Abstract
The superoxide radical (O2−) has long been considered a major cause of aging. O2− in cytosolic, extracellular, and mitochondrial pools is detoxified by dedicated superoxide dismutase (SOD) isoforms. We tested the impact of each SOD isoform in Caenorhabditis elegans by manipulating its five sod genes and saw no major effects on life span. sod genes are not required for daf-2 insulin/IGF-1 receptor mutant longevity. However, loss of the extracellular Cu/ZnSOD sod-4 enhances daf-2 longevity and constitutive diapause, suggesting a signaling role for sod-4. Overall, these findings imply that O2− is not a major determinant of aging in C. elegans.
Keywords: Aging, Caenorhabditis elegans, free radical, superoxide dismutase, insulin/IGF-1 signaling, genetics
Many forms of pathology lead to elevated levels of damage to biological macromolecules (Halliwell and Gutteridge 2007). This is also true of aging, the poorly understood biological process that leads to progressive deterioration and death. One strategy to discover the underlying mechanisms of aging has been to seek the causes of its associated molecular damage. An important early theory, proposed by Harman (1956), postulates that the cause might be oxygen free radicals. Harman later developed the theory, proposing a central role for the superoxide (O2−) radical, issuing from the mitochondrial electron transport chain (Harman 1972). During the last few decades, much effort has been invested in tests of this nexus of theories (for review, see Muller et al. 2007). Despite this, the importance of O2− as a cause of aging remains uncertain.
In this study, we take a genetic approach to critically test the role of O2− in aging in a short-lived animal, the nematode Caenorhabditis elegans, by manipulating expression of genes encoding the antioxidant enzyme superoxide dismutase (SOD). This enzyme catalyzes the dismutation reaction
H2O2 (hydrogen peroxide) may then be broken down into H2O and O2 by catalase or glutathione peroxidase. O2− does not readily cross cellular membranes and, consequently, there are distinct extracellular, cytosolic, and mitochondrial O2− pools (Missirlis et al. 2003; Muller et al. 2004). In principle, one or more of these three O2− pools may play a role in aging.
Instrumental in lowering levels of O2− in each pool is a dedicated, compartment-specific SOD isoform. Cytosolic and extracellular O2− is consumed by distinct Cu/ZnSOD isoforms, while O2− in the mitochondrial matrix is consumed by MnSOD (Weisiger and Fridovich 1973). Most eukaryotes have a single SOD isoform for each compartment, but C. elegans, unusually, possesses two isoforms for each compartment. The two cytosolic Cu/ZnSOD isoforms are encoded by sod-1 and sod-5 (Larsen 1993; Giglio et al. 1994; Jensen and Culotta 2005). The sod-4 gene encodes two predicted extracellular Cu/ ZnSOD isoforms, products of alternative splicing of mRNA (Fujii et al. 1998). Two mitochondrial MnSOD isoforms are encoded by sod-2 and sod-3 (Giglio et al. 1994; Suzuki et al. 1996; Hunter et al. 1997).
This superabundance of SOD isoforms has been a technical hurdle to investigations of the role of SOD and O2− in aging in C. elegans, and some of the sod genes have been barely studied. In situ gel SOD activity assays of a sod-1 deletion mutant imply that this gene encodes the major cytosolic Cu/ZnSOD (Jensen and Culotta 2005), leaving the function of sod-5 unclear. sod-3 mRNA levels are elevated in the dauer larva (Honda and Honda 1999), suggesting that this gene may play a special role in antioxidant defense in this long-lived, stress-resistant diapausal stage, but the role of sod-2 has remained obscure. In this study, we describe in detail the function of each of the five sod genes, characterizing their expression, and the phenotypic effects of manipulating their expression. This has allowed us to assess the effect on life history, especially aging, of each of the three major O2− pools, thereby critically testing the role of SOD and, by inference, O2−, in longevity assurance and aging.
O2− can affect living organisms in a variety of ways. It can cause molecular damage that might contribute to aging; thus, one expectation of our study was that lowering SOD activity and increasing O2− levels might accelerate aging, and vice versa. H2O2 derived from O2− can also act a secondary messenger—for example, in receptor tyrosine kinase signaling pathways (Finkel 1998)—and as an activator of heat-shock factor. O2− can also be deployed as a chemical weapon in immune defense against bacterial pathogens in higher animals, and probably in C. elegans as well (Chavez et al. 2007). There is even evidence that in C. elegans O2− can increase life span, perhaps by activating stress defense processes (Cypser and Johnson 2002; Schulz et al. 2007).
A powerful approach to investigate mechanisms of aging is the mutational analysis of genes with effects on life span. In C. elegans, mutations affecting the insulin/IGF-1 signaling (IIS) pathway can strikingly increase adult life span. For example, mutation of daf-2, which encodes an insulin/IGF-1 receptor, can increase adult life span by more than twofold (Kenyon 2005). Severe daf-2 loss of function can also cause constitutive formation of dauer larvae, which are developmentally arrested, long-lived, diapausal third-stage larvae (Riddle and Albert 1997).
