Abstract
Growth factor-induced migration of endothelial cell monolayers enables embryonic development, wound healing, and angiogenesis. Although collective migration is widespread and therapeutically relevant, the underlying mechanism by which cell monolayers respond to growth factor, sense directional signals, induce motility, and coordinate individual cell movements is only partially understood. Here we used RNAi to identify 100 regulatory proteins that enhance or suppress endothelial sheet migration into cell-free space. We measured multiple live-cell migration parameters for all siRNA perturbations and found that each targeted protein primarily regulates one of four functional outputs: cell motility, directed migration, cell–cell coordination, or cell density. We demonstrate that cell motility regulators drive random, growth factor-independent motility in the presence or absence of open space. In contrast, directed migration regulators selectively transduce growth factor signals to direct cells along the monolayer boundary toward open space. Lastly, we found that regulators of cell–cell coordination are growth factor-independent and reorient randomly migrating cells inside the sheet when boundary cells begin to migrate. Thus, cells transition from random to collective migration through a modular control system, whereby growth factor signals convert boundary cells into pioneers, while cells inside the monolayer reorient and follow pioneers through growth factor-independent migration and cell–cell coordination.
Keywords: Collective migration, functional genomics, fibroblast growth factor, RNAi, signaling modules
Cell migration requires spatial and temporal coordination of protrusive, adhesive, and contractile activities to generate forward movement (Lauffenburger and Horwitz 1996; Ridley et al. 2003). In addition to having cell-autonomous migration activities, migratory cells often exist in two-dimensional monolayers, mechanically linked through cell–cell junctions. In this multicellular context, cells must coordinate migration with neighboring cells to ensure efficient collective movement. To orchestrate this balance, cells employ signaling systems that coordinate inputs from receptors, cell–cell interactions, and cell–matrix adhesion (Pignatelli 1998; Gupton and Waterman-Storer 2006; Ogita and Takai 2008). While significant genetic, biochemical, and pharmacological work has been done to dissect signaling systems underlying individual cell migration (Lauffenburger and Horwitz 1996; Ridley et al. 2003), less is known about the genes and mechanisms driving cellular movements within multicellular sheets. Collective cell movement has been observed in cell systems such as Dictyostelium aggregation during slug formation, border cell migration during Drosophila oogenesis, tubular branching during tracheal morphogenesis, and lateral line migration during zebrafish development (Meili and Firtel 2003; Montell 2003; Ghabrial and Krasnow 2006; Perlin and Talbot 2007). Coordinated movement of cells in two-dimensional sheets is also critical for the development and maintenance of organ systems. During embryogenesis, germ layers must migrate and close to form precursors for adult organ systems (Chaffer et al. 2007; Rohde and Heisenberg 2007). When epithelial sheets in the skin, digestive tract, or blood vessels are damaged, surrounding cell layers migrate to fill open space and restore sheet integrity (Heath 1996; Vasioukhin and Fuchs 2001). During cancer development, growing tumors release angiogenic growth factors that cause the extension of new blood vessels through endothelial sheet movements (Folkman 2007). Some cancers, such as melanoma, have also been shown to metastasize in sheets (Hegerfeldt et al. 2002).
In several examples of sheet migration, fibroblast growth factor (FGF) has been shown to play a central role. In Drosophila, tubular branching during tracheal morphogenesis is induced by FGF (Ghabrial and Krasnow 2006). In mice, knockout of FGF receptor 1 (FGFR1) leads to a loss of mesoderm and endoderm migration and embryonic lethality (Deng et al. 1994; Yamaguchi et al. 1994; Ciruna and Rossant 2001). FGF also induces healing of gastric lesions in the rat stomach by promoting epithelial closure (Konturek et al. 1993). Similarly, in wounded skin, keratinocytes induce the production of FGF7 by 160-fold, which accelerates re-epithelialization (Werner et al. 1992). In addition, expression of FGF by bladder carcinoma cells in nude mice is sufficient to induce angiogenesis and vascularization of the developing tumor (D’Amore and Smith 1993; Fernig and Gallagher 1994; Slavin 1995; Bikfalvi et al. 1997; Parsons-Wingerter et al. 2000). The ubiquitous importance of FGF signaling for sheet migration was an incentive for using FGF as a trigger for endothelial sheet migration in our experimental system.
