Abstract
MicroRNAs (miRNAs) modulate gene expression by inhibiting mRNA translation and promoting mRNA degradation, but little is known of their potential roles in organ formation or function. miR-133a-1 and miR-133a-2 are identical, muscle-specific miRNAs that are regulated during muscle development by the SRF transcription factor. We show that mice lacking either miR-133a-1 or miR-133a-2 are normal, whereas deletion of both miRNAs causes lethal ventricular-septal defects in approximately half of double-mutant embryos or neonates; miR-133a double-mutant mice that survive to adulthood succumb to dilated cardiomyopathy and heart failure. The absence of miR-133a expression results in ectopic expression of smooth muscle genes in the heart and aberrant cardiomyocyte proliferation. These abnormalities can be attributed, at least in part, to elevated expression of SRF and cyclin D2, which are targets for repression by miR-133a. These findings reveal essential and redundant roles for miR-133a-1 and miR-133a-2 in orchestrating cardiac development, gene expression, and function and point to these miRNAs as critical components of an SRF-dependent myogenic transcriptional circuit.
Keywords: microRNA, heart development, serum response factor, myocyte enhancer factor-2, cyclin D2
Studies in vertebrate and invertebrate model organisms have revealed a network of transcription factors that orchestrates the complex morphogenetic and molecular events of heart development (Olson 2006). Key among these cardiogenic transcription factors are myocyte enhancer factor-2 (MEF2) and serum response factor (SRF), which directly activate the expression of numerous genes encoding proteins involved in muscle differentiation, morphogenesis, growth, and contractility (Miano et al. 2007; Niu et al. 2007; Potthoff and Olson 2007). In addition, recent studies have revealed that MEF2 and SRF regulate the expression of several microRNAs (miRNAs) in the embryonic and adult heart, which may mediate at least a subset of the actions of these cardiogenic transcription factors (Zhao et al. 2005; Liu et al. 2007).
miRNAs are ∼22-nucleotide (nt) regulatory RNAs that associate with the 3′-untranslated regions of mRNAs through imperfect base-pairing, resulting in translational inhibition or mRNA degradation (Valencia-Sanchez et al. 2006). There are estimated to be up to 1000 miRNAs encoded by the human genome. Individual miRNAs can target dozens or even hundreds of mRNAs with varying efficiencies, and individual mRNAs can be targeted by multiple miRNAs, providing a rich layer of regulatory complexity to gene expression programs. Although implicated in diverse developmental processes in fruit flies and nematodes, the functions of miRNAs during vertebrate development are only beginning to be explored, and there have been only a few reports of loss-of-function mutations of miRNAs in mice (Thai et al. 2007; van Rooij et al. 2007; C. Xiao et al. 2007; Zhao et al. 2007; Ventura et al. 2008; Wang et al. 2008).
MEF2 and SRF cooperatively regulate the expression of two bicistronic miRNA clusters encoding miR-133a-1/miR-1-2 and miR-133a-2/miR-1-1 in cardiac and skeletal muscle (Zhao et al. 2005; Liu et al. 2007). A third bicistronic miRNA cluster comprised of miR-206 and miR-133b is expressed specifically in skeletal muscle but not in the heart (McCarthy 2008). miR-133a-1 and miR-133a-2 are identical in sequence, whereas miR-133b differs by only 2 nt at the 3′ terminus (Fig. 1). Similarly, miR-1-1 and miR-1-2 are identical and differ from miR-206 by 4 nt (McCarthy 2008).
Figure 1.
Genomic organization of the miR-133 family. miR-133a-1 and miR-133a-2 have identical sequences, whereas miR-133b differs by 2 nt at the 3′ terminus. Each of the three miR-133 miRNAs is transcribed as a bicistronic transcript with miR-1-2, miR-1-1, or miR-206 as indicated. Genomic distances between the miR coding regions in the mouse genome and expression patterns of each miR cluster are shown.
miR-1-2 is the only one of these six muscle-specific miRNAs to be analyzed by loss-of-function mutation in mice (Zhao et al. 2007). Approximately half of miR-1-2 mutant mice die from ventricular-septal defects (VSDs) between late embryogenesis and birth. A subset of miR-1-2-null mice that survive to adulthood shows subtle electrophysiologic defects, but are otherwise remarkably normal, without evidence of cardiac dysfunction (Zhao et al. 2007). Since miR-1-1 and miR-1-2 are identical, and miR-1-1 expression is unaltered in miR-1-2-null mice, these mice represent only a partial miR-1 gene deletion and do not reveal the complete loss-of-function phenotype of miR-1.
Numerous functions have been ascribed to miR-133, based largely on overexpression studies in cultured muscle cells or partial knockdown experiments (Chen et al. 2006; Boutz et al. 2007a, b; Care et al. 2007; J. Xiao et al. 2007; Xu et al. 2007; Luo et al. 2008), but no complete loss-of-function phenotypes for miR-133 have been reported, and much remains to be learned about the functions of miR-133 in vivo. In the present study, we analyzed the functions of miR-133a-1 and miR-133a-2 in vivo through loss-of-function mutations in the mouse. We show that mice lacking either gene are normal, whereas deletion of both genes results in late embryonic or neonatal lethality due to VSDs, accompanied by abnormalities in cardiomyocyte proliferation, apoptosis, and aberrant expression of smooth muscle genes in the heart. The surviving mutant mice display severe deficits in cardiac contractility and die from heart failure and sudden death. These defects can be attributed, at least in part, to the inappropriate expression of SRF and cyclin D2, which serve as direct targets for translational repression by miR-133a-1 and miR-133a-2. We conclude that miR-133a-1 and miR-133a-2 redundantly regulate the gene expression programs required for normal cardiac growth and function.
Results
Targeted deletion of miR-133a-1 and miR-133a-2
The miR-133 family contains three miRNAs: miR-133a-1, miR-133a-2, and miR-133b, which are transcribed as bicistronic transcripts together with miR-1-2, miR-1-1, and miR-206, respectively (Fig. 1). miR-133a-1 and 133a-2 are identical in sequence and differ from miR-133b by only 2 nt at the 3′ end. The miR-1-2/miR-133a-1 and miR-1-1/miR-133a-2 genes are expressed specifically in cardiac and skeletal muscle, whereas the miR-206/miR-133b gene is expressed only in skeletal muscle (McCarthy 2008; van Rooij et al. 2008).
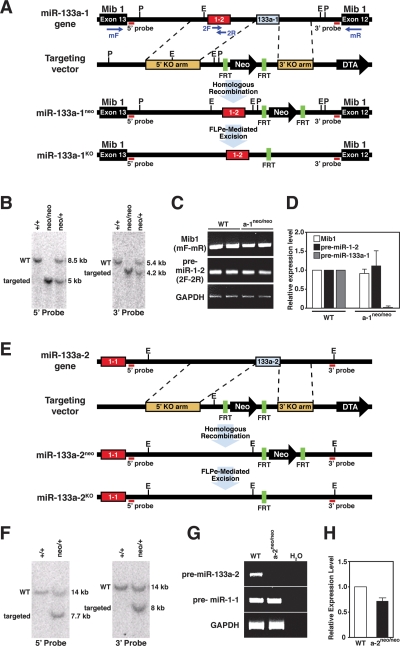
To explore the functions of the two miR-133a genes in vivo, we generated mice lacking miR-133a-1 and miR-133a-2 by homologous recombination. The miR-133a-1 coding region is located on mouse chromosome 18, ∼2.5 kb downstream from the miR-1-2 stem–loop sequence within intron 12 of the mind bomb 1 (Mib1) locus, which is transcribed from the opposing DNA strand (Fig. 2A). Our targeting strategy for miR-133a-1 deleted the pre-miR stem–loop sequence (68 base pair [bp]) and replaced it with a neomycin resistance cassette flanked by two FRT sites, allowing for FLPe recombinase-mediated excision (Fig. 2A). Targeted embryonic stem (ES) cell clones were used to generate chimeric mice, which transmitted the mutant allele through the germline, yielding mice heterozygous for the miR-133a-1neo allele (Fig. 2B). Breeding of these animals yielded miR-133-a1neo/neo mice, which were viable.
Figure 2.