One possibility is that increased SOD levels and reduced damage from O2− contribute to the longevity of IIS mutants. daf-2 mutants do show increased SOD and catalase activity levels, and resistance to oxidative stress (Vanfleteren 1993; Honda and Honda 1999). sod-3 mRNA and protein levels are elevated in daf-2 mutants (Honda and Honda 1999; Yanase et al. 2002; Dong et al. 2007), suggesting a possible role for mitochondrial MnSOD in longevity assurance. However, a more critical test is to examine the effects of alteration of SOD activity on life span.
In this study, we first examine the biology of the five C. elegans sod genes and show that sod-1 and sod-2 encode the major Cu/ZnSOD and MnSOD isoforms in reproductive development, while sod-5 and sod-3 encode minor, auxiliary isoforms mainly expressed in dauer larvae. We then critically test the importance of SOD and, by extension, O2− in C. elegans aging and in the daf-2 Age phenotype by means of sod gene deletion and overexpression.
Results and Discussion
To understand the respective roles of the five sod genes, we characterized their expression using several techniques, including RT-PCR, Western blot analysis, SOD activity assays, and analysis of expression of sod∷gfp transgenes. Expression was studied mainly in wild-type third-stage (L3) larvae and dauer larvae, daf-2(m577) mutants at the L3 stage, and mutants with deletions in each of the five sod genes (Supplemental Fig. S1A; for a detailed account of sod gene expression, see the Supplemental Material).
We report that sod-1 and sod-2 encode the major cytosolic Cu/ZnSOD and MnSOD isoforms, respectively. sod-1 contributes ∼80% of total SOD activity and is ubiquitously expressed (Supplemental Fig. S4C), and SOD-1 protein is localized to the cytosol and mitochondrial intermembrane space (Fig. 3A, below). By contrast, sod-5 and sod-3 are, respectively, minor cytosolic Cu/ZnSOD and MnSOD isoforms whose expression is up-regulated in dauer larvae (Supplemental Figs. S1B–E, S4D,F). In wild-type L3s, sod-5∷gfp expression is largely restricted to the ASI, ASK, and ASG amphid neurons (Supplemental Fig. S3B,C), which influence longevity and dauer larva formation (Bargmann and Horvitz 1991; Alcedo and Kenyon 2004).
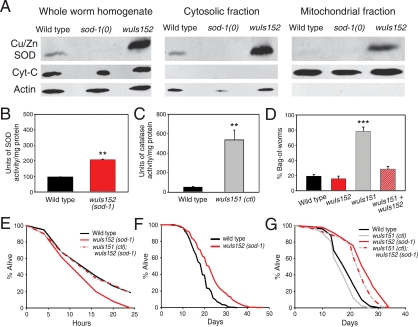
Figure 3.
Overexpression of sod-1 cytosolic Cu/ZnSOD increases life span. (A) Western blots of protein extracts of wild-type, sod-1 mutant, and sod-1 overexpresser lines using anti-Cu/ZnSOD antibodies. Cytochrome C (Cyt-C) and actin were used as mitochondrion- and cytosol-specific markers, respectively. Increased SOD-1 protein was also detected in wuIs154 transgenics (data not shown). (B) Total SOD activity in protein extracts from lines overexpressing sod-1. (**) P < 0.01, Student's t-test. (C) Total catalase activity in line overexpressing catalase. (**) P < 0.01, Student’s t-test. (D) Catalase overexpression causes high levels of mortality from internal hatching of larvae (bagging), and this is suppressed by overexpression of sod-1. (***) P < 0.0001, Student’s t-test. (E) sod-1 overexpression increases sensitivity to oxidative stress (40 mM paraquat), and this is suppressed by overexpression of catalase. (F) Overexpression of sod-1 increases life span. (G) Elevated catalase does not further extend longevity of the sod-1 overexpresser.
It has been shown previously that in daf-2 mutants, there is an elevation in levels of SOD activity (Vanfleteren 1993) and large fold increase in sod-3 mRNA (Honda and Honda 1999; Yanase et al. 2002). We confirmed this but also saw increases in expression of sod-1 and sod-5 (Supplemental Figs. S1B, S4B–D; Supplemental Material). Although sod-3 is the most highly up-regulated of the sod genes in terms of fold change in expression, its relative contribution to SOD levels remains very small, both in terms of overall SOD activity and MnSOD protein levels (Supplemental Fig. S1B–E).