Although growth factor-induced sheet migration has been observed in a number of experimental systems, several key questions remain open. First, while sheet migration is known to involve proliferation, cell migration, and cell–cell adhesion, it is not known whether these or possibly other relevant processes are controlled by a single interconnected upstream signaling system or are regulated separately by distinct signaling pathways or modules (Fig. 1A). Second, it is not known if growth factor-triggered sheet migration results from the activation of a specific process, such as proliferation or polarization, or whether growth factor globally regulates all functional processes. Third, it is not known whether forces driving sheet migration involve directional sensing. A directed migration model postulates that cells move into open space by following an extracellular directional signal while an alternative cell diffusion model postulates that sheets move through the induction of random cell motility that then leads to filling of an available open space (Bindschadler and McGrath 2007). Fourth, it is not known how cells positioned inside the sheet coordinate their movement with cells at the sheet boundary, thus preventing sheet rupture and loss of sheet cohesion when boundary cells migrate into open space. One model proposes that cells respond to a common diffusive directional signal and, therefore, move in a coordinated fashion (Matsubayashi et al. 2004). A second possibility is that cell–cell adhesion provides a mechanical connection to coordinate cell movement within the sheet.
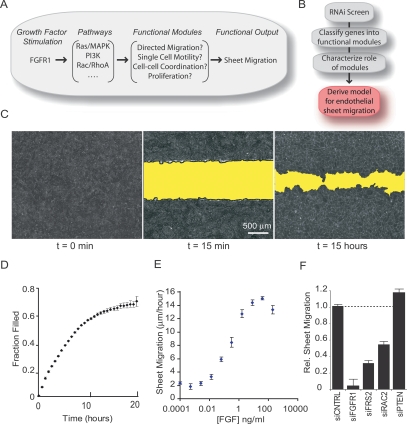
Figure 1.
FGF-induced sheet migration in HUVEC endothelial cells. (A) Schematic view of how growth factor-induced signaling pathways may regulate functional modules to produce sheet migration. (B) Experimental workflow for investigating sheet migration. (C) Fluorescent microscopy images of confluent HUVEC cells stained with AlexaFluor594-conjugated wheat germ agglutinin before (left panel) and 15 min after (middle panel) cell removal. (Right panel) After 15 h in the presence of serum, cells were fixed and stained with fluorescein phalloidin. (D) Kinetic analysis of sheet migration was performed by continuous imaging of fluorescently stained monolayers over 20 h (n = 4). (E) Sheet migration rates for HUVEC monolayers treated with varying concentrations of FGF (n = 4). (F) Sheet migration rates for cells transfected with siRNA pools targeting predicted positive (FGFR1, FRS2, and RAC2) and negative (PTEN) regulators of sheet migration (n = 4). Values were normalized against cells transfected with control siRNA. Standard error bars are shown.
We set out to answer these questions by using RNAi to target all putative human signaling proteins and identified a set of 100 known and novel regulators of endothelial sheet migration (Fig. 1B). We found that these regulators can be grouped into independent functional modules (proliferation, cell motility, directed migration, and cell–cell coordination) according to their effects on relevant live-cell migratory parameters. Markedly, we show that growth factor signaling feeds primarily into the directed migration module with little effect on either the cell motility or cell–cell coordination modules. This strongly argues for a model where growth factors are necessary for directed migration of cells at the sheet boundary without affecting random, diffusive migration inside the sheet. Using mixed cell experiments where subpopulations of endothelial cells are deficient in specific control proteins, we show that boundary cells selectively respond to growth factor to become pioneers. In contrast, neighboring cells inside the sheet follow these pioneers through growth factor-independent drag forces that orient previously randomly moving cells. This follower behavior requires both, the cell motility and cell–cell coordination modules. Thus, our study introduces a modular control structure where regulatory proteins function within independent modules that together control the emergent behavior of endothelial sheet migration.
Results
FGF induces endothelial sheet migration
To quantify sheet migration, we measured endothelial sheet movement into a stripped band of cell-free space. Confluent monolayers of human umbilical vein endothelial cells (HUVEC) were used as a model endothelium, and cell-free space was created with a Delrin tip to remove a band of cells from the center of a well (Fig. 1C). Sheet migration rates were monitored by measuring the reduction of cell-free area as a function of time. Following cell removal, sheet movement was initially linear then slowed as the sheet approached full closure after ∼20 h (Fig. 1D). Based on this analysis, we measured the sheet migration rate at a fixed time point 15 h after cell stripping in order to maximize signal-to-noise, remain close to the linear range of the assay, and minimize contributions from cell proliferation (which takes ∼24 h).
We were interested in studying cell migration triggered by growth factor input and tested whether basic FGF is sufficient to induce endothelial sheet migration in the absence of serum. While sheet migration diminished drastically under serum-free conditions, uniform addition of FGF elicited rapid sheet migration at concentrations greater than 1 ng/mL FGF (sheet migration rate is defined as the filled area normalized by time and length of sheet margin) (Fig. 1E). We chose a concentration of 1 ng/mL of FGF for the RNAi experiments to prevent saturation of the regulatory pathways.
