Abstract
In this study, we investigate whether the stable segregation of proteins and lipids within the yeast plasma membrane serves a particular biological function. We show that 21 proteins cluster within or associate with the ergosterol-rich membrane compartment of Can1 (MCC). However, proteins of the endocytic machinery are excluded from MCC. In a screen, we identified 28 genes affecting MCC appearance and found that genes involved in lipid biosynthesis and vesicle transport are significantly overrepresented. Deletion of Pil1, a component of eisosomes, or of Nce102, an integral membrane protein of MCC, results in the dissipation of all MCC markers. These deletion mutants also show accelerated endocytosis of MCC-resident permeases Can1 and Fur4. Our data suggest that release from MCC makes these proteins accessible to the endocytic machinery. Addition of arginine to wild-type cells leads to a similar redistribution and increased turnover of Can1. Thus, MCC represents a protective area within the plasma membrane to control turnover of transport proteins.
Introduction
The plasma membrane of fungal cells is laterally compartmented. Various membrane proteins fused to GFP are organized in specific surface patterns, whereas others are distributed homogeneously. Bagnat and Simons (2002) observed that Fus1-GFP, Gas1-derived GFP-glycosylphosphatidylinositol, and ergosterol are clustered at the tip of the shmoo, the mating projection of Saccharomyces cerevisiae. Proteins destined to the tip of the shmoo partition into this compartment and are thus retained and segregated from the rest of the membrane. Wachtler et al. (2003) reported that sterols are localized in distinct regions of the plasma membrane of Schizosaccharomyces pombe in a cell cycle–dependent manner. Membrane sterols are detected at the septum, the site of cell division, and at the growing tips. The phenomenon of sterol-rich domains in yeast plasma membrane was reviewed in Alvarez et al. (2007). Our earlier studies (Malínská et al., 2003, 2004; Grossmann et al., 2007) show that the plasma membrane proteins in S. cerevisiae are distributed in at least three different modes: either they are concentrated in discrete patches, each patch being ∼300 nm in diameter, they occupy a mesh-shaped compartment, which spreads between the patches, or they are homogenously dispersed throughout these two areas. The patchy compartment called membrane compartment of Can1 (MCC) contains, in addition to the arginine transporter Can1, two other proton symporters, Fur4 and Tat2, and three tetraspan proteins of unknown function, Sur7, Fmp45, and Ynl194c (Young et al., 2002; Malínská et al., 2003, 2004; Grossmann et al., 2007). In the mesh-shaped membrane compartment of Pma1, only the most abundant plasma membrane protein, the H+-ATPase, has been localized so far (Malínská et al., 2003). Finally, Hxt1 and Gap1 represent proteins that are homogenously distributed within the plasma membrane (Malínská et al., 2003; Lauwers et al., 2007).
In close vicinity to the plasma membrane and congruent with the MCC domain, the eisosome, a novel organelle postulated to be involved in endocytosis, has recently been described (Walther et al., 2006). The two cytosolic proteins and major constituents of the eisosome, Pil1 and Lsp1, were colocalized with the MCC marker Sur7 (Walther et al., 2006). In this study, to obtain a better understanding of the composition of the MCC, we identified several other proteins associated with this compartment. A collection of yeast strains expressing full-length GFP fusions (Huh et al., 2003) revealed that several gene products exhibit a punctuate pattern. Inspecting this collection, we identified 10 new proteins with patchy localization at the cell cortex. Including independently published data, we were thus able to allocate 21 proteins altogether, the distribution of which fitted the MCC pattern (Roelants et al., 2002; Young et al., 2002; Fadri et al., 2005; Walther et al., 2006, 2007; Luo et al., 2008). Nine of these proteins are members of the actual membrane compartment C, and 12 are putative cytosolic proteins, which gather in the immediate neighborhood of the MCC patches.
The existence of plasma membrane compartments containing distinct sets of proteins leads to questions concerning the relevance of this separation and the mechanism of its formation. Therefore, in the second part of our study, we performed a genome-wide visual screen for deletion mutants in which the formation of MCC is disturbed. Deviations from the original membrane pattern were observed in 28 mutants.
The genes affecting the wild-type plasma membrane compartmentation belong to two main groups: (1) genes involved in lipid biosynthesis and (2) genes involved in vesicle transport. The strongest deviations from the MCC pattern were manifested in nce102Δ and pil1Δ cells, lacking either the integral membrane protein Nce102, which itself is located within MCC, or the cytosolic Pil1 of eisosomes, respectively. In this study, which is focused on the MCC membrane compartmentation, we concentrated on Nce102 and its possible role in membrane organization.
Although compartmentation of the plasma membrane is a widespread phenomenon found in cells from bacteria to humans, physiological roles of this segregation are still debated (Munro, 2003; Douglass and Vale, 2005; Kenworthy, 2008). Our analysis of nce102Δ and pil1Δ mutants manifests a biological function regarding the recycling and/or degradation of plasma membrane proteins in S. cerevisiae. It is shown that Can1 and Fur4 are more rapidly internalized and degraded when dissociated from MCC patches. In accordance with this proposal, established markers of endocytosis locate exclusively outside MCC.
Results
Protein composition of stable cortical patches
Visual inspection of the yeast database of proteins fused to GFP (Huh et al., 2003), covering two thirds of all annotated ORFs, revealed potential patch formation of several proteins. These proteins were tested for colocalization with Sur7–monomeric red fluorescent protein (mRFP), an endogenous marker of MCC (Malínská et al., 2004). In addition to the known set of 11 proteins, 10 new proteins that colocalized with the MCC pattern were identified (Fig. 1 and Table I). The set includes nine integral plasma membrane proteins with either 12 or four predicted transmembrane domains, three of which are transporters for small molecules. The 12 other proteins are soluble, and their patchy appearance at the cell cortex indicates their accumulation at the cytoplasmic side of the plasma membrane. These proteins include the eisosomal components Pil1 and Lsp1, two protein kinases that regulate endocytosis (Pkh1 and Pkh2), Slm1, a protein involved in actin cytoskeleton formation, several flavodoxin-like proteins (Pst2, Rfs1, and Ycp4), and four proteins with an unknown function. A BLAST (basic local alignment search tool) analysis did not identify conserved domains in the 21 proteins that would indicate the existence of a specific targeting sequence motif. Because of the incomplete localization database, even further MCC-associated proteins can be expected.
Figure 1.
10 new proteins sharing the MCC localization. Cortical distributions of 10 proteins (left; green in merge) were colocalized with the MCC pattern marked with Sur7-mRFP (middle; red in merge). Tangential confocal sections are presented showing the cell surface and fluorescence intensity profiles (diagrams) measured along the arrows. Mean filter was applied on the plotted curves to reduce the noise present in the raw data. Red and green curves were normalized to the same maximum value. Bar, 5 μm.