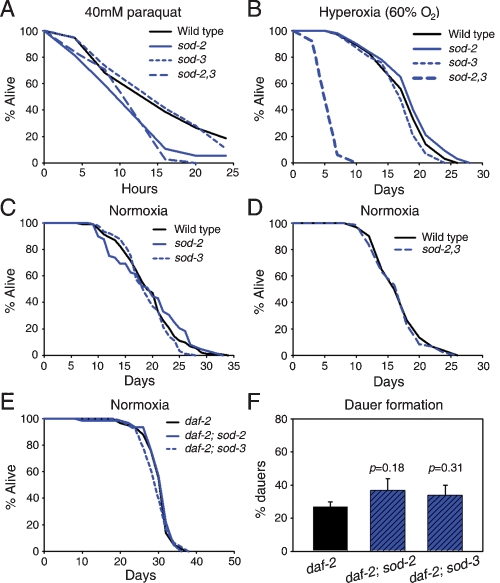
We examined the organismal effects of deletion alleles of sod-2 and sod-3 MnSOD genes on C. elegans. We find that sod-2(0) but not sod-3(0) results in delayed development (data not shown), delayed and reduced reproduction, and a slowed defecation cycle (Supplemental Fig. S5). However, sod-3(0) further reduces fertility of sod-2(0) animals (Supplemental Fig. S5B), implying functional redundancy between sod-2 and sod-3.
As expected, loss of MnSOD increases sensitivity to oxidative stress. sod-2(0) causes a moderate reduction in resistance to the O2− generator paraquat, while sod-3(0) does not, either alone or when added to sod-2(0) (Fig. 1A). In combination, sod-2(0) and sod-3(0) cause severe hypersensitivity to hyperoxia (60% oxygen), while each mutation alone has no effect (Fig. 1B). Thus, sod-2 and sod-3 are functionally redundant when defending against mild but not severe oxidative stress.
Figure 1.
Oxidative stress resistance and longevity of MnSOD mutants. (A) Survival in 40 mM paraquat. (B) Survival under hyperoxia (60% O2). (C,D) Life span of MnSOD single (C) and double (D) mutants under normoxia (20°C). (E) Effect of sod-2(0) or sod-3(0) on daf-2(m577) longevity (25°C). (F) Effect of sod-2(0) or sod-3(0) on daf-2(m577) Daf-c.
Next, we tested the effect of sod-2(0) and sod-3(0) on life span. Mean life span is unaffected by either mutation alone (Fig. 1C; Supplemental Table S1) or both mutations combined (Fig. 1D; Supplemental Table S1). Thus, surprisingly, complete absence of MnSOD has no effect on life span. We also find that activity of cytosolic and mitochondrial aconitase (an oxidation-sensitive iron–sulfur protein) is not detectably reduced in sod-2; sod-3 animals (data not shown), suggesting no major increase in oxidative damage to protein. To test for a role in daf-2 mutant longevity, we examined the impact of sod-2(0) or sod-3(0) on daf-2 longevity but there is none (Fig. 1E; Supplemental Table S2). Altogether, these results strongly imply, against expectation, that mitochondrial matrix O2− has no effect on aging and that MnSOD does not contribute to longevity assurance in C. elegans.
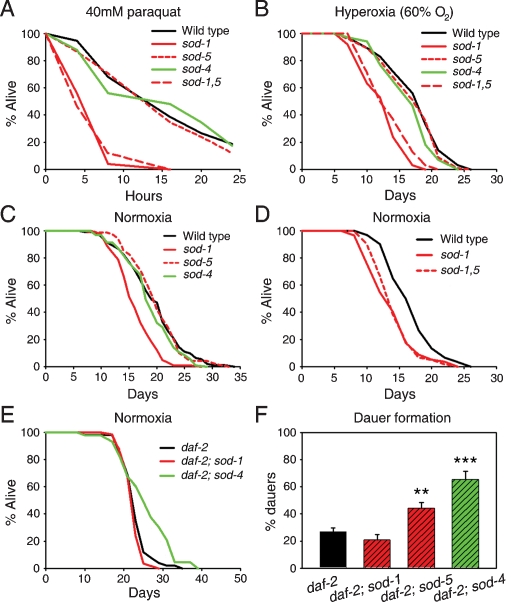
Next, we describe the effects of deletion of sod-1 and sod-5 cytosolic Cu/ZnSOD genes. sod-1(0) increases sensitivity to paraquat, but sod-5(0) does not, either alone or when added to sod-1(0) (Fig. 2A). By contrast, neither sod-1(0) nor sod-5(0) has a marked effect on sensitivity to mild hyperoxia (Fig. 2B). sod-1(0) also decreases mean life span, by 15%–31%, while sod-5(0) has no effect, either alone or when added to sod-1(0) (Fig. 2C,D; Supplemental Table S1). Moreover, addition of sod-2(0) does not further reduce the life span of sod-1 mutants (data not shown), supporting the view that O2− does not move between mitochondrial and cytosolic pools. Potentially, sod-1(0) shortens life span by accelerating the age increase in molecular damage. We therefore examined the effect of sod-1(0) on the age increase in damage to protein and lipid but could not detect any acceleration (Supplemental Material), perhaps because the impact of sod-1(0) is relatively subtle and difficult to detect.