We then performed control experiments and transfected cells with Dicer-generated pools of siRNAs against known migration regulators and assayed for changes in FGF-induced sheet migration. Targeting the FGFR1 and the actin regulator, RAC2, led to a ninefold and twofold reduction in sheet migration rates, respectively, while knockdown of phosphatase and tensin homolog (PTEN), a suppressor of PI3K signaling, resulted in a modest but significant increase in sheet migration (Fig. 1F). Using real-time PCR to quantify the remaining concentration for seven of the targeted mRNA transcripts, we found that transcript levels were on average reduced by 75%, with remaining mRNA concentrations ranging from 15% to 45% of control transfection (Supplemental Fig. 1A). These validation experiments showed that siRNA knockdowns were effective and reduced or enhanced sheet migration in a predictable manner, encouraging us to proceed with systematic siRNA perturbations.
Design, implementation, and validation of a 96-well formatted siRNA assay for sheet migration
Adapting the single chamber sheet migration assay to a high throughput multiwell format requires a method for generating cell-free bands in a 96-well microplate. We solved this problem by engineering a cell stripping tool that contains 96 individual spring-loaded Delrin tips mounted onto a scraping block equipped with a guide wall to ensure consistent horizontal cell-free bands in the center of each well (Fig. 2A,B). This system allows reproducible sheet migration measurements in a 96-well format (Supplemental Fig. 1B) suitable for medium- to high-throughput experimentation.
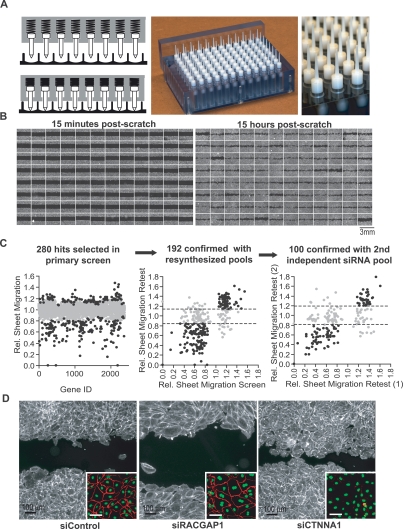
Figure 2.
Design, implementation, and validation of a 96-well formatted assay for sheet migration. (A) High throughput scratch tool with 96 individual spring loaded tips capable of generating uniform cell-free bands in a multiwell format. (Left panel) Diagrams of spring loaded tips with and without applied pressure. (Middle and right panels) Photographs of the scratch unit and the tips, respectively. (B) Montages of 96 images from a multiwell plate where each well was transfected with a different pool of siRNA. Fluorescent images were taken 15 min (left panel) and 15 h (right panel) after cell removal. (C) Results from siRNA screens targeting 2400 human signaling proteins. (Left panel) Sheet migration rates (gray dots) are the mean of duplicates and normalized to plate medians. A subset of genes (black dots) was selected for retest based on their deviation from median. (Middle panel) As a control, pools of siRNAs were resynthesized and retested. (Right panel) As a second control, repeating hits (black dots) were again selected and a second, independent siRNA pool was synthesized against the same targets and retested. One-hundred hits that showed consistent deviations in all experiments were considered confirmed hits. For both retest experiments, sheet migration rates were normalized to cells transfected with control siRNA, and each retest experiment was performed at least four times. (D) Examples of siRNA effects on sheet migration. Cells transfected with control, RACGAP1 and CTNNA1 siRNAs, were stained with fluorescein-phalloidin (main image) and VE-cadherin (inset).
We then used this platform to test for possible regulators among a set of Dicer-generated siRNA pools each targeting one of the 2400 putative, predicted, and known signaling proteins (using NCBI database from 2003). Protein targets were included in this set if they had homology with known signaling proteins from various model systems or contained characteristic domains such as kinase, phosphatase, small GTPase, PH, SH2, C1, C2, and EF-domains (>100 putative signaling domains were used) (Liou et al. 2005; Brandman et al. 2007; Galvez et al. 2007). The siRNA pools were generated by in vitro addition of recombinant Dicer protein to ∼300- to 500-base-pair (bp) segments of double-stranded RNA to ensure that each individual 20–22mer is present at a very low concentration, diluting potential off-target effects. On-target effects, on the other hand, are additive, so, cumulatively, diverse pools generate potent knockdown (Myers et al. 2006). In our experimental protocol, each of the 2400 siRNA transfections was performed in duplicate with a cumulative R-value of 0.85 between replicates (Supplemental Fig. 1C).