Generation of miR-133a-1 and miR-133a-2 mutant mice. (A) Strategy for targeting of miR-133a-1. The miR-1-2/miR-133a-1 gene is located within intron 12 of the mind bomb 1 gene (Mib1). The genomic structure, targeting vector, and targeted allele for miR-133a-1 are shown. Pre-miR-133a-1 (68 bp) was replaced with a neomycin resistance cassette flanked by FRT sites, which allowed for FLPe recombinase-mediated excision. Probes for Southern blot analysis and primer positions for RT–PCR are shown. (E) EcoRI; (P) PstI. (B) Southern blot analysis for wild-type (WT) and miR-133a-1 mutant mice. Genomic tail DNA from agouti offspring was digested with PstI and probed with the indicated 5′ probe. The wild-type band migrated at 8.5 kb, and the targeted mutant band migrated at 5 kb, indicative of proper transmission of the targeted allele. Tail DNA was also digested with EcoRI and probed with the indicated 3′ probe. The wild-type band migrated at 5.4 kb, and the targeted mutant band migrated at 4.2 kb, confirming germline transmission of the targeted allele. Genotypes of mice are shown on top. (C) Expression of Mib1 and pre-miR-1-2 in wild-type and miR-133a-1 mutant mice detected by RT–PCR. RNA was isolated from hearts of adult wild-type and miR-133a-1 mutant mice (n = 2 for each genotype). Primer positions from A are shown in parentheses at the left. Primers 2F and 2R for miR-1-2 were located within the pre-miR-1-2 sequences. GAPDH mRNA was detected as a loading control. (D) Expression of Mib1, pre-miR-1-2, and pre-miR-133a-1 in wild-type and miR-133a-1 mutant mice detected by real-time PCR. Expression levels of each gene in miR-133a-1 mutant mice were normalized to GAPDH before comparison with expression in wild-type mice (n = 3 for each genotype). The Y-axis represents relative expression level compared with wild-type mice. Error bars indicate SEM. Pre-miR-133a-1 is not detected in miR-133a-1 mutant mice. (E) Strategy for targeting of miR-133a-2. The genomic structure, targeting vector, and targeted allele for miR-133a-2 are shown. Pre-miR-133a-2 (108 bp) was replaced with the neomycin resistance cassette flanked by FRT sites, which allowed for FLPe recombinase-mediated excision. Probes for Southern blot analysis are shown. (E) EcoRI. (F) Southern blot analysis for wild-type and miR-133a-2 mutant mice. Tail DNA from agouti off spring was digested with EcoRI and probed with the indicated 5′ and 3′ probes. Wild-type bands migrated at 14 kb and targeted mutant bands migrated at 7.7 kb using the 5′ probe and 8 kb using the 3′ probe. Genotypes of mice are shown on top. (G) Expression of pre-miR-133a-2 and pre-miR-1-1 in adult wild-type and miR-133a-2 mutant mice detected by RT–PCR. Primers for pre-miR-133a-2 and pre-miR-1-1 were located within their pre-stem–loop sequences. GAPDH levels were detected as a loading control. (H) Expression levels of pre-miR-1-1 in wild-type and miR-133a-2 mutant mice detected by real-time PCR. Expression of pre-miR-1-1 in miR-133a-2 mutant mice was normalized to GAPDH before comparison with expression in wild-type mice (n = 3 for each genotype). The Y-axis represents relative expression levels compared with wild-type mice. Error bars indicate the SEM.
Expression of the pre-miR-1-2 stem–loop and mib1 transcript was unaltered in miR-133a-1neo/neo mice, as detected by RT–PCR with primers specific to the pre-miR-1-2 stem–loop (2F-2R) and primers spanning exons 12 and 13 of the mib1 gene (mF-mR) (Fig. 2C). The absence of the pre-miR-133a-1 stem–loop was confirmed in homozygous mutant mice by real-time PCR with primers specific for the pre-miR-133a-1 stem–loop (Fig. 2D). Heterozygous miR-133a-1neo/+ mice were bred to mice expressing the FLPe recombinase in the male germline (Rodriguez et al. 2000) in order to remove the neomycin resistance cassette, generating the miR-133a-1KO allele.
The miR-133a-2 and miR-1-1 coding regions are separated by ∼9 kb on chromosome 2 (Fig. 2E). We replaced a 108-bp genomic region encoding the pre-miR-133a-2 stem–loop with a neomycin resistance cassette flanked by two FRT sites (Fig. 2E). Targeting and removal of the neomycin resistance cassette by breeding to mice expressing FLPe recombinase were performed as described for the generation of miR-133a-1KO mice (Fig. 2E,F). miR-133a-2KO/KO mice were also viable. The pre-miR-133a-2 stem–loop was undetectable in heart tissue from homozygous miR-133a-2neo/neo mice with primers specific to pre-miR-133a-2 stem–loop sequence, whereas the expression of pre-miR-1-1 was unaffected in homozygous mutants, as shown by RT–PCR and real-time PCR (Fig. 2G,H).
miR-133a-1KO/KO and miR-133a-2KO/KO mice were obtained at Mendelian ratios from intercrosses of heterozygous mice (Supplemental Table 1) and showed no apparent defects in behavior, weight, or life span. Hearts of mutant mice also appeared normal by histological analysis (data not shown). We also performed thoracic aortic banding (TAB) on these mice to test their response to pressure overload and observed no difference in the cardiac hypertrophic response between wild-type and mutant mice (data not shown). There was an ∼50% decrease in mature miR-133a expression in hearts of miR-133a-1KO/KO and miR-133a-2KO/KO mice relative to wild-type littermates (Supplemental Fig. S1A).
Generation of miR-133a-1/133a-2 double-mutant (dKO) mice
To generate miR-133a-1/133a-2 dKO mice, we first generated double-heterozygous mutant mice, which were bred to obtain mice lacking both miR-133a-1 and 133a-2. During the course of mating, we obtained miR-133a-1neo/neo; 133a-2neo/+ mice and miR-133a-1neo/+; 133a-2neo/neo mice at Mendelian ratios, indicating that a single miR-133a allele was sufficient for normal muscle development.
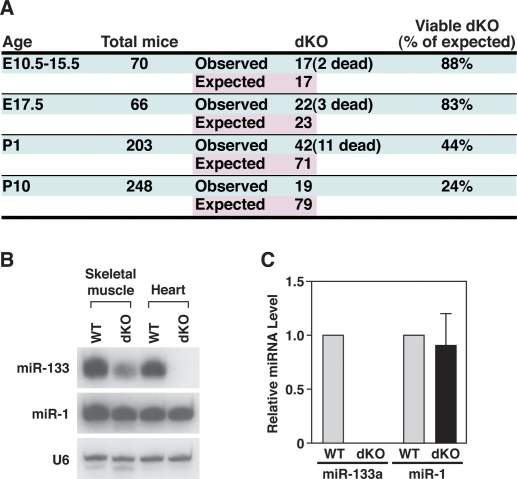
We intercrossed mice with single wild-type miR133a alleles to obtain dKO mice lacking both miR-133a alleles. dKO mice were viable, but underrepresented at birth, and 50% of these mice died between postnatal day 0 (P0) and P1 (Fig. 3A). However, ∼24% of dKO mice survived to adulthood (Fig. 3A). Analysis of offspring from timed matings of heterozygous mutant mice revealed dKO mutant embryos between embryonic day 10.5 (E10.5) and E17.5 (Fig. 3A) at a frequency slightly less than Mendelian with occasional dead dKO embryos.
Figure 3.
Expression of miR-133a and miR-1 in mutant mice. (A) Genotypes of offspring from miR-133a mutant intercrosses. Timed matings were set up from miR-133a-1neo/neo; miR-133a-2neo/+ intercrosses, or from miR-133a-1neo/+; miR-133a-2neo/neo intercrosses to obtain dKO embryos. dKO mice were also intercrossed with miR-133a-1neo/neo; miR-133a-2neo/+ mice to obtain dKO mice at P1 and P10. Mice were genotyped at the indicated ages. Numbers of total mice analyzed, dKO mice observed, and dKO mice expected, based on Mendelian inheritance, are shown. (B) Northern blot analysis of heart and skeletal muscle RNA from wild-type and mutant mice at P1. Ten micrograms of RNA from skeletal muscle and heart tissues were used in the Northern blots. 32P-labeled Star-Fire probes for miR-133a and miR-1 were used. U6 probe was used as a loading control. (C) Expression levels of miR-133a and miR-1 in hearts of wild-type and dKO mutant mice at P1 detected by real-time PCR. Expression of miR-133a and miR-1 was normalized to U6 (n = 3 for each genotype). Error bars indicate the SEM.