Figure 2.
Oxidative stress resistance and longevity of Cu/ZnSOD mutants. (A) Survival in 40 mM paraquat. (B) Survival under hyperoxia (60% O2). (C,D) Life span of Cu/ZnSOD single (C) and double (D) mutants under normoxia (20°C). (E) Effect of loss of Cu/ZnSOD on daf-2(m577) longevity (20°C). (F) Effect of loss of Cu/ZnSOD on daf-2(m577) Daf-c. (**) P < 0.01; (***) P < 0.0001, Student’s t-tests.
If the shorter life span of sod-1 mutants is due to accelerated aging, then overexpression of sod-1 should increase life span. To test this, we first examined the effect of expression of the sod-1∷gfp transgene on life span but saw no effect (data not shown). However, we subsequently discovered that fusion of GFP to SOD-1 reduces Cu/ZnSOD-specific activity (Supplemental Fig. S7; Supplemental Material). Next, we generated transgenic lines with multiple copies of the sod-1 gene, focusing initially on two lines bearing integrated transgene arrays, wuIs152 and wuIs154. Overall SOD activity is increased approximately twofold by wuIs152 (Fig. 3B) and wuIs154 (data not shown), and both lines show increased Cu/ZnSOD immunoreactivity (Fig. 3A; data not shown).
Against expectation, sod-1 overexpression increases sensitivity to paraquat (Fig. 3E). Potentially, this could result from elevated levels of H2O2 due to faster conversion of paraquat-generated O2− into H2O2. To test this, we generated an integrated transgene array, wuIs151, with multiple copies of the entire ctl-1 ctl-2 ctl-3 gene cluster, which produces a 10-fold increase in catalase activity (Fig. 3C). Catalase overexpression suppresses the increased paraquat sensitivity resulting from sod-1 overexpression (Fig. 3E), implying that this hypersensitivity is indeed due to elevated H2O2 levels.
wuIs152 caused a statistically significant (P < 0.05) increase in life span in five out of 11 trials, and in no instance did it decrease life span (Supplemental Table S3). Combined data for these 11 trials defines a 21.5% increase in mean life span (Fig. 3F, P < 0.0001). Although wuIs154 did not increase life span, increased life span was seen in three further lines with extrachromosomal arrays (wuEx125, wuEx122, and wuEx123) (Supplemental Table S3). From this we conclude that elevated SOD-1 can slightly increase life span and that the absence of an effect in wuIs154 likely reflects a life-shortening mutation associated with chromosomal insertion of the transgene array.
Given that SOD converts O2− into H2O2, it is possible that increased levels of SOD-1 lowers cytosolic O2− but increases cytosolic H2O2, replacing damage from O2− with damage from H2O2. To test this, we compared the effect on life span of elevated SOD-1, catalase, or both. Overexpression of catalase alone results in a high level of mortality due to internal hatching of larvae (bagging) (Fig. 3D), perhaps reflecting H2O2 deficiency. This bagging is suppressed by overexpression of sod-1 (Fig. 3D), perhaps due to restoration of H2O2. In the absence of bagging (prevented by the inhibitor of DNA replication fluorodeoxyuridine, FUdR) overexpression of catalase slightly shortens life span, either alone or in addition to overexpression of sod-1 (Fig. 3G; SupplementalTable S3). Taken together, these results imply that cytosolic Cu/ZnSOD and, by implication, cytosolic O2−, are weak determinants of longevity and aging, respectively. By contrast, H2O2 does not seem to contribute to aging.
We next investigated whether sod-1 and sod-5 might contribute to the daf-2(m577) longevity (Age) phenotype. daf-2(m577) is a weak, temperature-sensitive (ts) allele resulting in a moderate increase in life span at 20°C and a large increase at 25°C (Gems et al. 1998; Patel et al. 2008). m577 was selected because it is a class 1 allele showing fewer pleiotropic effects than class 2 alleles such as e1370, making epistasis results easier to interpret. sod-1(0) slightly shortens daf-2 life span at 25°C, perhaps reflecting a minor contribution of sod-1 to daf-2 Age, but not 20°C (Fig. 2E; Supplemental Table S2). sod-5(0) had little consistent effect (Supplemental Table S2).
