google Scholar]
- 14.Collado M, Medema RH, Garcia-Cao I, Dubuisson ML, Barradas M, Glassford J, Rivas C, Burgering BM, Serrano M, Lam EW. Inhibition of the phosphoinositide 3-kinase pathway induces a senescence-like arrest mediated by p27Kip1. J Biol Chem. 2000;275:21960–21968. doi: 10.1074/jbc.M000759200. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt M, Fernandez de Mattos S, van der Horst A, Klompmaker R, Kops GJ, Lam EW, Burgering BM, Medema RH. Cell cycle inhibition by FoxO forkhead transcription factors involves downregulation of cyclin D. Mol Cell Biol. 2002;22:7842–7852. doi: 10.1128/MCB.22.22.7842-7852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sevignani C, Calin GA, Siracusa LD, Croce CM. Mammalian microRNAs: a small world for fine-tuning gene expression. Mamm Genome. 2006;17:189–202. doi: 10.1007/s00335-005-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 18.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 19.Lawrie CH. MicroRNAs and haematology: small molecules, big function. Br J Haematol. 2007;137:503–512. doi: 10.1111/j.1365-2141.2007.06611.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci U S A. 2007;104:7080–7085. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, Valtieri M, Calin GA, Liu CG, Sorrentino A, Croce CM, Peschle C. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci USA. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 23.Ness SA, Kowenz-Leutz E, Casini T, Graf T, Leutz A. Myb and NF-M: combinatorial activators of myeloid genes in heterologous cell types. Genes Dev. 1993;7:749–759. doi: 10.1101/gad.7.5.749. [DOI] [PubMed] [Google Scholar]
- 24.Garzon R, Pichiorri F, Palumbo T, Iuliano R, Cimmino A, Aqeilan R, Volinia S, Bhatt D, Alder H, Marcucci G, Calin GA, Liu CG, Bloomfield CD, Andreeff M, Croce CM. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci U S A. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solar GP, Kerr WG, Zeigler FC, Hess D, Donahue C, de Sauvage FJ, Eaton DL. Role of c-mpl in early hematopoiesis. Blood. 1998;92:4–10. [PubMed] [Google Scholar]
- 26.Fox NE, Priestley GV, Papayannopoulou Th, Kaushansky K. Thrombopoietin (TPO) expands hematopoietic stem cells (HSCs) in vivo. J Clin Invest. 2002;110:389–394. doi: 10.1172/JCI15430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balduini A, D’Apolito M, Arcelli D, Conti V, Pecci A, Pietra D, Danova M, Benvenuto F, Perotti C, Zelante L, Volinia S, Balduini CL, Savoia A. Cord blood expanded CD41+cells: identification of novel components of megakaryocytopoiesis. J Thromb Hemost. 2006;4:848–860. doi: 10.1111/j.1538-7836.2006.01802.x. [DOI] [PubMed] [Google Scholar]
- 28.Vegiopoulos A, Garcia P, Emambokus N, Frampton J. Coordination of erythropoiesis by the transcription factor c-Myb. Blood. 2006;107:4703–4710. doi: 10.1182/blood-2005-07-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaefer A, Magócsi M, Stöcker U, Fandrich A, Marquardt H. Ca2+/Calmodulin-dependent and -independent Down-regulation of c-myb mRNA Levels in Erythropoietin-responsive Murine Erythroleukemia Cells: The role of calcineurin. J Biol Chem. 1996;271:13484–13490. doi: 10.1074/jbc.271.23.13484. [DOI] [PubMed] [Google Scholar]
- 30.Guerra J, Withers DA, Boxer LM. Myb binding sites mediate negative regulation of c-myb expression in T- cell lines. Blood. 1995;86:1873–1880. [PubMed] [Google Scholar]
- 31.Bloch A, Liu XM, Wang LG. Regulation of c-myb expression in ML-1 human myeloblastic leukemia cells by c-ets-1 protein. Adv Enzyme Regul. 1995;35:35–41. doi: 10.1016/0065-2571(94)00019-y. [DOI] [PubMed] [Google Scholar]
- 32.Bartunek P, Kralova J, Blendinger G, Dvorak M, Zenke M. GATA-1 and c-myb crosstalk during red blood cell differentiation through GATA-1 binding sites in the c-myb promoter. Oncogene. 2003;22:1927–1935. doi: 10.1038/sj.onc.1206281. [DOI] [PubMed] [Google Scholar]
- 33.Lauder A, Castellanos A, Weston K. c-Myb transcription is activated by protein kinase B (PKB) following interleukin 2 stimulation of Tcells and is required for PKB-mediated protection from apoptosis. Mol Cell Biol. 2001;21:5797–5805. doi: 10.1128/MCB.21.17.5797-5805.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bender TP, Thompson CB, Kuehl WM. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science. 1987;237:1473–1476. doi: 10.1126/science.3498214. [DOI] [PubMed] [Google Scholar]
- 35.Sano Y, Ishii S. Increased affinity of c-Myb for CREB-binding protein (CBP) after CBP-induced acetylation. J Biol Chem. 2001;276:3674–3682. doi: 10.1074/jbc.M006896200. [DOI] [PubMed] [Google Scholar]
- 36.Verger A, Perdomo J, Crossley M. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 2003;4:137–142. doi: 10.1038/sj.embor.embor738. [DOI] [PMC free article] [PubMed] [Google Scholar]