Table I. Members of MCC and associated cytosolic proteins.
| Name | ORF | Molecular function | Biological process | TMDs | Localization references |
|---|---|---|---|---|---|
|
Integral MCC components |
|||||
| Can1 | YEL063C | H+-driven arginine permease | Basic amino acid transport | 12 | Malínská et al., 2003 |
| Fur4 | YBR021W | H+-driven uracil permease | Uracil transport | 12 | Malínská et al., 2004 |
| Tat2 | YOL020W | H+-driven tryptophan and tyrosine permease |
Aromatic amino acid transport | 12 | Grossmann et al., 2007 |
| Nce102 | YPR149W | Unknown | Nonclassical protein secretion | 4 | This study |
| Fmp45 | YDL222C | Unknown | Ascospore formation | 4 | Young et al., 2002 |
| Sur7 | YML052W | Unknown | Ascospore formation | 4 |
Malínská et al., 2003; Young et al., 2002 |
| Ygr131w | YGR131W | Unknown | Unknown | 4 | This study |
| Ylr414c | YLR414C | Unknown | Unknown | 4 | This study |
| Ynl194c | YNL194C | Unknown | Ascospore formation | 4 | Young et al., 2002 |
|
MCC-associated cytosolic proteins |
|||||
| Lsp1 | YPL004C | Protein kinase inhibitor activity |
Endocytosis (eisosome) | 0 | Walther et al., 2006 |
| Mdg1 | YNL173C | Unknown | Pheromone signaling | 0 | This study |
| Pil1 | YGR086C | Protein kinase inhibitor activity |
Endocytosis (eisosome) | 0 | Walther et al., 2006 |
| Pkh1 | YDR490C | Ser/Thr protein kinase | Endocytosis | 0 |
Roelants et al., 2002; Walther et al., 2007; Luo et al., 2008 |
| Pkh2 | YOL100W | Ser/Thr protein kinase | Endocytosis | 0 |
Roelants et al., 2002; Walther et al., 2007; Luo et al., 2008 |
| Pst2 | YDR032C | Unknown | Unknown | 0 | This study |
| Rfs1 | YBR052C | Unknown | Unknown | 0 | This study |
| Slm1 | YIL105C | Phosphoinositide binding | Actin cytoskeleton organization | 0 | This study; Fadri et al., 2005 |
| Slm2 | YNL047C | Phosphoinositide binding | Actin cytoskeleton organization | 0 | Fadri et al., 2005 |
| Ycp4 | YCR004C | Unknown | Unknown | 0 | This study |
| Ygr130c | YGR130C | Unknown | Unknown | 0 | This study |
| Ymr031c | YMR031C | Unknown | Unknown | 0 | This study |
TMD, transmembrane domain. Putative transmembrane domains are given as predicted by TmPred and transmembrane hidden Markov model (TMHMM).
Involvement of nonessential genes in MCC formation
To identify proteins involved in the plasma membrane compartmentation, we performed a visual genome-wide screen for deletion mutants that shows an alteration in or a complete loss of MCC compartmentation. The hexose/H+ symporter HUP1 of the unicellular alga Chlorella kessleri was selected for the genome-wide screen. When expressed in S. cerevisiae, HUP1 accumulates in MCC patches and serves as the most sensitive marker of MCC integrity. Moreover, when expressed in yeast under the control of alcohol dehydrogenase promoter, HUP1 is stably expressed under all growth conditions (Grossmann et al., 2006, 2007). The yeast strain collection of nonessential gene knockouts was transformed with the HUP1-GFP fusion (see Materials and methods). The transformation was successful in 91.3% (4,413/4,836) of mutants from the collection. The screen was performed by taking confocal images of the surface and a cross section of at least 30 cells each plus a differential interference contrast image. For 4,365 strains (98.9%), an analyzable GFP signal was obtained. An altered distribution of HUP1-GFP was detected in 28 strains (Table II). These strains were subsequently checked for the distributions of Can1-GFP, Sur7-GFP, and of the plasma membrane sterols by staining with filipin (Fig. 2 and Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200806035/DC1). A complete image dataset of the mutant phenotypes is available in the supplemental material.
Table II. List of mutants revealed by a genome-wide screen that are affected in MCC formation.
| Name | ORF | Main biological process |
Cellular compartment |
Strength of phenotype
|
|||
|---|---|---|---|---|---|---|---|
| HUP1 | Can1 | Sur7 | Filipin | ||||
| Level I | |||||||
| ERG2 | YMR202W | Ergosterol biosynthesis | ERc | +++ | +++ | ++ | ++ |
| ERG24 | YNL280C | Ergosterol biosynthesis | ERa | +++ | +++ | + | ++ |
| ERG6 | YML008C | Ergosterol biosynthesis | ERa | +++ | ++ | + | ++ |
| NCE102 | YPR149W | Nonclassical protein secretion | Plasma membraned | +++ | +++ | ++ | + |
| OCH1 | YGL038C | N-glycosylation | Golgia | +++ | +++ | + | + |
| PIL1 | YGR086C | Endocytosis (eisosome) | Cytoplasm–plasma membrane associatedb |
+++ | +++ | ++ | ++ |
| Level II | |||||||
| COG1 | YGL223C | Intra-Golgi vesicle-mediated transport | Golgia | ++ | +++ | + | − |
| GOS1 | YHL031C | Intra-Golgi vesicle-mediated transport | Golgib | ++ | + | + | − |
| OPI3 | YJR073C | Phosphatidylcholine biosynthesis | ERa | ++ | +++ | + | − |
| RVS161 | YCR009C | Polarization of the actin cytoskeleton, endocytosis, and cell polarity |
Cytoplasm–plasma membrane associatedb |
++ | ++ | + | − |
| SUR4 | YLR372W | Sphingolipid biosynthesis | ERa | +++ | +++ | + | − |
| TAF14 | YPL129W | RNA polymerase II transcription initiation and chromatin modification |
Nucleusa | + | + | + | − |
| VPS52 | YDR484W | Retrograde transport and endosome to Golgi |
Golgia | +++ | ++ | + | − |
| VPS54 | YDR027C | Retrograde transport and endosome to Golgi |
Golgia | +++ | +++ | + | − |
| Level III | |||||||
| CAX4 | YGR036C | N-glycosylation | ERa | +++ | ++ | − | − |
| ELP6 | YMR312W | Regulation of transcription | Nucleusc | + | + | − | − |
| ERG5 | YMR015C | Ergosterol biosynthesis | ERa | +++ | +++ | − | − |
| FYV6 | YNL133C | Unknown | Nucleusa | +++ | + | − | − |
| HNT3 | YOR154W | Unknown | Cytoplasma | +++ | +++ | − | − |
| MNN10 | YDR245W | N-glycosylation | Golgia | ++ | − | − | − |
| MNN11 | YER001W | N-glycosylation | Golgia | + | + | − | − |
| NOT3 | YIL038C | Regulation of transcription | Cytoplasma | + | + | − | − |
| PEP12 | YOR036W | Golgi to vacuole transport | Golgib | +++ | +++ | − | − |
| PEP7 | YDR323C | Golgi to vacuole transport | Endosomea | +++ | ++ | − | − |
| PER1 | YCR044C | GPI anchor biosynthesis | ERa | ++ | ++ | − | − |
| RPL13B | YMR142C | Translation | Cytoplasma | ++ | ++ | − | − |
| SLM1 | YIL105C | Actin cytoskeleton organization | Cytoplasm–plasma membrane associatedd |
++ | + | − | − |
| SWA2 | YDR320C | Vesicular transport | ERb | ++ | + | − | − |
GPI, glycosylphosphatidylinositol. The phenotypes are categorized as follows: wild type–like, −; weak, +; medium, ++; strong, +++. Examples are given in Fig. 2.
GFP localization database; University of California, San Fransisco.
Inferred from direct assays.
Inferred from site of activity.
This study.
Figure 2.
Distribution of MCC markers in selected knockout strains. Distributions of HUP1-GFP, Can1-GFP, Sur7-GFP, and filipin-stained sterols were monitored in the library of single gene deletion strains (see Materials and methods). Examples of detected phenotypes (classification of phenotypes: wild type [WT]–like, −; weak, +; medium, ++; strong, +++) on tangential confocal sections (HUP1, Can1, and Sur7) or wide-field images (filipin; transversal sections) are presented. Note the relatively high background fluorescence intensity between MCC patches in cells expressing HUP1-GFP (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200806035/DC1; Grossmann et al., 2006). For a full dataset of all mutant phenotypes listed in Table II, see supplemental material. Bars, 5 μm.