Finally, we examined the phenotypic effects of loss of the sod-4 extracellular Cu/ZnSOD. sod-4(0) does not affect sensitivity to oxidative stress (Fig. 2A,B) or life span in otherwise wild-type animals (Fig. 2C). Surprisingly, sod-4(0) enhances daf-2 Age at both 20°C and 25°C (Fig. 2E; Supplemental Table S2). One possibility is that this reflects an effect on signaling. SOD-4, like mammalian Cu/ZnSOD, might generate H2O2, which then crosses into the cell and promotes insulin signaling by inhibiting redox-sensitive, signal-quenching phosphatases (Goldstein et al. 2005). In C. elegans, treatment with H2O2 increases PIP3 levels and promotes cytosolic retention of DAF-16 (Weinkove et al. 2006). If sod-4(0) does reduce IIS, then it should enhance other daf-2 mutant traits, including constitutive dauer larva formation (Daf-c). We therefore tested the effect of loss of each sod gene on daf-2(m577) Daf-c and report that sod-4(0) significantly enhances Daf-c (Figs. 1F, 2F). Thus, sod-4 may contribute to IIS.
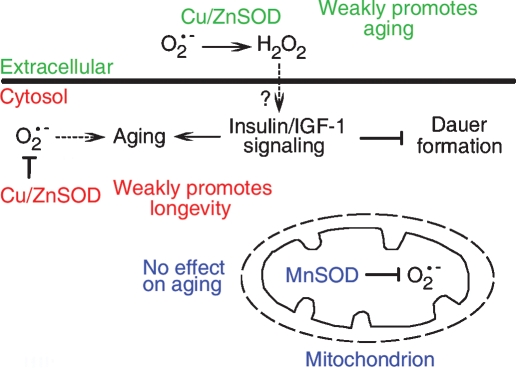
O2− has long been viewed as a possible major determinant of aging (Harman 1956, 1972). In this study, we explored the importance of the three major O2− pools on aging in C. elegans. Overall, our results imply that O2− is not a major determinant of aging in this model organism, either in wild-type animals or long-lived daf-2(m577) mutants; however, it remains possible that SOD contributes substantially to longevity in other contexts (e.g., under dietary restriction). Our findings point to the novel conclusion that each O2− pool is different in terms of its effect on aging. In wild-type C. elegans, the cytosolic O2− pool contributes weakly to aging while mitochondrial O2−, long considered a likely determinant of aging, and extracellular O2− have no detectable effect on normal aging. In daf-2 mutants, extracellular O2− appears to promote longevity; however, it is more likely that SOD-4 converts O2− into H2O2, which then weakly activates IIS, thereby shortening life span. The effects of SOD and O2− on aging therefore vary according to cell compartment and to genotype (summarized in Fig. 4).
Figure 4.
Influence of SOD and O2− on aging. This scheme shows a synthesis of conclusions drawn from the present study. Different SOD isoforms (and by deduction, the corresponding O2− pools) have different effects on aging. Extracellular Cu/ZnSOD weakly inhibits dauer formation and promotes aging, potentially by generating H2O2, which crosses into the cell and stimulates insulin/IGF-1 signaling by inhibiting redox-sensitive phosphatases. Cytosolic Cu/ZnSOD weakly promotes longevity, perhaps by protecting against molecular damage (sod-1 does not influence daf-2 Daf-c). Mitochondrial MnSOD has no detectable effect on aging. Arrow with dotted line implies a weak effect.
Our findings paint a clearer picture of the role of the various SOD isoforms in C. elegans. sod-1 and sod-2 encode the major Cu/ZnSOD and MnSOD isoforms, corresponding to the equivalent isoforms in other eukaryotes. By contrast, sod-5 and sod-3 encode inducible, auxiliary Cu/ZnSOD and MnSOD isoforms. The presence of these supernumerary isoforms, like that of the stress-resistant dauer larva stage in which they are up-regulated, may reflect the hostile soil environment in which C. elegans has evolved.
We examined the effect on life span of loss of SOD in each of the three major cellular compartments. The oxidative damage theory of aging predicts that loss of SOD should cause accelerated aging, particularly cytosolic Cu/ZnSOD and MnSOD, both of which contribute to scavenging of mitochondrially generated O2− (the former in the mitochondrial intermembrane space). In fact, only loss of sod-1 shortened life span, and then only modestly. However, overexpression of sod-1 did increase life span slightly (Fig. 3F; Supplemental Table S3), implying a small contributory role of O2− to aging.
Loss of MnSOD isoforms had no effect on life span, either in a daf-2(+) or daf-2(m577) background (Fig. 1C–E; Supplemental Tables S1, S2). This strongly implies that O2− within the mitochondrial matrix is not a significant cause of aging in C. elegans. An alternative possibility is that other mechanisms protect C. elegans mitochondria against O2−; however, the oxygen hypersensitivity of sod-2; sod-3 mutants argues against this. Very recently, another study also reported, using different mutant alleles, that sod-2(0); sod-3(0) does not affect life span in an otherwise wild-type background (Honda et al. 2008), confirming our findings. These investigators also observed that in a daf-2(e1370) mutant, sod-2(0) slightly shortened life span and lessened Daf-c while sod-3(0) had the opposite effect. daf-2 alleles fall into two phenotypic classes: Class 2 alleles are more pleiotropic than class 1 alleles and show more complex epistatic interactions with other mutations (Gems et al. 1998; Patel et al. 2008). The difference in effects of loss of MnSOD on daf-2(m577) (this study) and daf-2(e1370) (Honda et al. 2008) is interesting since m577 is a class 1 allele and e1370 a class 2 allele and suggests that MnSOD exerts a selective influence on class 2-specific defects.