We performed Northern blot analysis on hearts and skeletal muscle of wild-type and dKO mice at P1. As shown in Figure 3B, no miR-133a expression was detectable in hearts of dKO mice, confirming the targeting strategies completely eliminated expression of the miR-133a genes. Although miR-133b is highly homologous to miR-133a-1 and miR-133a-2, it is expressed specifically in skeletal muscle, but not in the heart. Accordingly, a weak miR-133 signal, representing the expression of miR-133b, was detected in skeletal muscle of dKO animals (Fig. 3B). Importantly, expression of miR-1 was not affected in miR-133a dKO mice (Fig. 3B). Quantification of miR-1 and miR-133a expression in hearts of wild-type and dKO mice at P1 by real-time PCR confirmed the results of the Northern blot (Fig. 3C). Northern blot analysis was also performed on heart and skeletal muscle of dKO and compound heterozygous adult mice, and the same results were obtained (Supplemental Fig. S1B). Moreover, expression of Mib1 in dKO mice was unaltered, as indicated by real-time PCR and Western blot analysis (Supplemental Fig. S1C).
Abnormalities in cardiac structure in miR-133a-1/133a-2-null mice
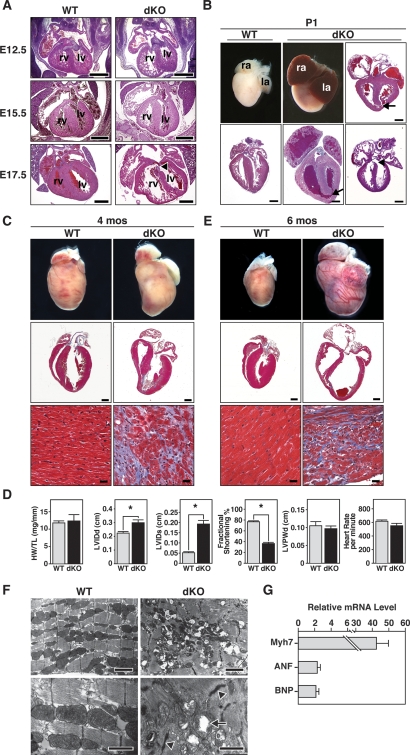
The miR-133a-1/miR-1-2 and miR-133a-2/miR-1-1 genes are expressed throughout the ventricular myocardium and interventricular septum from E8.5 until adulthood (Zhao et al. 2005; Liu et al. 2007). Histologic analysis of embryos between E12.5 and E15.5 indicated that the hearts of the dKO embryos developed normally except that the compact zone of the free wall of the right ventricle was thinned by one-half compared with controls, and the right ventricular chamber was dilated (Fig. 4A). At E17.5, the ventricular free walls of dKO mice were thinner especially at the apex, and, again, the right ventricular chamber was dilated (Fig. 4A).
Figure 4.
Abnormalities of embryonic and adult dKO mutant hearts. (A) Sections of wild-type and dKO hearts during embryogenesis. At E12.5, E15.5, and E17.5, dKO hearts were normal, except for dilatation of the RV and thinning of the RV-free wall. Arrowhead indicates VSD near the atrioventricular valve. (rv) Right ventricle; (lv) left ventricle. Bar, 500 μm. (B) Whole hearts and longitudinal sections of wild-type and dKO hearts at P1. Arrows point to VSD at the apex of heart and arrowhead to VSD near atrioventricular valve. Sections of three different dKO hearts are shown. (ra) Right atrium; (la) left atrium. Bar, 500 μm. (C) Hearts of wild-type and dKO mutant mice at 4 mo of age. Whole-mount pictures of the hearts are shown in the top panel. The middle panels show histological sections stained with Masson’s trichrome. The bottom panels show the interventricular septum at high magnification. Note extensive fibrosis of dKO heart, especially at the junction of the interventricular septum, where VSDs were frequently observed. (Middle panel) Bar, 1 mm. (Bottom panel) Bar, 20 μm. (D) Analyses of cardiac function by echocardiography. Four-month-old male miR-133a dKO mice and their control littermates (n = 11 for each group) were analyzed. (HW/TL) Heart weight-to-tibia length ratio; (LVIDd) left ventricular internal diameter at end-diastole; (LVIDs) left ventricular internal diameter at end-systole; (LVPWd) left ventricle posterior wall thickness at end-diastole. Asterisks indicate statistical significance. The P-values for the following measurements are HW/TL: P = 0.7684; LVIDd: P = 0.0065; LVIDs: P = 3.9e-007; fractional shortening: P = 2.0e-011; LVPWd: P = 0.5424; heart rate: P = 0.1102. (E) Hearts of wild-type and dKO mutant mice that died suddenly at 6 mo of age. Whole-mount pictures and Masson’s trichrome-stained sections of hearts of wild-type and dKO mice at the time of death are shown. The bottom panels show the interventricular septum at high magnification. Note severe ventricular dilatation and fibrosis of dKO hearts. (Middle panel) Bar, 1 mm. (Bottom panel) Bar, 20 μm. (F) Transmission electron micrographs of adult wild-type and dKO mutant mice at 4 mo of age show disorganized sarcomeres and mitochondrial abnormalities in the mutant. Arrowheads point to abnormal Z-lines and arrows point to mitochondria in dKO mutant heart. (Top panels) Bar, 2 μm. (Bottom panel) Bar, 1 μm. (G) Transcripts for the indicated markers of cardiac stress were measured by real-time PCR in RNA samples from wild-type and dKO mice at 4 mo of age. Expression levels in dKO mice are expressed relative to expression in wild-type mice (n = 3 for each genotype). Error bars indicate the SEM.
DKO mice that died at P1 showed enlarged hearts with a rounded apex relative to wild-type littermates (Fig. 4B). The atria were also dilated, engorged with blood, and frequently contained thrombi, suggesting a possible defect in cardiac contractility (Fig. 4B). All dKO mice that died at P1 exhibited large VSDs near the apex (Fig. 4B), likely representing the cause of death. This type of VSD is similar to, although more severe than, the trabecular (muscular) VSD in humans. We also observed VSDs near the atrioventricular valve and hypocellularity of the wall of the ventricular septum near the apex of the heart in a subset of mutants at P1. At the apex, the ventricular septum was fenestrated with blood-filled channels, and the thickness of the remaining septum measured 25% less than wild type. Since VSDs were not seen in dKO mice that survived to adulthood, we assume all animals with these defects died soon after birth. There were no obvious abnormalities in skeletal muscle in dKO mice (data not shown), presumably reflecting redundant functions of miR-133b.
At 4 mo of age, dKO hearts showed extensive fibrosis, but no evidence of cardiomyocyte hypertrophy (Fig. 4C). Extensive fibrosis was also observed in a subset of dKO hearts at 2 mo of age (data not shown). There was no significant difference in heart weight/tibia length ratios or in cardiomyocyte sizes between dKO mice and their littermates (Fig. 4D; data not shown). We assessed cardiac function by echocardiography in dKO mice that survived to adulthood. At 2 and 4 mo of age, dKO mice showed a pronounced reduction in cardiac contractility at systole, as indicated by a dramatic increase in left ventricular internal diameter at systole (LVIDs) and impaired fractional shortening (FS) (Fig. 4D). There was no significant difference in heart rate or LV posterior wall thickness in diastole (LVPWd) between wild-type and dKO mice, confirming that the dKO hearts were not hypertrophic (Fig. 4D). All adult dKO mice displayed dilated cardiomyopathy by 5–6 mo of age with thinning of the ventricular walls, dilation of the ventricular chambers, atrial thrombi, and extensive cardiac fibrosis (Fig. 4E). At least half of these mice suffered sudden death.
Transmission electron microscopy of adult hearts from wild-type and miR-133a dKO mice showed pronounced sarcomere fragmentation and disorganization, as well as disrupted Z-discs in the mutant mice (Fig. 4F). Mitochondria were also randomly distributed, and vacuolization and loss of cristae in mitochondria were apparent throughout dKO hearts. Similar defects in sarcomeric organization and mitochondria were also observed in dKO hearts at P1 (data not shown).
Markers of cardiac stress, such as β-myosin heavy chain (Myh7), atrial natriuretic factor (ANF), and b-type natriuretic protein (BNP), were up-regulated in adult dKO hearts, consistent with the heart failure phenotype (Fig. 4G).