27 out of 28 strains affected in HUP1 distribution also showed an altered Can1 pattern. The Sur7 pattern was affected in 14 strains, and seven deletions affected sterol distribution (Table II). Thus, although HUP1 is a heterologous protein, its association with the MCC is controlled by the same factors as Can1. The accumulation of Sur7 and sterols in MCC patches is less sensitive, defining at least three levels of MCC formation: a core of six proteins (level I), a second level of eight (important for efficient SUR7 association with the patches; level II), and a third level of 14 that is required only for the association of the two transporters (level III). The core includes ergosterol biosynthetic genes, the eisosome marker Pil1, the Nce102 protein, and the Golgi protein Och1. A homogenous distribution of all the plasma membrane markers was observed in pil1Δ cells. Only a few enlarged patches are formed in the plasma membrane of this mutant. Similarly, dissipation of Lsp1 and Sur7 patches in pil1Δ cells was reported by Walther et al. (2006). In the nce102Δ strain, HUP1 and Can1 were homogenously distributed, and the Sur7 patches were more diffuse (Fig. 2). Among the nine membrane proteins that are MCC constituents, Nce102 was the only one affecting MCC integrity. The NCE102 deletion also severely affects the number and distribution of eisosomes as judged from the Pil1-GFP pattern, whereas in the PIL1 deletion, the Nce102-GFP fusion protein is completely homogeneous (Fig. S1 B).
Gene ontology term analysis revealed that proteins involved in vesicle-mediated transport (9/28 strains showing altered compartmentation [32%]; background frequency of 4.9%; p-value of 4.7 × 10−4) and lipid biosynthesis (8/28 [27%]; background of 1.5%; p-value of 5.0 × 10−7) were significantly overrepresented among the genes detected in the screen. This strongly suggests that lipids and the lipid composition of the plasma membrane play a major role in lateral compartmentation. To test whether the immediate lipid milieu of Can1 is changed in the mutants exhibiting an altered distribution, we checked whether Can1 is more accessible to increasing concentrations of Triton X-100. As shown in Fig. 3, Can1-GFP solubilized with lower concentrations of detergent in the mutants as compared with the wild type. This agrees with the behavior of Can1 after treating the cells with uncouplers; the protein disperses (Grossmann et al., 2007), and, at the same time, it is more efficiently extractable by Triton X-100 (Fig. 3). Thus, the transporters appear to be recruited to a preexisting core MCC compartment with a specific lipid composition. As a control, we tested the Triton X-100 extractability of Gap1, a protein that is homogeneously distributed in wild-type cells (Lauwers et al., 2007). The data show that there is no difference in the extractability between the wild-type and the Nce102 and Pil1 deletion mutants (Fig. 3).
Figure 3.
Extractability of the transport proteins Can1 and Gap1 by Triton X-100. Membranes were isolated from exponentially growing cells as described in Materials and methods. Aliquots corresponding to 50 μg of membrane protein were treated with increasing concentrations of Triton X-100. The nonsolubilized proteins were resolved by SDS-PAGE and detected by specific antibodies on Western blots. The figure is representative of three independent experiments. WT, wild type.
Nce102 is necessary for the formation of MCC patches
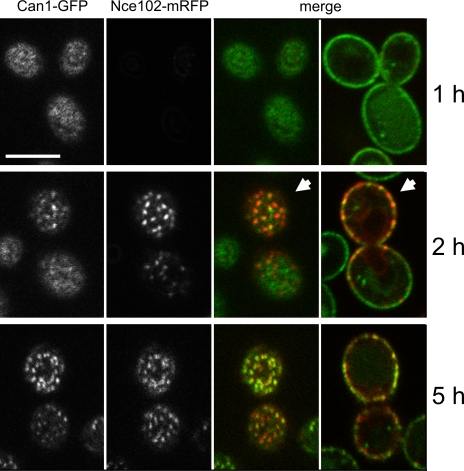
To demonstrate directly the specific role of Nce102 in pattern formation, we tested whether the homogeneous distribution of Can1-GFP observed in nce102Δ can be restored to wild-type levels by regulated expression of the NCE102 gene. The nce102Δ strain expressing Can1-GFP was transformed with a vector containing NCE102 under a galactose-inducible promoter (pGal1). The nce102Δ phenotype persisted when the cells were grown in medium containing 2% raffinose or 2% glucose. When 2% galactose was used as a carbon source, the Nce102 protein was fully expressed in <2 h, and under these conditions the normal distribution of Can1 was restored (Fig. 4). This demonstrates that Nce102 is required for the de novo formation and probably also for the maintenance of the MCC compartment. However, the association of Can1 with MCC is delayed as compared with the appearance of Nce102-mRFP. After a 2-h induction, the mutant phenotype is rescued in buds and younger cells.
Figure 4.
Nce102 is required for MCC localization of Can1. Plasma membrane distribution of Can1-GFP was observed in nce102Δ cells expressing Nce102-mRFP under the control of a galactose-inducible promoter. After the induction, gradual restoration of the wild type–like patchy distribution of Can1-GFP was followed on tangential confocal sections. On transversal sections of the same cells, it is notable that the pattern of bud membrane was restored earlier (arrowheads). Bar, 5 μm.
Organization of MCC patches in relation to cell growth
As shown in Fig. 1, Nce102 clearly localizes to the MCC of mother cells. However, in young buds, Nce102-GFP appears to be distributed homogenously (Fig. 5 A). Only after the bud diameter reaches about one third of the mother (Fig. 5 A, state III) do discernible patches become apparent. In contrast, Sur7-GFP and Pil1 are organized in patches in the buds from the time of their emergence (Fig. 5 A). One of the main differences between young bud and mother cell membranes is the rate of growth, which is determined by the arrival of secretory cargo. A similar situation exists in growing shmoos, the mating projections induced by mating factors. In agreement with this similarity between shmoos and buds, we observed a homogeneous distribution of Nce102-GFP and patches of Sur7-GFP and Pil1-GFP in shmoos (Fig. 5 B). Interestingly, Can1-GFP, which is hard to visualize in young buds, can be seen in shmoos, and its distribution correlates with that of Nce102-GFP. These data implicate an interdependence of Can1 and Nce102. However, these two proteins can be separated from each other by membrane depolarization. As reported previously, Can1 patches can be dissipated by the addition of FCCP (carbonyl cyanide p-trifluoromethoxyphenylhydrazone), a potent protonophore (Grossmann et al., 2007). The addition of FCCP to cells expressing Can1-GFP and Nce102-mRFP causes dissipation of the Can1 patches only, whereas Nce102 remains in the MCC (Fig. 6). This indicates that the interaction between Nce102 and Can1 is mainly stabilized by electrostatic forces. Thus, Nce102 behaves similarly to Sur7, which has previously been shown to not respond to uncouplers either (Grossmann et al., 2007).
Figure 5.
Nce102 is homogenously distributed in membranes of buds and shmoos. (A) Development of the membrane distribution of Nce102-GFP, Sur7-GFP, and Pil1-GFP in mother cells and buds of increasing size (I–IV). (B) Localization of the proteins examined in A and Can1-GFP in cells (genetic background: BY4741 MATa; OD600 = 0.25) treated with 30 μg/ml α factor for 2 h. 3D reconstructions of confocal z stacks are presented. Bars, 2 μm.