Loss of function of many genes involved in mitochondrial function result in a Clk or Mit phenotype, which includes a delayed reproductive schedule, lowered fertility, slowed defecation cycle, and increased life span (Wong et al. 1995; Rea 2005). In this study, we found that sod-2(0) results in all these traits (Supplemental Fig. S5) except the increase in life span (Fig. 1C,D). RNAi of sod-2 enhances clk-1(qm30) mutant longevity (Yang et al. 2007), suggesting a possible cryptic effect of sod-2 on aging via a Clk/Mit-type mechanism.
Loss of sod-4 enhances the daf-2 Age and Daf-c phenotypes (Fig. 2E,F), suggesting that sod-4 may play a role in IIS regulation of dauer formation and life span. We postulate that SOD-4-generated H2O2 promotes IIS by inhibiting IIS antagonistic phosphatase enzymes, as occurs in mammals (Goldstein et al. 2005).
While our findings imply that SOD and O2− have, at most, minor effects on aging in C. elegans, the question remains: What is the relevance of these findings to higher animals? The effects on life span of altering SOD gene expression vary between C. elegans, Drosophila, and the mouse (for review, see Muller et al. 2007). Loss of MnSOD causes early lethality in the fly and the mouse but not the worm, while loss of cytosolic Cu/ZnSOD causes only small decreases life span in the worm and the mouse but a large (∼80%) decrease in life span in the fly. Overexpressing cytosolic Cu/ZnSOD slightly increases life span in C. elegans (this study), and in Drosophila overexpression of cytosolic Cu/ZnSOD and MnSOD seem to increase life span, although findings vary (Muller et al. 2007); however, overexpression of cytosolic Cu/ZnSOD does not increase life span in the mouse (Huang et al. 2000). By contrast, in the filamentous fungus Podospora anserina, reduction of mitochondrial ROS production results in a dramatic retardation of aging: Instead of aging rapidly after a brief period of growth, hyphae grow continuously and, apparently, indefinitely (Dufour et al. 2000).
It has been suggested that damage from reactive oxygen species might represent a public (i.e., evolutionarily conserved) rather than a private (i.e., lineage-specific) mechanism of aging (Martin et al. 1996). Our findings imply that the role of O2− in aging is to some extent public and to some extent private: Cytosolic O2− appears to contribute to aging in C. elegans and Drosophila but not mice. By contrast, loss of MnSOD is lethal to the fly and the mouse but has no effect on aging in the worm. Thus, O2− in some cellular compartments seems to contribute to some degree to aging in some species, but in other contexts (e.g., C. elegans) it appears unimportant to aging.
Materials and methods
Nematode culture
Nematodes were cultured on NGM agar seeded with E. coli OP50 as described previously (Sulston and Hodgkin 1988). Strains were maintained at 20°C unless otherwise noted. Mutant alleles and transgenic arrays used or generated in this study are listed in the Supplemental Material.
Oxidative stress resistance assays
For paraquat, young adults were placed overnight on agar plates containing 40 μM fluorodeoxyuridine (FUdR), and then picked into microtiter wells with 100 μl of 40 mM paraquat (Sigma, 856177) in M9 buffer. Viability was assayed over a 20-h period. For hyperoxia, young adults were picked onto plates containing 40 μM FUdR and placed in a sealed chamber under 60% O2 (22°C). Animals were briefly removed from the O2 chamber to score viability every 2–3 d.
Life span measurements
Life span assays were performed as described previously (Gems et al. 1998), at either 20°C or 25°C (see Supplemental Tables S1–S3). Survivorship of populations was compared statistically using the log rank test, performed using JMP 7.0.1 (SAS).
Construction of transgenic lines
Reporter constructs for each of the sod genes are full-length, translational fusions of GFP to the C terminus (see Supplemental Figure S2). For sod and ctl (catalase) overexpression, gDNA fragments were generated by PCR and microinjected directly. For primer sequences for reporter and overexpression constructs, see the Supplemental Material. Unless otherwise noted, pRF4 [rol-6(su1006)] was used as a marker of transformation. Integrated lines were generated by X-irradiation, and backcrossed to wild type (N2) at least five times before further study.