Figure 2B shows an example of a stripped 96-well microplate where each well is transfected with an siRNA pool targeting a different putative regulatory protein. Immediately after stripping, most wells have nearly identical cell-free bands. After 15 h, however, the remaining cell-free spaces vary significantly in size (Fig. 2B, right panel), highlighting siRNA-mediated changes in sheet migration. A quantitative analysis of sheet migration rates after siRNA transfection revealed that 88% of the 2400 protein knockdowns were within 20% of plate median values (Fig. 2C; Supplemental Fig. 1A). We selected the 280 siRNA targets with the strongest deviations using both an absolute and relative threshold to account for possible deviations in knockdown efficiency between plates (Fig. 2C).
Rather than maximizing the number of possible hits, our goal was to minimize the number of false positives. To eliminate the possibility of well-specific contaminants in the original library, we resynthesized each siRNA pool and tested the effect on sheet migration. Of the original 280 hits selected in the primary screen, 192 siRNA pools showed the same effect on migration rate with the second preparation (Fig. 2C). Although diced pools of siRNA are less likely to trigger off-target knockdowns compared with chemically synthesized oligonucleotides (Myers et al. 2006), we tested for off-target effects by generating a second pool of siRNA against an independent coding region or UTR sequence of the same message. Based on this more stringent test, 100 siRNA pools showed consistent effects on endothelial sheet migration with two independent targeting regions (Fig. 2C).
As a first survey of the identified gene products, we generated an interaction map using the PubMed database for protein–protein interactions. We noticed several major classes of functionally related proteins, including genes in the FGFR1 signaling pathway as well as cytoskeletal and adhesion regulators (Supplemental Fig. 2A). We also classified the hits using Database for Annotation, Visualization and Integrated Discovery (DAVID). Within DAVID, we selected the Simple Modular Architecture Research Tool (SMART) domain database and the Kyoto Encyclopedia of Genes and Genomes (KEGG) as references and found that sheet migration hits were statistically enriched (P < 0.05) for Rho GTPase (RHO), guanylate kinase (GuKc), serine/threonine phosphatase (PP2Ac), serine/threonine kinase (S_TK), Ras, and protein tyrosine phosphatase (PTP) domains when compared against all genes in the library (Supplemental Fig. 2B, black bars). Additionally, these genes were represented in signaling pathways controlling the actin cytoskeleton (Actin), focal adhesions (FAs), adherens junctions (AJs), leukocyte transendothelial migration (LTM), and tight junctions (TFs). These pathways and domain categories are both intuitively important for sheet migration (Supplemental Fig. 2B, gray bars; see Supplemental Table 1A for a list of the genes within each category; see Supplemental Table 1B for the nucleotide accession number used to generate each siRNA pool).
When surveying images from the primary screen we also recognized a few distinct phenotypes (Fig. 2D). A pool of siRNA targeting the Rho GTPase-activating protein RACGAP1, for example, led to a noticeable increase in cell size and a high frequency of binucleate cells (Fig. 2D, inset), consistent with one of its proposed roles in the initiation of cytokinesis (Zhao and Fang 2005). Similarly, cells transfected with siRNA against α-catenin (CTNNA1) adopted a rounded morphology with bright cortical actin and a loss of cadherin junctions (Fig. 2D, inset), highlighting a possible dual role for CTNNA1 in both the establishment of cell junctions and regulation of the actin cytoskeleton (Drees et al. 2005).
Live-cell tracking defines distinct functional processes
We then measured live-cell motility parameters for individual cells in a monolayer to distinguish functional processes important for sheet migration. These parameters were measured by staining confluent cells with a live-cell nuclear dye, and imaging every 20 min in a temperature- and CO2-controlled chamber for at least 4 h. We then adapted a particle tracking algorithm to determine cell trajectories (Fig. 3A; Crocker and Grier 1996). Markedly, we found that HUVEC cells migrate effectively and can pass each other within a confluent monolayer, requiring that endothelial cell–cell junctions are dynamic and not fixed structures (Fig. 3B; Supplemental Movie 1). We confirmed that, even though these cells moved readily past each other within the monolayer, the monolayer was completely connected by AJs as indicated by anti-VE-cadherin staining (Fig. 2D). The finding of random cell migration within an intact endothelial sheet may at least in part be explained by an earlier observation that cadherin junctions are not fixed structures but exhibit dynamic flow-like movements (Kametani and Takeichi 2007). Interestingly, while the direction of cell migration in the monolayer was random, we found that the relative movement between neighboring cells was coordinated over a few cell diameters. The coordinated directional movement can be highlighted by coloring tracks according to the direction of movement (Fig. 3B). This movement is reminiscent of viscous flow in fluid dynamics, which is based on reversible interactions between molecules. Here, dynamic cohesive interactions between cells are likely responsible for creating an analogous coordinated flow-like movement in cell monolayers.
Figure 3.