Figure 6.
Can1-GFP dissociates from Nce102-mRFP upon membrane depolarization. Surface views of cells expressing Can1-GFP and Nce102-mRFP before (top) and 2 min after the addition of 50 μM FCCP to the medium (bottom) are shown. Bar, 2 μm.
MCC transporters are more prone to degradation in pil1Δ and nce102Δ cells
We checked whether the compartmentation has any effect on protein stability and/or its turnover. Walther et al. (2006) reported a defect in endocytosis of the mating factor receptor Ste3 in the pil1Δ mutant, which was taken as an indication of the role of Pil1 in endocytosis. Because of the fact that Pil1 is located in close vicinity to the patches containing Can1, we also tested whether the endocytosis of arginine permease is dependent on the presence of Pil1, which would point to a more general role of Pil1 in this process. The expression of CAN1 is highly dependent on the growth phase, and its gene product is subjected to continuous turnover. At high extracellular arginine concentration, the permease is endocytosed and subsequently targeted to the vacuole (Opekarová et al., 1998). Its endocytosis follows the classical clathrin–actin-mediated pathway because Can1 turnover is severely inhibited in the sla2/end4 mutant at the nonpermissive temperature (unpublished data). Can1-GFP turnover was monitored in wild-type, nce102Δ, and pil1Δ cells in the presence of cycloheximide. The basal turnover of Can1-GFP was faster in the two mutants (Fig. 7 A); i.e., when it was not localized in the patches of the MCC compartment. This result has also been obtained with another proton symporter accumulated in MCC, the uracil permease Fur4 (Fig. 7 B; Dupre and Haguenauer-Tsapis, 2003; Bultynck et al., 2006). Like Can1, Fur4 is also homogenously distributed in the plasma membrane of nce102Δ and pil1Δ mutants (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200806035/DC1).
Figure 7.
Degradation of MCC transporters is accelerated in mutants affected in the domain formation. (A and B) Exponentially growing cultures of wild-type (WT), nce102Δ, and pil1Δ cells expressing Can1-GFP (A) and Fur4-GFP (B) were treated with cycloheximide. At the given time points, total membranes were isolated from the culture aliquots (see Materials and methods). The membrane proteins were resolved by SDS-PAGE, and Can1-GFP and Fur4-GFP were detected by anti-GFP antibody on Western blots. 2.5 μg of the total protein was loaded into each lane. Black lines indicate that intervening lanes have been spliced out.
Similar although more rapid effects were observed when the fluorescence of cells expressing Can1-GFP was followed after the addition of 5 mM arginine (Fig. 8 A and Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200806035/DC1). Also, Can1-GFP was internalized faster in the two mutants as compared with the wild-type cells. After 90 min of incubation, stronger vacuolar staining is apparent in the mutants as compared with the wild type. Furthermore, we observed that in the wild-type cells, the Can1-GFP pattern becomes more dispersed upon the addition of arginine (Fig. 8 A, WT surface). In the presence of its abundant substrate, the transporter's distribution is similar to that observed in the mutants from our screen (i.e., Can1 is dissipated from the patches to the surrounding area). Accordingly, the Triton X-100 extractability of Can1-GFP from membranes of cells treated with 5 mM arginine for 10 min is changed (Fig. 8 B) and is comparable with that found in the membranes from the two mutants (Fig. 3). However, the addition of cycloheximide does not cause a spreading of Can1-GFP (unpublished data). Induced spreading of a transport protein was observed previously for HUP1-GFP; patchy accumulation of this transporter was more distinct in a low glucose medium (Grossmann et al., 2006).
Figure 8.
Can1 is released from MCC patches before endocytosis. (A) Can1-GFP was localized in the wild-type (WT), nce102Δ, and pil1Δ cells before (top) and 90 min after the addition of 5 mM arginine (middle). Arginine-induced loss of patchy Can1-GFP pattern on the surface confocal sections (left) and the amount of the internalized protein on transversal sections could be easily followed. Note the significantly more intensive vacuolar staining in the mutants lacking the MCC patches as compared with wild type. The whole experiment is documented in Fig. S3 (available at http://www.jcb.org/cgi/content/full/jcb.200806035/DC1). (B) Extractability of Can1-GFP in Triton X-100 was detected in the membranes of wild-type cells before and 10 min after the addition of 5 mM arginine. Anti-GFP antibody was used for the detection of the protein on Western blots. Bar, 5 μm.
Classical endocytosis occurs apart from MCC
The aforementioned results imply that the internalization of Can1 takes place when the protein has left the MCC microdomain. Therefore, we investigated how typical endocytosis proteins are located in relation to MCC.
The inspection of the localization database revealed a group of cortically clustered cytosolic proteins. All of these proteins belong to the group of cortical actin-binding proteins that are involved in endocytosis (Toret et al., 2008). For example, Rvs161 is a cytosolic protein, which during endocytosis clusters at the endocytic site and remains there for several seconds (Kaksonen et al., 2005). We document its movement in Videos 1 and 2 (available at http://www.jcb.org/cgi/content/full/jcb.200806035/DC1). Its patches are transient, but, once formed, they do not move parallel to the membrane.
We monitored the colocalization of Rvs161-GFP and Sur7-mRFP for 180 s (36 frames; one frame per 5 s). Being aware of the thickness of a confocal section (∼700 nm), artifactual overlaps of the fluorescence signals were avoided by analyzing tangential confocal sections that revealed the cell surface. Fig. 9 B shows a merged image of all time frames for Rvs161-GFP compared with Sur7-mRFP. The overlap with MCC patches is minimal (Fig. 9 B and Fig. S4 A, available at http://www.jcb.org/cgi/content/full/jcb.200806035/DC1). Similarly, mutually exclusive localization of MCC patches and generally accepted early endocytosis markers Ede1 (Fig. 9 C and Fig. S4 B) and Sla2 (not depicted) were observed. This strongly supports the observation that Can1 is more rapidly degraded when it is not confined to MCC, as is the case in the pil1Δ and nce102Δ mutants. This is also consistent with the observation that in comparison to basal turnover in the presence of cycloheximide, Can1 is internalized and degraded considerably faster in response to the addition of 5 mM arginine followed by the rapid spreading of permease (Fig. 8 B).
Figure 9.
Sites of classical endocytosis do not colocalize with MCC. (B and C) The plasma membrane distributions of Rvs161 (B) and Ede1 (C), markers of late and early endocytic steps, respectively, were tested for colocalization with the MCC marker Sur7. For comparison, localization of the MCC resident Nce102 was analyzed (A). Tangential confocal sections showing the cell surface are presented. Because of a high mobility of Rvs161 patches, a maximum intensity projection of 36 frames (5 s per frame) instead of a single frame is shown in B. In this arrangement, a higher number of Rvs161 patches could be localized toward the stable Sur7 pattern at the same time. The rate of colocalization was quantified by fluorescence intensity profiles (top diagrams and arrows in merge) and 2D scatter plots of the whole full resolution images (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200806035/DC1). For easy orientation in the scatter plots, real pixel colors were used. Note the diagonal orientation of the Nce102-derived scatter plot demonstrating the colocalization of red and green fluorescence signals and a clear separation of red and green pixels in the two other cases. Examples of Sur7 patches adjacent to endocytic sites are highlighted (arrowheads). Bar, 5 μm.