SOD and catalase activity assays
SOD activity was measured using an assay involving the inhibition of superoxide-induced lucigenin chemiluminescence by SOD, as described previously (Lenaerts et al. 2002). Catalase activity was assayed at 25°C according to a standard method (Aebi 1984), adapted for use in microtiter plate format.
Dauer formation assays
Eggs from strains bearing daf-2(m577) were collected by hypochlorite lysis and cultured at 23.5°C. The proportion of dauer larvae was scored 72 h later.
Acknowledgements
We thank David Hoogewijs for assistance with mRNA level estimations, and Peter Piper, Jennifer Tullet, and David Weinkove for useful discussion and comments on the manuscript. This work was supported by grants from the Wellcome Trust and the European Union (to D.G.), and Ghent University (GOA 12050101), the Fund for Scientific Research-Flanders (G.0025.06), and the European Community (LSHM-CT-2004-512020) (to J.R.V.). P.B. acknowledges a grant from the Institute for the Promotion of Innovation by Science and Technology in Flanders (IWT). Some strains were obtained from the Caenorhabditis Genetics Center, which is supported by the National Institutes of Health National Center for Research Resources.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.504808.
References
- Aebi H. Catalase in vitro. Methods Enzymol. 1984;105:121–126. doi: 10.1016/s0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- Alcedo J., Kenyon C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron. 2004;41:45–55. doi: 10.1016/s0896-6273(03)00816-x. [DOI] [PubMed] [Google Scholar]
- Bargmann C.I., Horvitz H.R. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science. 1991;251:1243–1246. doi: 10.1126/science.2006412. [DOI] [PubMed] [Google Scholar]
- Chavez V., Mohri-Shiomi A., Maadani A., Vega L.A., Garsin D.A. Oxidative stress enzymes are required for DAF-16-mediated immunity due to generation of reactive oxygen species by Caenorhabditis elegans. Genetics. 2007;176:1567–1577. doi: 10.1534/genetics.107.072587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypser J.R., Johnson T.E. Multiple stressors in Caenorhabditis elegans induce stress hormesis and extended longevity. J. Gerontol. 2002;57:B109–B114. doi: 10.1093/gerona/57.3.b109. [DOI] [PubMed] [Google Scholar]
- Dong M.Q., Venable J.D., Au N., Xu T., Park S.K., Cociorva D., Johnson J.R., Dillin A., Yates J.R. Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science. 2007;317:660–663. doi: 10.1126/science.1139952. [DOI] [PubMed] [Google Scholar]
- Dufour E., Boulay J., Rincheval V., Sainsard-Chanet A. A causal link between respiration and senescence in Podospora anserina. Proc. Natl. Acad. Sci. 2000;97:4138–4143. doi: 10.1073/pnas.070501997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T. Oxygen radicals and signaling. Curr. Opin. Cell Biol. 1998;10:248–253. doi: 10.1016/s0955-0674(98)80147-6. [DOI] [PubMed] [Google Scholar]
- Fujii M., Ishii N., Joguchi A., Yasuda K., Ayusawa D. Novel superoxide dismutase gene encoding membrane-bound and extracellular isoforms by alternative splicing in Caenorhabditis elegans. DNA Res. 1998;5:25–30. doi: 10.1093/dnares/5.1.25. [DOI] [PubMed] [Google Scholar]
- Gems D., Sutton A.J., Sundermeyer M.L., Larson P.L., Albert P.S., King K.V., Edgley M., Riddle D.L. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglio M.-P., Hunter T., Bannister J.V., Bannister W.H., Hunter G.J. The manganese superoxide dismutase gene of Caenorhabditis elegans. Biochem. Mol. Biol. Int. 1994;33:37–40. [PubMed] [Google Scholar]
- Goldstein B.J., Mahadev K., Wu X. Redox paradox: Insulin action is facilitated by insulin-stimulated reactive oxygen species with multiple potential signaling targets. Diabetes. 2005;54:311–321. doi: 10.2337/diabetes.54.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B., Gutteridge J.M.C. Free radicals in biology and medicine. Oxford University Press; Oxford, UK: 2007. [Google Scholar]
- Harman D. Aging: A theory based on free radical and radiation chemistry. J. Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Harman D. The biologic clock: The mitochondria? J. Am. Geriatr. Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- Honda Y., Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. 1999;13:1385–1393. [PubMed] [Google Scholar]
- Honda Y., Tanaka M., Honda S. Modulation of longevity and diapause by redox regulation mechanisms under the insulin-like signaling control in Caenorhabditis elegans. Exp. Gerontol. 2008;43:520–529. doi: 10.1016/j.exger.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Huang T., Carlson E., Gillespie A., Shi Y., Epstein C. Ubiquitous overexpression of CuZn superoxide dismutase does not extend life span in mice. J. Gerontol. 2000;55:B5–B9. doi: 10.1093/gerona/55.1.b5. [DOI] [PubMed] [Google Scholar]