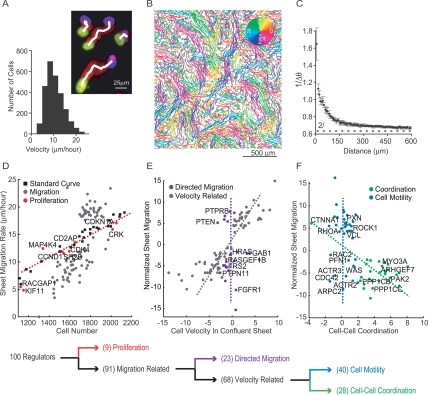
Sequential decision tree analysis to functionally cluster siRNA perturbations. (A) A series of superimposed images of migrating cells with the initial position in yellow, final position in blue, and intermediate positions in red (nuclear marker). The white line marks the output of the tracking algorithm. The histogram illustrates the distribution of individual cell velocities within the sheet. (B) Cell tracks from an intact monolayer imaged every 20 min for 16 h and colored according to the direction of movement. (C) Quantification of coordinated cell movement. The graph shows the inverted, angular difference between cell pairs as a function of distance between the pairs (five repeats; error bars represent standard error). The dashed line at 2/π represents the threshold for random movement. (D) Effect of cell density on sheet migration. White squares show migration rates measured for control cells plated at different densities. The red dashed line represents the best linear fit to these control measurements (r-squared = 0.97). Sheet migration rates are plotted as a function of cell number for validated hits (n = 4; gray and red dots). Gene perturbations that lie within two standard deviations of the control line were considered density-dependent and marked as red dots. (E) Remaining siRNA targets were tested for a possible role in directed cell migration. Sheet migration rate is plotted as a function of average individual cell velocity within an unperturbed monolayer for each migration-related siRNA perturbation (normalized values). The gray line approximates the correlation axis between cell velocity and sheet migration. Velocity-independent regulators, whose effect on single cell velocity was within two standard deviations of control values, are shown in purple (directed migration module). (F) Remaining velocity-related siRNA knockdowns were classified into cell–cell coordination and cell motility modules. Sheet migration rates are plotted as a function of directional correlation. Genes that enhance or suppress directional correlation are marked in green (cell–cell coordination module), while genes that affect sheet migration but have weak effects on directional correlation, are marked in blue (cell motility module). The decision tree is summarized below the individual plots. All data points represent an average of at least four independent experiments.
The velocity of cells within the sheet was measured from the average cell displacement over time and exhibited a relatively uniform distribution centered at ∼10 μm/h (Fig. 3A). A cell–cell coordination parameter was measured by calculating the average angular difference between the directions of a pair of cell trajectories and plotting the inverted value as a function of the distance between cell pairs (Fig. 3C). For cells closer than 100 μm, the direction of migration was highly correlated while cells further than 200 μm apart migrated almost randomly relative to one another (Fig. 3C).
Functional classification of siRNA perturbations
Using sheet migration, cell density, single cell velocity, and cell–cell coordination as parameters, we employed a hierarchical decision tree analysis to separate different siRNA phenotypes into functional modules. First, we tested the hypothesis that a subset of siRNAs may affect sheet migration indirectly through changes in cell density. Sheet migration rates for monolayers plated at varying cell densities were used to generate a standard curve (Fig. 3D, black squares and red dashed line, r-squared = 0.97). Cell density and sheet migration values for each siRNA perturbation were then compared with this curve. Although many siRNA affect both cell density and sheet migration, the majority of points deviate from the standard curve by more than two standard deviations (Fig. 3D, gray dots). Nine of the siRNA, however, were within these limits, arguing that these proteins affect sheet migration indirectly by altering proliferation rates and changing cell density (Fig. 3D, red dots).
The siRNAs with density-dependent migration changes include cyclin D (CCND1), cyclin-dependent kinase 4 (CDK4), cyclin-dependent kinase inhibitor 1a (CDKN1A), and kinesin family member 11 (KIF11), each with well documented roles in cell cycle progression or mitotic completion (Fig. 3D, red dots; Sawin et al. 1992; Sharp et al. 1999; Goshima and Vale 2003). Interestingly, even though Rho GTPases are considered key cytoskeletal regulators, the GTPase-activating protein RACGAP1 was classified as a density-dependent migration effector, consistent with a role as a cell division regulator rather than a regulator of migration (see evidence of binucleate cells in Fig. 2D). Surprisingly, our study also argues that MAP4K4, which was identified previously as a promigratory kinase in a cancer cell motility screen (Collins et al. 2006), instead indirectly affects migration by altering sheet density in HUVEC (Fig. 2D). For the subsequent analysis of the remaining genes, we corrected for the density effects and calculated a density-corrected migration rate.