All the experiments presented here were performed with the mutant alleles containing the neomycin resistance cassette. However, we repeated these experiments with mutant dKO mice that had the neomycin resistance cassettes removed and obtained comparable results (data not shown).
Aberrant cardiomyocyte proliferation and apoptosis in miR-133a dKO hearts
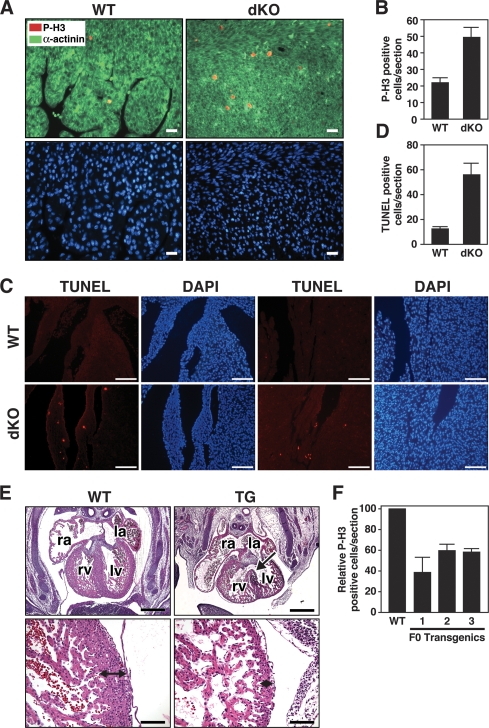
To further investigate the basis for cardiomyocyte hypoplasia in dKO mice, we analyzed cardiomyocyte proliferation by phospho-histone H3 staining in P1 animals. Cardiomyocytes from dKO mice showed a 2.5-fold increase in proliferation compared with wild type (Fig. 5A,B). Enhanced phospho-histone H3 staining was observed throughout the ventricles and the atria of the mutants. High-resolution microscopy confirmed that excessive proliferation in hearts of dKO mice was confined to cardiomyocytes, rather than cardiac fibroblasts (Supplemental Fig. 2), consistent with the myocyte-specific expression of miR-133a-1 and miR-133a-2 (Zhao et al. 2005; Liu et al. 2007).
Figure 5.
Abnormal cardiomyocyte proliferation and apoptosis in miR-133a dKO hearts. (A) Immunohistochemistry on heart sections of wild-type and dKO mutant mice at P1. Phospho-histone H3 (red), α-actinin (green), and Hoechst (blue) staining at 40× magnification show increased proliferation in the cardiomyocytes in dKO mutant mice. Bar, 20 μm. (B) Quantification of phospho-histone H3-positive cells was performed on three sections from each heart and averaged from six individual hearts. Error bars indicate the SEM. (C) TUNEL staining of wild-type and dKO hearts at P1 showed increased apoptosis in dKO mice. TUNEL-positive cells are located near the base (left panels) and apex (right panels) of the heart in dKO mice. DAPI staining indicates nuclei. Bar, 100 μm. (D) TUNEL-positive cells were quantified on multiple sections from each dKO heart. Error bars indicate the SEM. (E) Transgenic overexpression of miR-133a blocks proliferation of cardiomyocytes in vivo. Histological sections of wild-type and βMHC-miR-133a transgenic hearts at E13.5 are shown. The arrow in the top right panel points to a VSD. The bottom panels show higher magnifications of the left ventricular myocardium, which is about eight cells thick in wild type and only two cells thick in the transgenic. (Top panel) Bar, 500 μm. (Bottom panel) Bar, 100 μm. (F) Quantification of phospho-histone H3-positive cells in histological sections of hearts from wild-type and three independent F0 βMHC-miR-133a transgenic hearts at E13.5. Phospho-histone H3-positive cells were counted on three sections for each transgenic heart and normalized to the number of phospho-histone H3-positive cells of wild-type littermates. Error bars indicate the SEM.
An increase in apoptosis, detected by TUNEL staining, was also seen in dKO hearts at P1 (Fig. 5C,D). The increase in apoptosis was apparent at the apex and base of the interventricular septum and near the AV valves, likely accounting for the VSDs observed in dKO animals (Fig. 5D).
Transgenic overexpression of miR-133a-1 and miR-133a-2 in the developing heart
As an independent means of verifying the potential role of miR-133a in the control of cardiomyocyte proliferation in vivo, we overexpressed miR-133a in the developing hearts of transgenic mice under control of the β-myosin heavy chain (MHC) promoter, which directs high levels of cardiac expression by E9.0 (Ng et al. 1991). We obtained several F0 transgenic mice, but were only able to obtain one stable transgenic line, which expressed miR-133a in the heart at a level 15-fold higher than that of wild-type embryos at E12.5 (Supplemental Fig. 3). Transgenic offspring from this line all died by E15.5. Histological analyses of transgenic hearts at E13.5 showed cardiac abnormalities including VSDs, thin-walled ventricles, and enlarged atria, indicative of cardiac failure (Fig. 5E). The ventricular walls of transgenic embryos contained were only two to three cells thick, whereas in the nontransgenic littermates, the ventricular wall is composed of eight to nine cells in thickness at this stage (Fig. 5E). Because of the apparent embryonic lethality resulting from transgenic overexpression of miR-133a, which prevented us from obtaining stable transgenic lines, we generated multiple F0 transgenic embryos overexpressing miR-133a, all of which displayed the same cardiac defects at E13.5 (data not shown).
We observed no differences in TUNEL staining between wild-type and β-MHC-133a Tg hearts at E13.5 (data not shown). However, cardiomyocyte proliferation was diminished in transgenic hearts at E13.5 as assayed by phospho-histone H3 staining (Fig. 5F), suggesting that the reduction in the myocardial cell layer resulted from reduced myocyte proliferation rather than apoptosis. Thus, overexpression of miR-133a results in diminished proliferation of cardiomyocytes, a phenotype opposite that of dKO hearts and consistent with the conclusion that miR-133a suppresses cardiomyocyte proliferation.
Aberrant activation of a smooth muscle gene program in miR-133a dKO hearts
To begin to define the mechanistic basis of the neonatal lethality of miR-133a dKO mice, we compared the gene expression profiles of wild-type and dKO hearts at P1 by microarray analysis (Supplemental Table 2). Numerous genes were up- and down-regulated in dKO hearts. Because miRNAs typically diminish the expression of their mRNA targets (Baek et al. 2008; Selbach et al. 2008), we focused on those mRNAs that were up-regulated in dKO hearts, a subset of which would be expected to be direct targets of miR-133a. Indeed, numerous mRNAs that were up-regulated in dKO hearts are predicted targets for repression by miR-133a (Supplemental Table 2). Those up-regulated mRNAs that are not predicted targets of miR-133a likely respond through indirect mechanisms to the absence of miR-133a, possibly reflecting abnormalities in structure and function of dKO hearts. In contrast, relatively few of the mRNAs that were down-regulated in dKO hearts contained predicted target sequences for miR-133a.
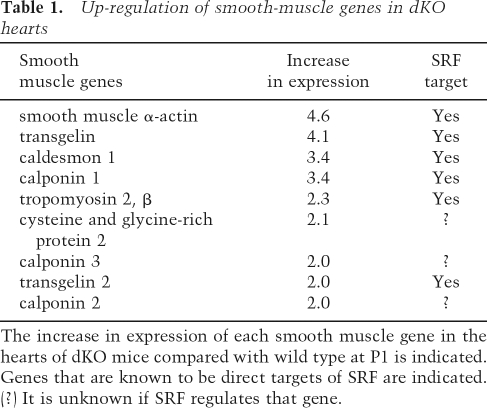
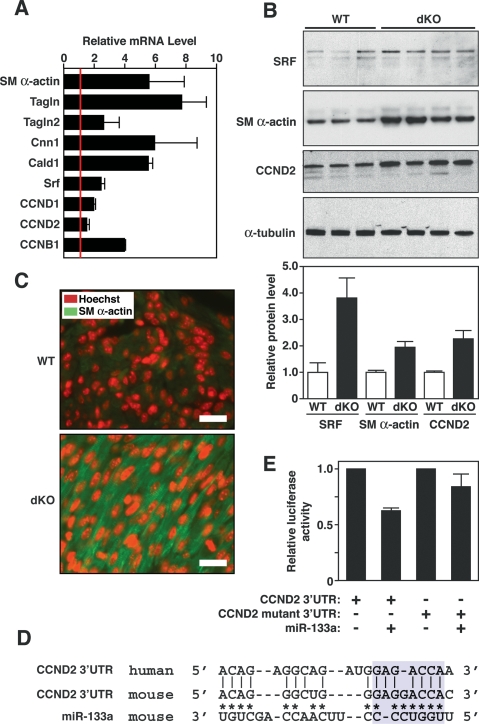
A disproportionate number of up-regulated genes in dKO hearts encoded smooth muscle-restricted proteins (Table 1). We confirmed the elevated cardiac expression of smooth muscle genes, including those encoding smooth muscle α-actin (SM α-actin), transgelins (TAGLN and TAGLN2, also known as SM22α and β), calponin I (CNN1), and caldesmon (CALD1) by real-time PCR (Fig. 6A). Western blot of cardiac extracts also showed elevated expression of SM α-actin in dKO hearts (Fig. 6B), and immunostaining of histological sections demonstrated SM α-actin to be expressed at a much higher level in cardiomyocytes of dKO hearts relative to wild-type hearts at P1 (Fig. 6C).