Discussion
More and more evidence indicates that the ability of the plasma membrane to form subcompartments through the generation of lateral microdomains is a widespread feature throughout all organisms. Membrane microdomains in living cells were first visualized by light microscopy in the budding yeast S. cerevisiae (Young et al., 2002; Malínská et al., 2003), and later they were also reported in plants (Sutter et al., 2006; Homann et al., 2007) and even in bacteria (Johnson et al., 2004; Matsumoto et al., 2006). Novel high resolution techniques of light microscopy can be used to visualize even very small microdomains in mammalian cells. Using stimulated emission-depletion microscopy, small syntaxin clusters of ∼70 nm in diameter have been resolved in vivo in neuroendocrine PC12 cells (Sieber et al., 2006). In mammalian cells, the clustering of specific membrane proteins is mainly discussed in relation to signaling (Tian et al., 2007; for review see Simons and Ikonen, 1997).
In this study, we focused on 300-nm patches of MCC, large plasma membrane domains observed in S. cerevisiae (Young et al., 2002; Malínská et al., 2003) and shown to be specific in lipid composition (Grossmann et al., 2007). By performing several colocalization experiments and incorporating the localization studies of others (Roelants et al., 2002; Young et al., 2002; Fadri et al., 2005; Walther et al., 2006, 2007; Luo et al., 2008), we listed a total of 21 proteins that mimic the distribution pattern of MCC (Table I). This list makes no claim to be complete, but it already presents a considerable number of proteins, which either reside within the membrane or are situated intracellularly, and are closely associated with the membrane compartment C. According to membrane topology predictions, only nine of these are integral membrane proteins, whereas the other 12 seem to be cytosolic (Table I). The first group contains three members of the major facilitator family (Can1, Fur4, and Tat2) and six proteins with four predicted transmembrane domains but with unknown functions. Among the cytosolic proteins, the primary components of eisosomes Pil1 and Lsp1 are listed. Eisosomes are large structures located internally next to the MCC patches; they were postulated to be static initiation sites for Ste3 endocytosis (Walther et al., 2006).
Approaching the question about the function of MCC, we performed a genome-wide screen to identify mutants exhibiting a defect in the plasma membrane compartmentation. Their classification and further analysis could reveal a mechanism for MCC formation as well as physiological consequences resulting from the loss of the protein arrangements in the patches.
We reported previously that the stable patchy distribution of Can1- and HUP1-GFP is affected in the ergosterol biosynthesis mutants erg6Δ and erg24Δ. The trafficking of the proteins is disturbed in these mutants; however, the fraction of the protein reaching the plasma membrane lost the patchy distribution (Malínská et al., 2003; Grossmann et al., 2006). In this study, we searched for further mutants exhibiting a similar defect in MCC sorting. An additional 26 mutants with a defect in the patterning of HUP1-GFP and/or Can1-GFP were identified. Less than half of these mutants also exhibited significant changes in the Sur7-GFP pattern, and even fewer were affected in the distribution of sterols. It goes without saying that such a screen is necessarily incomplete because only nonessential genes were analyzed, and important proteins might have been missed because of genetic redundancy.
Among the mutants found to be affected in MCC formation and design, two groups are overrepresented: mutants involved in vesicular transport and those involved in lipid metabolism. In fact, these two categories are functionally interconnected. Because of their hydrophobicity, the membrane proteins are trafficked in a complex with specific lipids within the vesicle membrane (Opekarová, 2004). An interaction of membrane proteins with specific lipids is essential for their correct targeting and activity. In S. cerevisiae, Pma1 targeting and stabilization require sphingolipids, especially their very long acyl chain component (Lee et al., 2002; Gaigg et al., 2005; Gaigg et al., 2006). However, the tryptophan permease Tat2 requires ergosterol specifically for the plasma membrane targeting (Umebayashi and Nakano, 2003). Similarly, HUP1 activity is decreased when the protein is heterologously expressed in a yeast mutant lacking ergosterol (Grossmann et al., 2006). The general amino acid permease Gap1 is not stable in the absence of sphingolipids, and it is inactive when forced to localize into the plasma membrane in endocytic mutants (Lauwers et al., 2007). A specific requirement for phosphatidyl ethanolamine was demonstrated for Can1 targeting (Opekarová et al., 2005). In a genome-wide visual screen for a role of sphingolipids and ergosterol in the cell surface delivery, Proszynski et al. (2005) demonstrated that they are indispensable for the plasma membrane delivery of a Fus-Mid-GFP marker protein (Proszynski et al., 2005). The specific lipid surrounding of membrane proteins is maintained even in the plasma membrane in the form of lateral membrane microdomains. For example, Tat2 remains associated with ergosterol that is enriched within the MCC patches (Grossmann et al., 2007).
The first notion of lipid-determined plasma membrane compartmentation is related to the observation of lipid rafts in mammalian cells (for reviews see Simons and Ikonen, 1997; Edidin, 2003; Jacobson et al., 2007). The raft fraction has operationally been defined as mild detergent-resistant membranes (Brown and Rose, 1992). However, all of the plasma membrane proteins of yeast analyzed so far, independent of their mode of localization, were associated with detergent-resistant membranes (Lauwers and André, 2006; Lauwers et al., 2007). Apparently, the observed MCC patches represent an organization of a specific type of rafts.
Our screen revealed only six proteins, the absence of which led to a clear effect on the distribution of all four markers (HUP1, Can1, Sur7, and ergosterol). In addition to the three members involved in the ergosterol biosynthesis (Erg24, Erg6, and Erg2), these are Och1, Nce102, and Pil1. Interestingly, the latter two colocalize with MCC; Nce102 is an integral part of MCC, and Pil1 is an eisosome component, which is in some way associated with the plasma membrane. Thus, Nce102 is the only protein component of MCC that controls the association of several transporters with this compartment. Pil1 is a primary component of eisosomes and was postulated to be involved in endocytosis of Ste3 (Walther et al., 2006). It is regulated by the protein kinases Pkh1 and Pkh2 (Walther et al., 2007; Luo et al., 2008), which are activated by sphingoid long-chain bases (Friant et al., 2001). Long-chain bases themselves are required for the internalization step of endocytosis (Zanolari et al., 2000). However, Pil1 down-regulates the Pkc1–mitogen-activated protein and Ypk1 pathways also implicated in endocytosis (Zhang et al., 2004).
The gene product of NCE102 has been suggested to be involved in nonclassical protein secretion (Cleves et al., 1996). The gene encodes a protein of ∼19 kD with four predicted membrane-spanning domains. As revealed in the screen, Nce102 expression is a prerequisite for the segregation of Can1 (and other specific transporters) into the MCC patches. The two proteins Pil1 and Nce102 have common features in that they are highly conserved throughout fungi (Ascomycota), their expression is cell cycle dependent (Spellman et al., 1998), and they are induced by stress (Gasch et al., 2000, 2001; Suzuki et al., 2003).
As indicated by Triton X-100 extraction, the lipid environment of Can1 changes when the protein is liberated from the MCC patches regardless of the mechanism of its release (membrane depolarization, mutations, or excess of substrate; Fig. 3). We addressed the question of whether the protein localization could affect its turnover. Indeed, we observed a faster Can1 internalization in both pil1Δ and nce102Δ mutants either upon the addition of an excess substrate (arginine) or when occuring after substrate-independent basal turnover/degradation in the presence of cycloheximide (Fig. 8 A and Fig. 7, respectively). The delayed rate of endocytosis in wild-type cells indicates that the proteins localized in MCC patches are protected against internalization.