We then tested the hypothesis that a subgroup of the remaining regulators is dedicated to directing sheet movement in the presence of open space. We determined whether such a directed migration module exists by identifying potential siRNA pools that enhance or suppress sheet migration into open space without affecting random cell motility within a sheet. Not surprisingly, most siRNA showed a strong positive correlation between sheet migration and individual cell velocity within the sheet, suggesting that these genes are general regulators of cell motility in the presence or absence of cell-free space (Fig. 3E, gray dots). A subset of siRNAs, however, had negligible effects on cell velocity within the sheet while exhibiting marked changes in sheet migration into open space (Fig. 3E, purple dots). These genes included a number of unknown proteins but also many of the players in the FGFR signaling pathway, including the receptor FGFR1, its adaptors FRS2 and GRB2-associated binding protein 1 (GAB1), and the downstream effector v-Ha-ras Harvey rat sarcoma viral oncogene homolog (HRAS). The existence of such a subset of proteins that selectively directs sheet migration into an open space strongly supports the hypothesis that cells have a directed migration control module.
The remaining siRNAs either increase or decrease individual cell velocities within a confluent monolayer. We tested whether siRNAs exist among this subset for which the effect on cell velocity could be explained by an indirect change in cell–cell coordination rather than a direct effect on the migration process. When comparing sheet migration with the cell–cell coordination parameter for these velocity regulators, we observed two distinct populations (Fig. 3F). The larger set of siRNA knockdowns had no significant effect on cell–cell coordination. We considered these to be cell-autonomous motility regulators that do not affect cell–cell coordination but enhance or reduce single cell velocity (Fig. 3F, blue dots). The remaining siRNA pools, however, likely acted indirectly on velocity by altering cell–cell coordination. This group of regulatory proteins, many of which are likely novel regulators of cell–cell adhesion, showed an interesting inverse relationship between sheet migration and cell–cell coordination. This suggests that highly coordinated movement results in slower cell velocities, possibly through drag forces imposed by cell–cell interactions (Fig. 3F, green dots, cell–cell coordination module). This drag hypothesis is supported by the strong reduction in cell–cell coordination and drastic increase of in-sheet velocity after knockdown of α-catenin, a known regulator of cell–cell adhesion important for the formation of cadherin junctions (Fig. 2D; Drees et al. 2005). Further supporting this interpretation, we found that directly targeting VE-cadherin, which was not included in our original siRNA library, also reduces cell–cell coordination (data not shown).
In contrast to cell–cell coordination regulators, the cell motility proteins, which include well known cytoskeletal and adhesion proteins, control the speed of individual cell movements but not the dynamics between cells. In addition to a number of novel proteins, this cell motility module includes many familiar players in cell migration, including Rho GTPases (RAC2, RHOA, and CDC42), Arp2/3 components (ACTR3, ACTR2, and ARPC2), FA proteins (PXN, ACTN1, and VCL), and stress fiber regulators (ROCK1). Together, our findings argue that sheet migration is controlled by a large number of proteins that primarily function to regulate cell density, single cell motility, directed migration, or cell–cell coordination (gene summary for each module listed in Supplemental Table 2).
Among these modules, we found the regulatory modules controlling directed cell movement into open space and cell–cell coordination within a cell monolayer to be particularly interesting for understanding sheet migration. To gain a mechanistic understanding of how these modules coordinate growth factor-triggered sheet migration, we made use of single siRNA perturbations to interfere with the modules and perform dynamic migration measurements.
Molecular components that direct sheet migration into cell-free space
The existence of a directed migration control module (Fig. 3E) argues strongly against a simple diffusive model for sheet migration whereby cells move randomly into unobstructed space. Because FGFR1 was identified as a member of this module, we compared the effect of titrating FGF concentration on random sheet migration against directed migration into cell-free space. While the cell velocity within the sheet was nearly constant in the absence or presence of FGF (Fig. 4A), the same increase in FGF caused a 10-fold increase in sheet migration.
Figure 4.
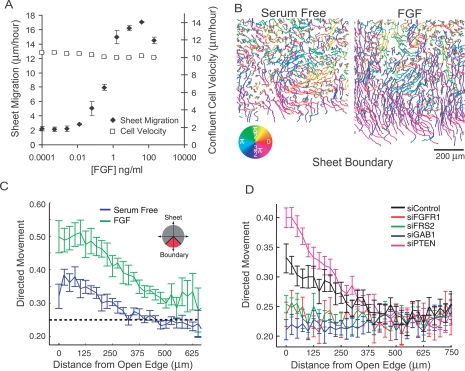
Molecular basis for directed migration into cell-free space. (A) Dose response curve relating sheet migration (black diamonds) to individual cell velocities in an unperturbed sheet (white squares) in response to varying concentrations of FGF (n = 3, error bars represent standard error of the mean). (B) Representative cell tracks near an open edge for monolayers treated with FGF (right panel) or serum-free media (left panel) colored according to the direction of movement. (C) Quantification of traces in (B) where fraction of cell movements oriented toward cell-free space (directed migration) is calculated as a function of distance from the open edge in the presence (green) or absence (blue) of FGF. (D) Same as C where monolayers are tracked in the presence of FGF and transfected with siRNA targeting various FGF-related signaling genes previously classified as being part of the directed migration module.