Table 1.
Up-regulation of smooth-muscle genes in dKO hearts
The increase in expression of each smooth muscle gene in the hearts of dKO mice compared with wild type at P1 is indicated. Genes that are known to be direct targets of SRF are indicated.
(?) It is unknown if SRF regulates that gene.
Figure 6.
Modulation of miR-133a targets in dKO hearts. (A) Expression of smooth muscle-specific genes and cyclin genes in hearts of wild-type and dKO mutant mice at P1 as detected by real-time PCR. Expression levels for each gene in dKO hearts were normalized to GAPDH and compared with wild-type hearts. Error bars indicate the SEM. (SM α-actin) Smooth muscle α-actin; (TAGLN) transgelin (also called SM22); (TAGLN2) transgelin 2 (also named SM22β); (CNN1) calponin I; (CALD1) caldesmon; (CCND1) cyclin D1; (CCND2) cyclin D2; (CCNB1) cyclin B1. (B) Expression of SRF, SM α-actin, and CCND2 in wild-type and dKO mutant hearts. Western blot analysis was performed on hearts from P1 wild-type (n = 3) and dKO mutant (n = 4) mice. α-Tubulin was detected as a loading control. Quantification of bands by densitometry showed a 3.5-fold and twofold increase in expression of SRF, SM α-actin, and CCN2 in dKO compared with wild-type hearts. (C) Increased SM α-actin expression in dKO hearts at P1. Histological sections of wild-type and dKO mutant hearts at P1 were stained for SM α-actin (green) and for nuclei with Hoechst (red). Pictures of wild-type and dKO hearts were taken under the same exposure parameters. Bar, 20 μm. Hoechst staining was reproducibly more intense in sections of dKO hearts compared with wild type, which may reflect greater DNA synthesis in the mutant. (D) Sequence alignment of the human and mouse cyclin D2 3′ UTR and miR-133a. Asterisks point to Watson-Crick base-pairing between mouse cyclin D2 3′ UTR and miR-133a. Base-pairing between miR-133a seed sequences with cyclin D2 3′ UTR is highlighted in blue. Mutations in cyclin D2 3′ UTR were introduced to disrupt base-pairing with the seed sequences. (E) Luciferase assay of cyclin D2 3′ UTR in Cos-1 cells. Wild-type and mutant cyclin D2 3′ UTR sequences were cloned into luciferase-reporter constructs and were cotransfected with a plasmid expressing miR-133a into Cos-1 cells. Forty-eight hours post-transfection, luciferase activity was measured and normalized to β-galactosidase activity. Error bars represent the SEM.
Many of the smooth muscle genes that are up-regulated in dKO hearts are controlled by SRF (Owens et al. 2004), a predicted target for translational repression by miR-133a (Chen et al. 2006). Accordingly, SRF mRNA and protein expression were increased in dKO hearts compared with wild-type hearts (Fig. 6A,B). SM α-actin is known to be up-regulated in response to cardiac stress (Black et al. 1991; van Bilsen and Chien 1993; Schaub et al. 1997). However, other smooth muscle genes are not typically up-regulated in the stressed myocardium, as we observed in miR-133a dKO mice. Thus, the activation of such a large collection of smooth muscle genes in dKO hearts cannot be attributed simply to a generalized cardiac stress response and, instead, points to a more specific role for miR-133a in the programming of cardiac versus smooth muscle gene expression. The smooth muscle-specific mRNAs encoding transgelin2 and calponin are also predicted targets of miR-133a, which would be expected to further augment the expression of smooth muscle proteins in dKO hearts.
Dysregulation of cell cycle-associated genes in miR-133a dKO hearts
In addition to the up-regulation of smooth muscle structural genes in dKO hearts, we noted the enhanced expression of numerous mRNAs involved in cell cycle control, including cyclin D1 (CCND1), cyclin D2 (CCND2), and cyclin B1 (CCNB1) (Fig. 6A). We confirmed the up-regulation of CCND2 protein in dKO hearts by Western blot (Fig. 6B).
Sequence alignment of the 3′ untranslated regions (UTRs) of human and mouse cyclin D2 mRNAs revealed “seed” sequences and flanking nucleotides that form Watson-Crick base-pairing with miR-133a (Fig. 6D). The predicted energy of the miR-133a/cyclin D2 mRNA interaction is −19.7 kcal/mol. miR-133a repressed a luciferase reporter gene linked to the 3′ UTR of cyclin D2 mRNA, whereas a mutation in the predicted miR-133a-binding site in the 3′ UTR prevented repression, confirming this mRNA as a target for miR-133a (Fig. 6E).
In light of the increased proliferation in dKO hearts, we examined expression of other potential targets of miR-133a that are known to play a role in cell cycle regulation. Poly-pyrimidine tract-binding protein 2 (PTBP2) and cdc42 mRNAs, both of which are predicted targets of miR-133a (Boutz et al. 2007a; Care et al. 2007), were up-regulated in dKO hearts (Supplemental Fig. 4A). However, other proposed targets such as RhoA, whsc2, caspase-9, and HERG (Care et al. 2007; J. Xiao et al. 2007; Xu et al. 2007), were unchanged in dKO mice (Supplemental Fig. 4A), suggesting that these targets might be regulated at post-transcriptional levels. We further analyzed protein expression of RhoA and caspase-9 by Western blot and observed no significant change in protein levels of these genes between wild-type and dKO hearts at P1 (Supplemental Fig. 4B).
Discussion
The results of this study reveal essential roles of miR-133a-1 and miR-133a-2 in the control of cardiac gene expression and function, such that the combined absence of these miRNAs results in lethal VSDs in a subset of mutant mice and heart failure in those that survive to adulthood. Many of the phenotypic abnormalities of miR-133a-1/133a-2-null mice can be ascribed to the up-regulation of two specific mRNA targets encoding SRF and cyclin D2, which contribute to ectopic expression of smooth muscle genes in the heart, aberrant cardiomyocyte proliferation and apoptosis, and consequent cardiac dysfunction.
VSDs, the most common form of congenital heart disease, arise from abnormalities in cardiomyocyte proliferation and survival (Hoffman and Kaplan 2002). Accordingly, we observed excessive cardiomyocyte proliferation and apoptosis in regions of miR-133a dKO hearts where VSDs were observed. The presence of lethal VSDs in only a subset of mutant mice suggests that miR-133a plays a modulatory role during a “window” of cardiac development, perhaps by titrating the expression of mRNAs involved in cardiac morphogenesis and growth. If embryos are able to pass this critical stage, possibly as a result of stochastic variations in expression of critical miR-133 targets, the functions of miR-133 may become less critical until later stages of postnatal development when sustained and efficient hemodynamic output is essential for life.
miR-133a-1 and miR-133a-2 are identical in sequence and coexpressed in cardiac and skeletal muscle. Because mice with only a single wild-type miR-133a allele appear normal, we conclude that development of cardiac and skeletal muscle, as well as homeostatic functions of these adult tissues, require no more than 25% of the amount of miR-133a normally expressed in these muscles. Whether conditions of stress might heighten the requirement for higher miR-133a levels, as has been observed for other miRNAs (Leung and Sharp 2007), is an interesting question for the future. The fact we did not observe skeletal muscle abnormalities in miR-133a dKO animals may reflect the expression of miR-133b in skeletal muscle, whereas this miRNA is not expressed in the heart.