To test mutual localization of sites of endocytosis and the MCC patches, we used GFP fusions of Rvs161 and two other endocytic markers, Ede1 and Sla2, for visualization of numerous endocytic events over time. We detected no overlap of these marker proteins with MCC patches, which must mean that endocytosis occurs outside the MCC. This statement was independently strengthened by the observation of a gradual release of Can1-GFP from its compartment when endocytosis was induced by an excess of arginine. Collectively, the separation of the endocytosis machinery from MCC and the release of Can1-GFP before endocytosis support the idea of MCC as a protective area providing Can1 with higher stability. As soon as the transporter is released from the protective surrounding, it is exposed to internalization by classical mechanisms. This explains why endocytosis of Can1 occurs faster in mutants that are unable to build up the MCC patches, which is obviously a shelter for certain membrane constituents. The same features have been observed for the Fur4 transport protein. Our observations do not necessarily contradict the findings of Walther et al. (2006), who reported a decreased rate of Ste3 endocytosis in pil1Δ cells. Proteins released from the MCC patches could be occasional competitors in endocytosis for the proteins homogenously distributed in the membrane, like Ste3 (Oestreich et al., 2007); the latter would always be exposed to internalization forces. This might be the reason why the endocytosis of Ste3 is retarded in mutants devoid of MCC patches (pil1Δ). This explanation does not hold for the artificial endocytic substrate FM4-64, the use of which has been reported to have several side effects. For example, we observed that the addition of FM4-64 to cells expressing Can1-GFP causes a rapid loss of the permease patterning comparable with that after the addition of uncoupling agents (unpublished data). Very recently, a retardation of Ste2-GFP and FM4-64 internalization was reported in the sur7Δ strain of Candida albicans. However, in this case, the eisosomes remained intact (Alvarez et al., 2008).
So far, we can only speculate on the molecular mechanism of the MCC shelter function. Because Pil1 down-regulates the Pkc1–mitogen-activated protein and Ypk1 pathways also involved in endocytosis (Zhang et al., 2004), it is conceivable that endocytosis is inhibited at places of Pil1 accumulation. In other words, Pil1 clustering underneath MCC patches directs the endocytic activity outside this specialized membrane area. As follows from our results, Nce102 anchors Can1, Fur4, and others within this area and thus protects them from internalization. The anchoring interaction seems to be mediated by Coulombic forces, as Can1 but not Nce102 (Fig. 6) is released from the MCC patches after membrane depolarization. With respect to endocytosis, two functionally distinct compartments coexist within the plasma membrane: MCC patches with the combined protecting potential of Nce102 and Pil1 and a remaining unprotected area (Fig. 10). This phenomenon can serve as an example of a spatially confined regulatory mechanism.
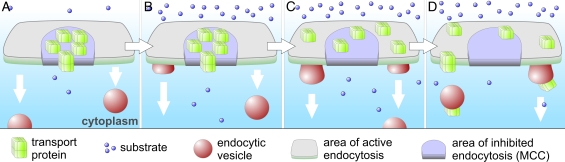
Figure 10.
Model of spatially confined protein turnover. In the presence of low substrate concentrations, specific transporters are concentrated in MCC and protected against internalization (A). After the excess of substrate is supplied (B), the transporters are released from the MCC patches to the surrounding membrane (C) and subjected to endocytosis (D).
Materials and methods
Strains and growth conditions
Plasmid amplification was performed in the Escherichia coli host XL1-blue (Bullock et al., 1987). The bacterial strains were grown at 37°C in 2TY medium (1% tryptone, 1.6% yeast extract, and 0.5% NaCl) supplemented with 100 μg/ml ampicillin for a selection of transformants. S. cerevisiae strains used in this study are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200806035/DC1). Yeast cells were cultured in a rich medium YPD (1% yeast extract, 2% peptone, and 2% glucose) or in a synthetic minimal medium (SD; 0.67% yeast nitrogen base without amino acids [Difco; BD] and 2% glucose supplemented with essential amino acids). To induce Gap1 expression, cells were grown in medium containing 0.1% proline as the sole nitrogen source. Endocytosis of Can1 was induced by the addition of 5 mM arginine to the exponentially growing culture. To generate an inducible NCE102 expression system, the NCE102 ORF was cloned into the GAL1 expression vector p415 GAL1 (Mumberg et al., 1994) as a HindIII–XhoI fragment. NCE102 expression was repressed in SD media containing either 2% glucose or raffinose. Transcription was induced in SD medium containing 2% galactose as the sole carbon source.
High throughput transformation of a deletion strain collection
The high throughput transformation of a yeast knockout collection (initial screen, yeast homozygous diploid collection [BY4743; Open Biosystems]; reconfirmation, yeast haploid collection [BY4741 MATa; Euroscarf]) was based on the lithium acetate (LiAc)/single-stranded carrier DNA/polyethylene glycol method described previously by Gietz and Woods (2002). The protocol was adapted to a 96-well format as follows: yeast strains were inoculated into a 96-well deep-well plate containing 1.5 ml YPD and were grown for 3 d at 30°C. The medium was supplemented with 50 mM MgCl2 to reduce vapor pressure and avoid evaporation and, thus, cross-contamination by capillary force. The medium above the cells was removed, and 160-μl aliquots of transformation mix consisting of 12 μl single-stranded carrier DNA (0.2 mg/ml; Sigma-Aldrich), 3 μl of plasmid DNA (0.1 μg/μl), 130 μl polyethylene glycol 3350 (50% wt/vol), and 15 μl LiAc/Tris-EDTA (1.0 M LiAc; 0.1 M Tris, pH 7.5, and 1.0 mM EDTA, pH 8.0) was added to the sediment. Cells were resuspended by pipetting and incubated at 30°C for 5 h. Aliquots of 15 μl DMSO were added to each well before incubating the plates at 42°C for 35 min (heat shock). Subsequently, 1 ml of selective SD medium (supplemented with 0.2% casamino acids, 0.002% tryptophan, and 0.002% adenine; all wt/vol) lacking uracil was added, and 150 μl of these dilutions was inoculated to 1.5-ml portions of selective SD medium in a fresh 96-well deep-well plate. After 3 d, transformants were visible at the wells' bottom.
Fluorescence microscopy and visual screening
Wide-field images were acquired using a fluorescence microscope (Axiovert 200M; Carl Zeiss, Inc.) with a 100× NA 1.4 Plan Apochromat objective coupled to a camera (AxioCam HRc; Carl Zeiss, Inc.). Confocal sections were scanned by a confocal microscope (LSM510 Meta; Carl Zeiss, Inc.). Fluorescence signals of filipin (wide field; excitation, 360–370 nm/detection, 397-nm long-pass filter), GFP (confocal; 488/505–550 nm), and mRFP (confocal; 543/580–615 nm) were detected. In double-labeling experiments, sequential scanning was used to avoid any cross talk of fluorescence channels. Except for the high throughput screening, living cells were immobilized by a thin slice (∼0.5 mm) of 1% agarose (23°C) dissolved in 50 mM potassium phosphate buffer, pH 5.5. For the screening, transformants were grown in 96-well microplates containing 200 μl of selective medium. Before microscopic observation, the cells were transferred to 96-well microplates with glass bottoms (GE Healthcare).
Filipin staining
Exponentially growing cells were washed in 50 mM potassium phosphate buffer, pH 5.5, diluted to A600 = 0.3, stained with 5 μg/ml filipin (Sigma-Aldrich) for 5 min, washed again in the same buffer, concentrated by brief centrifugation, and observed.