To investigate how FGF directs cells during sheet migration, we tracked individual cells within a migrating sheet. As expected, in regions distal to the open edge, cells moved randomly with similar speeds in both serum-starved and FGF treated cells. Near the sheet boundary, however, FGF-treated cells persistently migrate toward the cell-free space (Fig. 4B). To quantify this behavior, we counted the fraction of time cells spend migrating toward the monolayer edge (plus or minus 45°) and plotted the average value as a function of cell distance from the open edge (Fig. 4C). In this analysis, a value of 0.25 reflects a random orientation since a 90° window represents one-quarter of a circle. Addition of FGF caused a significant increase in directed movement throughout the cell monolayer that was maximal near the sheet boundary, dropping off gradually at positions further from the edge. In contrast, serum-starved cells showed weaker orientation overall but maintained some directed movements near the sheet boundary (Fig. 4C). Next, we tracked cells transfected with siRNA pools against four members of the directed migration module, namely FGFR1, FRS2, GAB1, and PTEN, and compared their capacity for directed migration relative to control cells. Consistent with their role in directed migration, the first three genes, which slow sheet migration, showed a reduced orientation toward the cell-free band, while PTEN, which enhances sheet migration, showed a more pronounced orientation into cell-free space (Fig. 4D).
Pioneer and follower cells
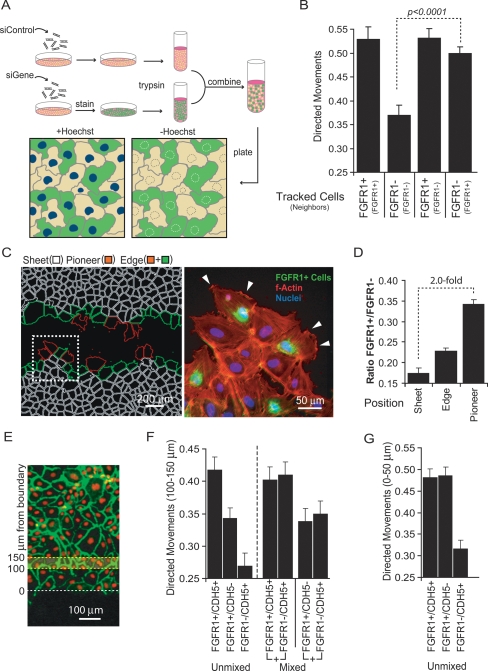
While our experiments show that growth factor signaling is required for directed movement into open space, it is not clear how cells behind the sheet margin can sense a directional signal. Earlier studies have also observed that a subset of epithelial boundary cells advance more effectively into open space than neighbors (Omelchenko et al. 2003), but it is not clear whether this behavior is growth factor regulated. Nevertheless, mosaic studies of Drosophila tracheal progenitor cells suggest that a single FGFR-expressing cell is sufficient for sheet extension and tracheal branch formation (Ghabrial and Krasnow 2006). Based on our data and these earlier observations, a plausible model for growth factor-triggered directed migration is based on pioneer behavior, where boundary cells lacking complete cell–cell contact polarize toward cell-free space in response to growth factor inputs. Differences among cells at the sheet boundary may result from changes to the sensitivity or strength of growth factor signaling in single cells. The most responsive cells could then function as pioneers, moving into cell-free space and guiding or pulling neighboring cells, thereby creating an oriented, viscous flow.
To test this pioneer cell model, we investigated whether cells lacking growth factor signaling can follow growth factor-competent cells into cell-free space. To do this, we performed coculture experiments by mixing cells transfected with either control or FGFR1-specific siRNA pools. Because cells were marked with live-cell dyes, we could track the behavior of each population independently within the same monolayer (Fig. 5A). In control experiments where we mixed cells of the same type, we found that, as in Figure 4C, cells with competent FGFR1 signaling (FGFR1+) showed oriented movements into cell-free space, while FGFR1-knockdown cells (FGFR1−) exhibit nearly random movements (Fig. 5B). Interestingly, however, in mixed populations, both the cells with and without FGFR1 moved in a highly oriented fashion at the sheet edge, suggesting that FGFR1 signaling acts cell-nonautonomously to rescue directional movement in neighboring cells without FGFR1 signaling (Fig. 5B). In a control experiment, this phenomenon was also observed when the staining patterns were reversed, suggesting that the experimental protocol did not affect directional choices (Supplemental Fig. 3A,B). When exogenous FGF was removed, however, all directional movement ceased, indicating that endogenous FGF production does not contribute significantly to polarized migration into open space (Supplemental Fig. 3B). We also tested the role of FGF in sparsely plated HUVEC and found a smaller but significant dose-dependent increase in cell migration velocity (Supplemental Fig. 4) again suggesting that FGF plays a role in triggering polarized movement in cells with incomplete cell–cell contacts.