Repression of SRF and smooth muscle gene expression by miR-133a
SRF, a direct target for regulation by miR-133 (Chen et al. 2006), was up-regulated in dKO hearts, as were numerous smooth muscle structural genes that are regulated by SRF. Consistent with the conclusion that dysregulation of SRF expression contributes to the heart failure phenotype of adult miR-133a dKO mice, SRF functions as a key transcriptional regulator of pathological cardiac remodeling leading to heart failure (Sprenkle et al. 1995; Nelson et al. 2005), and elevated expression of SRF in the heart has been shown to be sufficient to cause heart failure (Zhang et al. 2001).
SRF regulates numerous genes involved in myogenesis and cell growth, which raises the interesting question as to why smooth muscle-restricted SRF target genes are selectively activated in the hearts of miR-133a dKO mice. Smooth muscle genes are transiently expressed in the heart during embryogenesis (Ruzicka and Schwartz 1988; McHugh 1995; Li et al. 1996). Thus, the absence of miR-133a may arrest the heart in a primitive stage of development. Alternatively, given the dependence of SRF activity on cofactor availability (Wang et al. 2004; Pipes et al. 2006), it is possible the absence of miR-133a also results in up-regulation of additional cofactors or signaling pathways that selectively modulate the expression of smooth muscle genes.
We speculate that diminished cardiac function in dKO mice that survive to adulthood results, at least in part, from the inappropriate expression of SRF-dependent smooth muscle genes, such as SM α-actin. In this regard, cardiac function is highly sensitive to the ratios of different actin isoforms. SM α-actin possesses a reduced ability to activate myosin ATPase compared with cardiac α-actin (Strzelecka-Golaszewska and Sobieszek 1981), and elevated expression of SM α-actin in the heart has been shown to result in a hypocontractile phenotype (Kumar et al. 1997), as observed in miR-133a dKO mice. Aberrant expression of muscle contractile proteins is also likely to contribute to the sarcomere disarray seen in hearts of adult dKO mice.
Inhibition of miR-133 expression in vivo by an antagomir causes cardiac hypertrophy (Care et al. 2007). These findings have led to the notion that miR-133 acts as an inhibitor of cardiac growth (Care et al. 2007). On the contrary, adult dKO mice showed no evidence of cardiac hypertrophy, but instead, displayed dilated cardiomyopathy with ventricular wall thinning and severe fibrosis. Perhaps the transient knockdown of miR-133a with an antagomir has different consequences from genetic deletion in which the gene is eliminated throughout the life of the organism. Residual expression of miR-133a in antagomir-treated animals might also account for the different findings of these studies.
Control of cardiomyocyte proliferation by miR-133a
Based on overexpression studies in cultured skeletal myoblasts, miR-133 has been proposed to promote cell proliferation by repressing SRF, a positive regulator of myocyte differentiation (Niu et al. 2007). In contrast, we found excessive cardiomyocyte proliferation in miR-133a dKO mice, and diminished proliferation in response to transgenic overexpression of miR-133a, indicating that miR-133 functions as an inhibitor of cardiomyocyte proliferation in vivo. Given that SRF can function as either a positive or negative regulator of cardiomyocyte proliferation and differentiation, depending on cofactor availability and extracellular signaling (Shin et al. 2002; Pipes et al. 2006), the influence of miR-133a on downstream functions of SRF may vary depending on these and other variables.
In addition to repressing expression of SRF, miR-133a represses cyclin D2 expression. Thus, in the absence of miR-133a, elevated expression of cyclin D2 would be predicted to promote cell proliferation, possibly synergistically with SRF. Cyclin D2 has been shown to promote cardiomyocyte cell cycle progression (Pasumarthi et al. 2005). Since miR-133a does not appear to repress the expression of proapoptotic genes, enhanced apoptosis in dKO hearts may be a consequence of inappropriate cell cycle progression of cells that would otherwise be post-mitotic.
miR-133: an integral component of the transcriptional circuitry of the heart
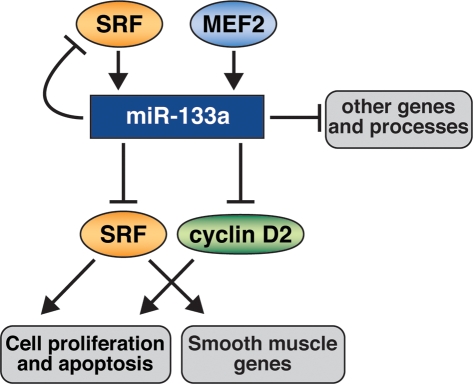
miRNAs are thought to confer precision and robustness to physiological and pathological processes and to commonly function as components of cellular networks by buffering extremes in gene expression. Our results reveal an integral role for miR-133 in the transcriptional circuits controlled by SRF in the heart (Fig. 7). The regulation of miR-133 by SRF and the targeting of SRF by miR-133 provide a negative feedback loop to precisely titrate the actions of SRF such that elevations of SRF activity enhance the expression of miR-133a with consequent dampening of SRF expression. Conversely, diminished activity of SRF would result in reduced expression of miR-133a, thereby elevating the expression of SRF. Thus, the opposing actions of SRF and miR-133 establish a finely tuned balance to control cardiac growth and differentiation.
Figure 7.
A model for the functions of miR-133a in the heart. SRF and MEF2 activate miR-133a expression, which directly or indirectly represses genes involved in many aspects of heart development and function, including sarcomeric structures, cell proliferation, apoptosis, and the smooth muscle gene program. miR-133a directly targets SRF, which provides a negative feedback loop to precisely modulate SRF activity.
miRNAs commonly have numerous mRNA targets of varying affinities. Nevertheless, there are an increasing number of examples in which the functions of a miRNA can be ascribed to one or a few targets. While our findings support the conclusion that SRF and cyclin D2 are critical downstream mediators of the actions of miR-133a in the heart, this miRNA has numerous predicted targets. The abnormalities arising from the absence of miR-133a-1 and 133a-2 are therefore likely to be attributable to the summation of changes in expression of numerous target mRNAs. Given the involvement of miRNAs in a variety of disease states and especially in cardiovascular disorders (van Rooij and Olson 2007), it will be especially interesting to investigate the potential role of miR-133a in adult heart disease.
Materials and methods
Gene targeting and mouse breeding
Both miR-133a-1 and miR-133a-2 targeting vectors were constructed using the pGKNEO-F2DTA vector, which contains a neomycin resistance gene driven by pGK promoter, flanked by FRT sites, and a diphtheria toxin gene cassette. Both targeting strategies were designed to replace the pre-miR sequences with the neomycin resistance cassette flanked by FRT sites. For the targeting vector of miR-133a-1, a 2-kb fragment upstream of pre-miR-133a-1 and a 4.4-kb fragment immediately downstream from the pre-miR-133a-1 were generated as the 5′ arm and the 3′ arm by TAKARA Taq LA PCR amplification (TAKARA) of 129SvEv genomic DNA. The pre-miR-1-2 sequence was not included in the 5′ arm. For the targeting vector of miR-133a-2, a 3.7-kb fragment upstream of the pre-miR-133a-2 and a 2.3-kb fragment downstream were generated as the 5′-arm and the 3′-arm. Both targeting vectors were linearized and electroporated into 129SvEv-derived ES cells. Four-hundred-eighty (for miR-133a-1) and 380 (for miR-133a-2) ES cell clones were isolated and analyzed for homologous recombination by Southern blotting. Three clones with a properly targeted miR-133a-1 or miR-133a-2 allele were injected into 3.5-d C57BL/6 blastocysts, and high-percentage chimeric male mice were crossed to C57BL/6 females to achieve germline transmission of the targeted alleles. Heterozygous miR-133a-1neo/+ or miR-133a-2neo/+ mice were intercrossed with hACTB:FLPe transgenic mice (Rodriguez et al. 2000) to remove the neomycin resistance cassette. Southern blotting was performed to confirm the removal of the neomycin resistance cassette.