- Hunter T., Bannister W.H., Hunter G.J. Cloning, expression, and characterization of two manganese superoxide dismutases from Caenorhabditis elegans. J. Biol. Chem. 1997;272:28652–28659. doi: 10.1074/jbc.272.45.28652. [DOI] [PubMed] [Google Scholar]
- Jensen L.T., Culotta V.C. Activation of CuZn superoxide dismutases from Caenorhabditis elegans does not require the copper chaperone CCS. J. Biol. Chem. 2005;280:41373–41379. doi: 10.1074/jbc.M509142200. [DOI] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: Insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Larsen P.L. Aging and resistance to oxidative stress in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 1993;90:8905–8909. doi: 10.1073/pnas.90.19.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenaerts I., Braeckman B., Matthijssens F., Vanfleteren J. A high-throughput microtiter plate assay for superoxide dismutase based on lucigenin chemiluminescence. Anal. Biochem. 2002;311:90–92. doi: 10.1016/s0003-2697(02)00397-4. [DOI] [PubMed] [Google Scholar]
- Martin G.M., Austad S.N., Johnson T.E. Genetic analysis of ageing: Role of oxidative damage and environmental stresses. Nat. Genet. 1996;13:25–34. doi: 10.1038/ng0596-25. [DOI] [PubMed] [Google Scholar]
- Missirlis F., Hu J., Kirby K., Hilliker A., Rouault T., Phillips J. Compartment-specific protection of iron-sulfur proteins by superoxide dismutase. J. Biol. Chem. 2003;278:47365–47369. doi: 10.1074/jbc.M307700200. [DOI] [PubMed] [Google Scholar]
- Muller F.L., Liu Y., Van Remmen H. Complex III releases superoxide to both sides of the inner mitochondrial membrane. J. Biol. Chem. 2004;279:49064–49073. doi: 10.1074/jbc.M407715200. [DOI] [PubMed] [Google Scholar]
- Muller F.L., Lustgarten M.S., Jang Y., Richardson A., Van Remmen H. Trends in oxidative aging theories. Free Radic. Biol. Med. 2007;43:477–503. doi: 10.1016/j.freeradbiomed.2007.03.034. [DOI] [PubMed] [Google Scholar]
- Patel D.S., Garza-Garcia A., Nanji M., McElwee J.J., Ackerman D., Driscoll P.C., Gems D. Clustering of genetically defined allele classes in the Caenorhabditis elegans DAF-2 insulin/IGF-1 receptor. Genetics. 2008;178:931–946. doi: 10.1534/genetics.107.070813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S.L. Metabolism in the Caenorhabditis elegans Mit mutants. Exp. Gerontol. 2005;40:841–849. doi: 10.1016/j.exger.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Riddle D.L., Albert P.S., eds . Genetic and environmental regulation of dauer larva development. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. [PubMed] [Google Scholar]
- Schulz T.J., Zarse K., Voigt A., Urban N., Birringer M., Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Sulston J., Hodgkin J. Methods. In: Wood W.B., editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1988. pp. 587–606. [Google Scholar]
- Suzuki N., Inokuma K., Yasuda K., Ishii N. Cloning, sequencing and mapping of a manganese superoxide dismutase gene of the nematode Caenorhabditis elegans. DNA Res. 1996;3:171–174. doi: 10.1093/dnares/3.3.171. [DOI] [PubMed] [Google Scholar]
- Vanfleteren J.R. Oxidative stress and ageing in Caenorhabditis elegans. Biochem. J. 1993;292:605–608. doi: 10.1042/bj2920605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinkove D., Halstead J.R., Gems D., Divecha N. Long-term starvation and ageing induce AGE-1/PI 3-kinase-dependent translocation of DAF-16/FOXO to the cytoplasm. BMC Biol. 2006;4:1. doi: 10.1186/1741-7007-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisiger R.A., Fridovich I. Superoxide dismutase. Organelle specificity. J. Biol. Chem. 1973;248:3582–3592. [PubMed] [Google Scholar]
- Wong A.E., Boutis P., Hekimi S. Mutations in the clk-1 gene of Caenorhabditis elegans affect developmental and behavioral timing. Genetics. 1995;139:1247–1259. doi: 10.1093/genetics/139.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanase S., Yasuda K., Ishii N. Adaptive responses to oxidative damage in three mutants of Caenorhabditis elegans (age-1, mev-1 and daf-16) that affect life span. Mech. Ageing Dev. 2002;123:1579–1587. doi: 10.1016/s0047-6374(02)00093-3. [DOI] [PubMed] [Google Scholar]
- Yang W., Li J., Hekimi S. A measurable increase in oxidative damage due to reduction in superoxide detoxification fails to shorten the life span of long-lived mitochondrial mutants of Caenorhabditis elegans. Genetics. 2007;177:2063–2074. doi: 10.1534/genetics.107.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]