Isolation of crude membranes
Usually, 100 OD600 units of early logarithmic cells were washed twice by 10 mM NaN3/NaF buffer to block endocytosis and were resuspended in 1 ml of ice-cold TNE-I buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 5 mM EDTA) supplemented with protease inhibitors (1 mM PMSF, 4 μM leupeptin, and 2 μM pepstatin). The samples were immediately frozen in liquid nitrogen and stored at −80°C. To isolate crude membranes, the cells were broken with glass beads in a FastPrep instrument (Thermo Fisher Scientific). Unbroken cells and larger cell debris were removed by low speed centrifugation in a centrifuge (Eppendorf) at 2,300 rpm (1 min/2 × 5 min). Crude membranes were pelleted by centrifugation at 14,000 rpm for 75 min and resuspended in TNE-I buffer.
Determination of detergent resistance
Aliquots corresponding to 50 μg of membrane protein in 100 μl TNE-I were treated with increasing concentrations of Triton X-100 (0–0.8%) at room temperature for 30 min. The nonsolubilized material was pelleted by centrifugation (Eppendorf microfuge; 14,000 rpm at 4°C for 30 min) and washed by 100 μl of the corresponding buffers under the same conditions. The pellets were resuspended in 40 μl of sample buffer and dissociated at 90°C for 2 min. Samples of 5 μl were resolved by SDS-PAGE, and Can1-GFP was detected by a specific anti-GFP antibody on a Western blot.
Online supplemental material
Table S1 lists all strains used in this study. Fig. S1 A presents confocal cross sections of selected deletion strains shown in Fig. 2. Fig. S1 B presents further analysis of pil1Δ and nce102Δ cells. In Fig. S2, the localization of Fur4-GFP in these mutants is compared with wild type. Fig. S3 shows the full image set of the experiment presented in Fig. 8 A. Fig. S4, Video 1, and Video 2 contain whole full resolution data used for the 2D scatter plot analyses presented in Fig. 9. Supplemental material also shows a full dataset of all mutant phenotypes listed in Table II. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200806035/DC1.
Supplementary Material
Acknowledgments
We are very grateful to I. Fuchs for excellent technical assistance; to J. Stolz for critical reading of the manuscript and for many stimulating discussions; and to I. Lager, S. Germann, A. Seemann, and A. Lausser for their assistance with the genome-wide screen. We also thank E. Lauwers and B. André as well as M. Schwab, W. Seufert, H. Tschochner, and their laboratories for their help and support.
G. Grossmann, M. Loibl, W. Stahlschmidt, I. Weig-Meckl, and W. Tanner were financially supported by the Deutsche Forschungsgemeinschaft (Priority Program 1108 and TA 36/18-1). M. Opekarová and J. Malinsky were supported by the Grant Agency of the Czech Republic (grants 204/06/0009 [project 204/08/J024] and 204/07/0133, respectively). W.B. Frommer was funded by the National Institutes of Health (National Institute of Diabetes and Digestive and Kidney Diseases; grant 1RO1DK079109-01) and the National Science Foundation 2010 (grant MCB-0618402).
Abbreviations used in this paper: LiAc, lithium acetate; MCC, membrane compartment of Can1; mRFP, monomeric red fluorescent protein.
References
- Alvarez, F.J., L.M. Douglas, and J.B. Konopka. 2007. Sterol-rich plasma membrane domains in fungi. Eukaryot. Cell. 6:755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez, F.J., L.M. Douglas, A. Rosebrock, and J.B. Konopka. 2008. The eisosome protein Sur7 regulates plasma membrane organization and prevents intracellular cell wall growth in Candida albicans. Mol. Biol. Cell. 10.1091/mbc.E08-05-0479. [DOI] [PMC free article] [PubMed]
- Bagnat, M., and K. Simons. 2002. Lipid rafts in protein sorting and cell polarity in budding yeast Saccharomyces cerevisiae. Biol. Chem. 383:1475–1480. [DOI] [PubMed] [Google Scholar]
- Brown, D.A., and J.K. Rose. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 68:533–544. [DOI] [PubMed] [Google Scholar]
- Bullock, W.O., J.M. Fernandez, and J.M. Short. 1987. Xl1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. Biotechniques. 5:376–379. [Google Scholar]
- Bultynck, G., V.L. Heath, A.P. Majeed, J.M. Galan, R. Haguenauer-Tsapis, and M.S. Cyert. 2006. Slm1 and slm2 are novel substrates of the calcineurin phosphatase required for heat stress-induced endocytosis of the yeast uracil permease. Mol. Cell. Biol. 26:4729–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves, A.E., D.N. Cooper, S.H. Barondes, and R.B. Kelly. 1996. A new pathway for protein export in Saccharomyces cerevisiae. J. Cell Biol. 133:1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass, A.D., and R.D. Vale. 2005. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 121:937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupre, S., and R. Haguenauer-Tsapis. 2003. Raft partitioning of the yeast uracil permease during trafficking along the endocytic pathway. Traffic. 4:83–96. [DOI] [PubMed] [Google Scholar]
- Edidin, M. 2003. Lipids on the frontier: a century of cell-membrane bilayers. Nat. Rev. Mol. Cell Biol. 4:414–418. [DOI] [PubMed] [Google Scholar]
- Fadri, M., A. Daquinag, S. Wang, T. Xue, and J. Kunz. 2005. The pleckstrin homology domain proteins Slm1 and Slm2 are required for actin cytoskeleton organization in yeast and bind phosphatidylinositol-4,5-bisphosphate and TORC2. Mol. Biol. Cell. 16:1883–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friant, S., R. Lombardi, T. Schmelzle, M.N. Hall, and H. Riezman. 2001. Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. EMBO J. 20:6783–6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaigg, B., B. Timischl, L. Corbino, and R. Schneiter. 2005. Synthesis of sphingolipids with very long chain fatty acids but not ergosterol is required for routing of newly synthesized plasma membrane ATPase to the cell surface of yeast. J. Biol. Chem. 280:22515–22522. [DOI] [PubMed] [Google Scholar]
- Gaigg, B., A. Toulmay, and R. Schneiter. 2006. Very long-chain fatty acid-containing lipids rather than sphingolipids per se are required for raft association and stable surface transport of newly synthesized plasma membrane ATPase in yeast. J. Biol. Chem. 281:34135–34145. [DOI] [PubMed] [Google Scholar]
- Gasch, A.P., P.T. Spellman, C.M. Kao, O. Carmel-Harel, M.B. Eisen, G. Storz, D. Botstein, and P.O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 11:4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch, A.P., M. Huang, S. Metzner, D. Botstein, S.J. Elledge, and P.O. Brown. 2001. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol. Biol. Cell. 12:2987–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D., and R.A. Woods. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350:87–96. [DOI] [PubMed] [Google Scholar]
- Grossmann, G., M. Opekarová, L. Novakova, J. Stolz, and W. Tanner. 2006. Lipid raft-based membrane compartmentation of a plant transport protein expressed in Saccharomyces cerevisiae. Eukaryot. Cell. 5:945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann, G., M. Opekarová, J. Malinsky, I. Weig-Meckl, and W. Tanner. 2007. Membrane potential governs lateral segregation of plasma membrane proteins and lipids in yeast. EMBO J. 26:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann, U., T. Meckel, J. Hewing, M.T. Hütt, and A.C. Hurst. 2007. Distinct fluorescent pattern of KAT1:GFP in the plasma membrane of Vicia faba guard cells. Eur. J. Cell Biol. 86:489–500. [DOI] [PubMed] [Google Scholar]