For the generation of the double-null mice, miR-133a-1neo/+ and miR-133a-2neo/+ mice were first intercrossed to generate double-heterozygous mice. Double-heterozygous mice were then intercrossed to generate dKO, miR-133a-1neo/neo; miR-133a-2neo/+, and miR-133a-1neo/+; miR-133a-2neo/neo mice. dKO mice were also generated from intercrosses from miR-133a-1neo/neo; miR-133a-1neo/+ mice or from miR-133a-1neo/+; miR-133a-2neo/neo mice. Similar breeding strategies were used to generate dKO mice without the neomycin resistance cassettes. Mouse genotypes were determined by PCR on tail DNA using primers specific to the miR-133a-1 and/or miR-133a-2 locus. Primer sequences for PCR genotyping are available upon request. All experimental procedures involving animals in this study were reviewed and approved by the Institutional Animal Care and Research Advisory Committee at the University of Texas Southwestern Medical Center.
RNA analyses
Total RNA was purified from tissues using TRIzol reagent according to the manufacturer’s instructions. RNA was treated with Turbo RNase-free DNase (Ambion, Inc.) prior to the reverse transcription step. RT–PCR was performed using random hexamer primers. Quantitative real-time PCR was performed using TaqMan probes (ABI) or SybrGreen probes. Primers for SybrGreen probes were described in Liu et al. (2007). Quantitative real-time PCR on miRNA was performed using the TaqMan miRNA assay kits (ABI) according to the manufacturer’s protocol.
For microarray, P1 hearts were pooled from five wild-type and five dKO animals prior to RNA isolation. Microarray analysis was performed by the University of Texas Southwestern Microarray Core Facility using the Mouse Genome 430 2.0 Array (Affimetrix) as described (Montgomery et al. 2007). Gene Ontology analysis was performed as described previously (Montgomery et al. 2007).
Northern blot
RNA from heart or skeletal muscle tissues was electrophoresed on a 20% polyacrylamide (7.6 M urea) gel in 1× TBE. Ten micrograms of RNA was denatured for 5 min at 70°C in a buffer containing 50% formamide and 10 mM EDTA (pH 8.0) before loading. After electrophoresis, RNA was then transferred onto a Hybond N membrane (Amersham) in 0.5× TBE buffer at 80 V for 1 h. Hybridization was performed at 39°C according to a standard protocol. 32P-labeled Star-Fire oligonucleotide probes (IDT) against mature miR-133a, miR-1, and U6 were used in the hybridization.
Western blot
Fifty micrograms of total protein extracts from P1 hearts was loaded on SDS-PAGE gels for Western blotting. Western blotting was performed by a standard protocol. Antibodies against SRF (Santa Cruz Biotechnologies; sc335, 1:1,000), smooth muscle α-actin (Sigma; A5228 1:400), cyclin D2 (Abcam; ab3085, 1:100), and α-tubulin (Sigma; T9026, 1:1,000) were used as described previously. Antibodies against caspase-9 and RhoA were described previously (Care et al. 2007; Xu et al. 2007). Antibody against Mib1 is a kind gift from Dr. P.J. Gallagher (Jin et al. 2002). Quantification of Western blot was performed by densitometry using the Storm 820 PhosphorImager.
Plasmids, cell culture, and luciferase assays
A 1-kb fragment of the cyclin D2 3′ UTR containing the miR-133a-binding site was cloned into pMIR-REPORT vector (Ambion). Mutagenesis of the miR-133a-binding site, cell culture, and luciferase assay were performed as described previously (van Rooij et al. 2007).
Histology
Tissues were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned at 5-μm intervals. Hematoxylin and eosin and Masson’s trichrome stains were performed using standard procedures.
Electron microscopy
Four-month-old miR-133a dKO and control littermate mice were anesthetized and transcardially perfused with 0.1 M phosphate buffer (pH 7.3), followed by 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cacodylate buffer. Hearts were removed, and left ventricles were sliced into 1-mm3 cubes and fixed overnight at 4°C. Samples were processed by the University of Texas Southwestern Medical Center Electron Microscopy Core facility. Sections were imaged on Jeol 1200 EX TEM at the indicated magnification.
Immunohistochemistry
Phospho-histone H3 staining was performed on paraffin-embedded sections as described previously (Xin et al. 2006). Rabbit anti-phosphohistone H3 (Upstate Cell Signaling Solutions) was used at a 1:200 dilution. Alexa Fluor 555-conjugated secondary goat anti-rabbit antibody (Invitrogen) was applied at a 1:500 dilution. Sections were costained with an antibody against mouse sarcomeric α-actinin (Sigma; clone EA-53, 1:100 dilution) and Alexa Fluor 488-conjugated goat anti-mouse antibody (Invitrogen). A TUNEL assay was performed according to a standard protocol (Roche). Phospho-histone H3-positive or TUNEL-positive cells were counted on at least three levels per heart and averaged among at least six animals for each genotype group. Staining of SM α-actin was performed on paraffin-embedded sections using a Cy3-conjugated anti-α-smooth muscle actin antibody (Sigma; Clone 1A4, 1:100). Hoechst staining was performed to visualize nuclei. Slides were mounted with Vectashield mounting medium with or without DAPI.
Transthoracic echocardiography
Cardiac function and heart dimensions were evaluated by two-dimensional echocardiography on conscious mice as described previously (Kim et al. 2008). M-mode tracings were used to measure anterior and posterior wall thicknesses at end diastole and end systole. LVID was measured as the largest anteroposterior diameter in either diastole (LVIDd) or systole (LVIDs). Left ventricular posterior wall dimensions at end diastole (LVPWd) and heart rate were also measured. Echocardiography and data analysis were performed by a single observer blinded to mouse geneotypes. FS was calculated according to the following formula: FS(%) = [(LVIDd − LVIDs)/LVIDd] × 100.
Statistical analysis
Gene expression measured by real-time PCR was normalized to GAPDH expression level and calculated as relative change to wild-type samples. Differences in morphological, physiological, and biochemical parameters between wild-type and mutant animals (or groups) were analyzed by two-sided Student’s t-test using MATLAB R12. The significance level α = 0.05.
Bioinformatics and miRNA target analysis
Analysis of predicted targets of miR-133a was based on the TargetScan prediction program (http://targetscan.org) and the Human miRNA Targets prediction program (http://cbio.mskcc. org/cgi-bin/mirnaviewer/mirnaviewer.pl). Candidate target genes were analyzed by three independent methods: luciferase reporter assay, real-time PCR, and Western blot analysis.
Acknowledgments
We thank John McAnally for generation of transgenic mice. We are grateful to John Shelton and Cheryl Nolen for technical help. We thank Kunhua Song, Mei Xin, Nik Munshi, and Rusty Montgomery for helpful discussions. We thank Jose Cabrera for graphics and Jennifer Brown for editorial assistance. We are grateful to Dr. P.J. Gallagher (Indiana University) for the Mib1 antibody. This work was supported by grants from the NIH, the Donald W. Reynolds Clinical Cardiovascular Research Center, the Robert A. Welch Foundation, and the Pogue Family Fund to E.N.O. N.L. was supported by a grant from the American Heart Association.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1738708.
References
- Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black F.M., Packer S.E., Parker T.G., Michael L.H., Roberts R., Schwartz R.J., Schneider M.D. The vascular smooth muscle α-actin gene is reactivated during cardiac hypertrophy provoked by load. J. Clin. Invest. 1991;88:1581–1588. doi: 10.1172/JCI115470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz P.L., Chawla G., Stoilov P., Black D.L. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes & Dev. 2007a;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz P.L., Stoilov P., Li Q., Lin C.H., Chawla G., Ostrow K., Shiue L., Ares M., Black D.L. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes & Dev. 2007b;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Care A., Catalucci D., Felicetti F., Bonci D., Addario A., Gallo P., Bang M.L., Segnalini P., Gu Y., Dalton N.D., et al. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- Chen J.F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M., Conlon F.L., Wang D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman J.I., Kaplan S. The incidence of congenital heart disease. J. Am. Coll. Cardiol. 2002;39:1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- Jin Y., Blue E.K., Dixon S., Shao Z., Gallagher P.J. A death-associated protein kinase (DAPK)-interacting protein, DIP-1, is an E3 ubiquitin ligase that promotes tumor necrosis factor-induced apoptosis and regulates the cellular levels of DAPK. J. Biol. Chem. 2002;277:46980–46986. doi: 10.1074/jbc.M208585200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Phan D., van Rooij E., Wang D.Z., McAnally J., Qi X., Richardson J.A., Hill J.A., Bassel-Duby R., Olson E.N. The MEF2D transcription factor mediates stress-dependent cardiac remodeling in mice. J. Clin. Invest. 2008;118:124–132. doi: 10.1172/JCI33255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Crawford K., Close L., Madison M., Lorenz J., Doetschman T., Pawlowski S., Duffy J., Neumann J., Robbins J., et al. Rescue of cardiac α-actin-deficient mice by enteric smooth muscle γ-actin. Proc. Natl. Acad. Sci. 1997;94:4406–4411. doi: 10.1073/pnas.94.9.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.K., Sharp P.A. microRNAs: A safeguard against turmoil? Cell. 2007;130:581–585. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Li L., Miano J.M., Cserjesi P., Olson E.N. SM22 α, a marker of adult smooth muscle, is expressed in multiple myogenic lineages during embryogenesis. Circ. Res. 1996;78:188–195. doi: 10.1161/01.res.78.2.188. [DOI] [PubMed] [Google Scholar]
- Liu N., Williams A.H., Kim Y., McAnally J., Bezprozvannaya S., Sutherland L.B., Richardson J.A., Bassel-Duby R., Olson E.N. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc. Natl. Acad. Sci. 2007;104:20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Lin H., Pan Z., Xiao J., Zhang Y., Lu Y., Yang B., Wang Z. Downregulation of MIRNA-1/MIRNA-133 contributes to re-expression of pacemaker channel genes HCN2 and HCN4 in hypertrophic heart. J. Biol. Chem. 2008;283:20045–20052. doi: 10.1074/jbc.M801035200. [DOI] [PubMed] [Google Scholar]
- McCarthy J.J. MicroRNA-206: The skeletal muscle-specific myomiR. Biochim. Biophys. Acta. 2008;1779:682–691. doi: 10.1016/j.bbagrm.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh K.M. Molecular analysis of smooth muscle development in the mouse. Dev. Dyn. 1995;204:278–290. doi: 10.1002/aja.1002040306. [DOI] [PubMed] [Google Scholar]
- Miano J.M., Long X., Fujiwara K. Serum response factor: Master regulator of the actin cytoskeleton and contractile apparatus. Am. J. Physiol. Cell Physiol. 2007;292:C70–C81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- Montgomery R.L., Davis C.A., Potthoff M.J., Haberland M., Fielitz J., Qi X., Hill J.A., Richardson J.A., Olson E.N. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes & Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T.J., Balza R., Xiao Q., Misra R.P. SRF-dependent gene expression in isolated cardiomyocytes: Regulation of genes involved in cardiac hypertrophy. J. Mol. Cell. Cardiol. 2005;39:479–489. doi: 10.1016/j.yjmcc.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Ng W.A., Grupp I.L., Subramaniam A., Robbins J. Cardiac myosin heavy chain mRNA expression and myocardial function in the mouse heart. Circ. Res. 1991;68:1742–1750. doi: 10.1161/01.res.68.6.1742. [DOI] [PubMed] [Google Scholar]
- Niu Z., Li A., Zhang S.X., Schwartz R.J. Serum response factor micromanaging cardiogenesis. Curr. Opin. Cell Biol. 2007;19:618–627. doi: 10.1016/j.ceb.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E.N. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens G.K., Kumar M.S., Wamhoff B.R. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- Pasumarthi K.B., Nakajima H., Nakajima H.O., Soonpaa M.H., Field L.J. Targeted expression of cyclin D2 results in cardiomyocyte DNA synthesis and infarct regression in transgenic mice. Circ. Res. 2005;96:110–118. doi: 10.1161/01.RES.0000152326.91223.4F. [DOI] [PubMed] [Google Scholar]
- Pipes G.C., Creemers E.E., Olson E.N. The myocardin family of transcriptional coactivators: Versatile regulators of cell growth, migration, and myogenesis. Genes & Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- Potthoff M.J., Olson E.N. MEF2: A central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- Rodriguez C.I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., Dymecki S.M. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Ruzicka D.L., Schwartz R.J. Sequential activation of α-actin genes during avian cardiogenesis: Vascular smooth muscle α-actin gene transcripts mark the onset of cardiomyocyte differentiation. J. Cell Biol. 1988;107:2575–2586. doi: 10.1083/jcb.107.6.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub M.C., Hefti M.A., Harder B.A., Eppenberger H.M. Various hypertrophic stimuli induce distinct phenotypes in cardiomyocytes. J. Mol. Med. 1997;75:901–920. doi: 10.1007/s001090050182. [DOI] [PubMed] [Google Scholar]
- Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Shin C.H., Liu Z.P., Passier R., Zhang C.L., Wang D.Z., Harris T.M., Yamagishi H., Richardson J.A., Childs G., Olson E.N. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell. 2002;110:725–735. doi: 10.1016/s0092-8674(02)00933-9. [DOI] [PubMed] [Google Scholar]
- Sprenkle A.B., Murray S.F., Glembotski C.C. Involvement of multiple cis elements in basal- and α-adrenergic agonist-inducible atrial natriuretic factor transcription. Roles for serum response elements and an SP-1-like element. Circ. Res. 1995;77:1060–1069. doi: 10.1161/01.res.77.6.1060. [DOI] [PubMed] [Google Scholar]
- Strzelecka-Golaszewska H., Sobieszek A. Activation of smooth muscle myosin by smooth and skeletal muscle actins. FEBS Lett. 1981;134:197–202. doi: 10.1016/0014-5793(81)80601-1. [DOI] [PubMed] [Google Scholar]
- Thai T.H., Calado D.P., Casola S., Ansel K.M., Xiao C., Xue Y., Murphy A., Frendewey D., Valenzuela D., Kutok J.L., et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez M.A., Liu J., Hannon G.J., Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes & Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- van Bilsen M., Chien K.R. Growth and hypertrophy of the heart: Towards an understanding of cardiac specific and inducible gene expression. Cardiovasc. Res. 1993;27:1140–1149. doi: 10.1093/cvr/27.7.1140. [DOI] [PubMed] [Google Scholar]
- van Rooij E., Olson E.N. MicroRNAs: Powerful new regulators of heart disease and provocative therapeutic targets. J. Clin. Invest. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E., Sutherland L.B., Qi X., Richardson J.A., Hill J., Olson E.N. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- van Rooij E., Liu N., Olson E.N. MicroRNAs flex their muscles. Trends Genet. 2008;24:159–166. doi: 10.1016/j.tig.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Ventura A., Young A.G., Winslow M.M., Lintault L., Meissner A., Erkeland S.J., Newman J., Bronson R.T., Crowley D., Stone J.R., et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wang D.Z., Hockemeyer D., McAnally J., Nordheim A., Olson E.N. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- Wang S., Aurora A.B., Johnson B.A., Qi X., McAnally J., Hill J.A., Richardson J.A., Bassel-Duby R., Olson E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Calado D.P., Galler G., Thai T.H., Patterson H.C., Wang J., Rajewsky N., Bender T.P., Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- Xiao J., Luo X., Lin H., Zhang Y., Lu Y., Wang N., Zhang Y., Yang B., Wang Z. MicroRNA miR-133 represses HERG K+ channel expression contributing to QT prolongation in diabetic hearts. J. Biol. Chem. 2007;282:12363–12367. doi: 10.1074/jbc.C700015200. [DOI] [PubMed] [Google Scholar]
- Xin M., Davis C.A., Molkentin J.D., Lien C.L., Duncan S.A., Richardson J.A., Olson E.N. A threshold of GATA4 and GATA6 expression is required for cardiovascular development. Proc. Natl. Acad. Sci. 2006;103:11189–11194. doi: 10.1073/pnas.0604604103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Lu Y., Pan Z., Chu W., Luo X., Lin H., Xiao J., Shan H., Wang Z., Yang B. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J. Cell Sci. 2007;120:3045–3052. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- Zhang X., Azhar G., Chai J., Sheridan P., Nagano K., Brown T., Yang J., Khrapko K., Borras A.M., Lawitts J., et al. Cardiomyopathy in transgenic mice with cardiac-specific overexpression of serum response factor. Am. J. Physiol. Heart Circ. Physiol. 2001;280:H1782–H1792. doi: 10.1152/ajpheart.2001.280.4.H1782. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Samal E., Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Ransom J.F., Li A., Vedantham V., von Drehle M., Muth A.N., Tsuchihashi T., McManus M.T., Schwartz R.J., Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]