- Huh, W.K., J.V. Falvo, L.C. Gerke, A.S. Carroll, R.W. Howson, J.S. Weissman, and E.K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691. [DOI] [PubMed] [Google Scholar]
- Jacobson, K., O.G. Mouritsen, and R.G. Anderson. 2007. Lipid rafts: at a crossroad between cell biology and physics. Nat. Cell Biol. 9:7–14. [DOI] [PubMed] [Google Scholar]
- Johnson, A.S., S. van Horck, and P.J. Lewis. 2004. Dynamic localization of membrane proteins in Bacillus subtilis. Microbiology. 150:2815–2824. [DOI] [PubMed] [Google Scholar]
- Kaksonen, M., C.P. Toret, and D.G. Drubin. 2005. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 123:305–320. [DOI] [PubMed] [Google Scholar]
- Kenworthy, A.K. 2008. Have we become overly reliant on lipid rafts? Talking point on the involvement of lipid rafts in T-cell activation. EMBO Rep. 9:531–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauwers, E., and B. André. 2006. Association of yeast transporters with detergent-resistant membranes correlates with their cell-surface location. Traffic. 7:1045–1059. [DOI] [PubMed] [Google Scholar]
- Lauwers, E., G. Grossmann, and B. André. 2007. Evidence for coupled biogenesis of yeast Gap1 permease and sphingolipids: essential role in transport activity and normal control by ubiquitination. Mol. Biol. Cell. 18:3068–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M.C., S. Hamamoto, and R. Schekman. 2002. Ceramide biosynthesis is required for the formation of the oligomeric H+-ATPase Pma1p in the yeast endoplasmic reticulum. J. Biol. Chem. 277:22395–22401. [DOI] [PubMed] [Google Scholar]
- Luo, G., A. Gruhler, Y. Liu, O.N. Jensen, and R.C. Dickson. 2008. The sphingolipid long-chain base-Pkh1/2-Ypk1/2 signaling pathway regulates eisosome assembly and turnover. J. Biol. Chem. 283:10433–10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malínská, K., J. Malinsky, M. Opekarová, and W. Tanner. 2003. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol. Biol. Cell. 14:4427–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malínská, K., J. Malinsky, M. Opekarová, and W. Tanner. 2004. Distribution of Can1p into stable domains reflects lateral protein segregation within the plasma membrane of living S. cerevisiae cells. J. Cell Sci. 117:6031–6041. [DOI] [PubMed] [Google Scholar]
- Matsumoto, K., J. Kusaka, A. Nishibori, and H. Hara. 2006. Lipid domains in bacterial membranes. Mol. Microbiol. 61:1110–1117. [DOI] [PubMed] [Google Scholar]
- Mumberg, D., R. Müller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro, S. 2003. Lipid rafts: elusive or illusive? Cell. 115:377–388. [DOI] [PubMed] [Google Scholar]
- Oestreich, A.J., B.A. Davies, J.A. Payne, and D.J. Katzmann. 2007. Mvb12 is a novel member of ESCRT-I involved in cargo selection by the multivesicular body pathway. Mol. Biol. Cell. 18:646–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opekarová, M. 2004. Regulation of transporter trafficking by the lipid environment. In Molecular Mechanisms Controlling Transmembrane Transport. E. Boles and R. Krämer, editors. Springer-Verlag Berlin Heidelberg, Berlin. 235–253.
- Opekarová, M., T. Caspari, B. Pinson, D. Bréthes, and W. Tanner. 1998. Post-translational fate of CAN1 permease of Saccharomyces cerevisiae. Yeast. 14:215–224. [DOI] [PubMed] [Google Scholar]
- Opekarová, M., K. Malínská, L. Novakova, and W. Tanner. 2005. Differential effect of phosphatidylethanolamine depletion on raft proteins: further evidence for diversity of rafts in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1711:87–95. [DOI] [PubMed] [Google Scholar]
- Proszynski, T.J., R.W. Klemm, M. Gravert, P.P. Hsu, Y. Gloor, J. Wagner, K. Kozak, H. Grabner, K. Walzer, M. Bagnat, et al. 2005. A genome-wide visual screen reveals a role for sphingolipids and ergosterol in cell surface delivery in yeast. Proc. Natl. Acad. Sci. USA. 102:17981–17986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants, F.M., P.D. Torrance, N. Bezman, and J. Thorner. 2002. Pkh1 and pkh2 differentially phosphorylate and activate ypk1 and ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol. Biol. Cell. 13:3005–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber, J.J., K. Willig, R. Heintzmann, S.W. Hell, and T. Lang. 2006. The SNARE motif is essential for the formation of syntaxin clusters in the plasma membrane. Biophys. J. 90:2843–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature. 387:569–572. [DOI] [PubMed] [Google Scholar]
- Spellman, P.T., G. Sherlock, M.Q. Zhang, V.R. Iyer, K. Anders, M.B. Eisen, P.O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 9:3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter, J.U., P. Campanoni, M. Tyrrell, and M.R. Blatt. 2006. Selective mobility and sensitivity to SNAREs is exhibited by the Arabidopsis KAT1 K+ channel at the plasma membrane. Plant Cell. 18:935–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, C., Y. Hori, and Y. Kashiwagi. 2003. Screening and characterization of transposon-insertion mutants in a pseudohyphal strain of Saccharomyces cerevisiae. Yeast. 20:407–415. [DOI] [PubMed] [Google Scholar]
- Tian, T., A. Harding, K. Inder, S. Plowman, R.G. Parton, and J.F. Hancock. 2007. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat. Cell Biol. 9:905–914. [DOI] [PubMed] [Google Scholar]
- Toret, C.P., L. Lee, M. Sekiya-Kawasaki, and D.G. Drubin. 2008. Multiple pathways regulate endocytic coat disassembly in Saccharomyces cerevisiae for optimal downstream trafficking. Traffic. 9:848–859. [DOI] [PubMed] [Google Scholar]
- Umebayashi, K., and A. Nakano. 2003. Ergosterol is required for targeting of tryptophan permease to the yeast plasma membrane. J. Cell Biol. 161:1117–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachtler, V., S. Rajagopalan, and M.K. Balasubramanian. 2003. Sterol-rich plasma membrane domains in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 116:867–874. [DOI] [PubMed] [Google Scholar]
- Walther, T.C., J.H. Brickner, P.S. Aguilar, S. Bernales, C. Pantoja, and P. Walter. 2006. Eisosomes mark static sites of endocytosis. Nature. 439:998–1003. [DOI] [PubMed] [Google Scholar]
- Walther, T.C., P.S. Aguilar, F. Fröhlich, F. Chu, K. Moreira, A.L. Burlingame, and P. Walter. 2007. Pkh-kinases control eisosome assembly and organization. EMBO J. 26:4946–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, M.E., T.S. Karpova, B. Brügger, D.M. Moschenross, G.K. Wang, R. Schneiter, F.T. Wieland, and J.A. Cooper. 2002. The Sur7p family defines novel cortical domains in Saccharomyces cerevisiae, affects sphingolipid metabolism, and is involved in sporulation. Mol. Cell. Biol. 22:927–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanolari, B., S. Friant, K. Funato, C. Sütterlin, B.J. Stevenson, and H. Riezman. 2000. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 19:2824–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., R.L. Lester, and R.C. Dickson. 2004. Pil1p and Lsp1p negatively regulate the 3-phosphoinositide-dependent protein kinase-like kinase Pkh1p and downstream signaling pathways Pkc1p and Ypk1p. J. Biol. Chem. 279:22030–22038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.