Figure 5.
Pioneer and follower behavior. (A) Schematic representation of coculture experiments to investigate pioneer and follower behavior (green and orange colored cells used for illustration). Cell marking was based on one population stained with CellTracker (InVitrogen) and both populations stained with Hoescht. (B) FGFR1+ cells induce polarized movement in neighboring FGFR1− cells. FGFR1+ or FGFR1− cells were cocultured 1:1 with FGFR1+ or FGFR1− cells (four combinations). Cells in contact with the sheet margin in one population were tracked and their orientation measured (tracked cells listed first and neighbors listed in parentheses). A random orientation is 0.25. (C, left panel) Example of an image depicting pioneer (red) versus nonpioneer (green) locations at the sheet margin (defined as more versus less than half of the circumference in contact with the open space). White cells depict internal sheet cells. Cell boundaries were based on nuclear and F-actin stains. (Right panel) Pioneer cells are often FGFR1+ and are rich in actin ruffles. Picture is a close-up of the white box shown in the left panel with phalloidin staining shown in red, Hoescht staining shown in blue, and FGFR1+ cells shown in green. Lamellipodial ruffling is highlight with white arrows. (D) FGFR+ cells are enriched in pioneer positions. Bar graph shows the ratio of FGFR1+/FGFR1− cells in pioneer, edge and sheet positions. Plating ratio was 0.2 between FGFR1+:FGFR1− cells. (E) Diagram illustrating the positions from which directed motility measurements were taken for the “follower” experiments in F and G. (F) Follower behavior is lost inside sheets lacking VE-cadherin. Directed motility of cells was measured 150 μm from the sheet margin under various coculture conditions. Unmixed experiments represent homogenous cultures receiving the indicated siRNA treatment. Mixed population experiments show the directed motility for each population within a cocultured experiment (plated at a 1:1 ratio) (G) Directed motility for cells positioned at the sheet margin in monolayers treated with combined gene and/or control knockdowns.
To investigate this pioneer and follower behavior more closely, we fixed sheets after 12 h to look for spatial relationships between FGFR1+ and FGFR1− cells. Using Hoescht staining as a primary mask, we used image processing of actin staining to create an outline of individual cells (Fig. 5C). We noticed that cells along the sheet margin tend to behave differently, with some cells acting as pioneers, projecting large portions of their surface into the open space while others maintain maximal cell contact with the monolayer (Fig. 5C, pioneer cells labeled in red). Additionally, it appeared that pioneer cells can often create finger-like projections jutting into open space (Fig. 5C, white box). When we took high-resolution images, we noticed that FGFR1+ cells are concentrated at these pioneer positions and have a higher incidence of actin ruffles (Fig. 5C, white arrows). To quantify the relative distribution of FGFR1+ and FGFR1− cells, we calculated the ratio of FGFR1+ to FGFR1− cells for all cells in the monolayer (sheet), for all cells contacting open space (edge), and for all cells with more than half of their surface area contacting open space (pioneer) (Fig. 5D, blinded visual scoring). When compared with FGFR1− cells, we found a small FGFR1+ enrichment at the sheet edge but, more importantly, a more than twofold enrichment of FGFR+ cells at pioneer positions.
The FGFR-mediated directed movement at the sheet margin does not address how cells more than 150 μm away from the sheet margin bias their movements toward the open space (Fig. 4C). Because the distance of this polar migration is similar to the range over which cells coordinate movement within a confluent monolayer, it is suggestive that the same cell–cell coordination is responsible for both collective movements (Fig. 3C). This raises the hypothesis that follower behavior is mediated by a combination of cell–cell coordination and random migration and occurs independently of open space.
We tested this pioneer and follower hypothesis by knockdown of VE-cadherin (CDH5), which we identified previously as a regulator of cell–cell coordination. If cell–cell coordination is responsible for the follower behavior, knockdown of VE-cadherin should have little effect on cell polarization at boundary positions but significantly reduce directed movements 150 μm away from the cell-free area (Fig. 5E). Indeed, quantitative analysis of directed cell movements in the presence of siRNA against VE-cadherin (Fig. 5F, unmixed) shows that a reduction in cadherin cell–cell contact sites decreases orientation deep inside the monolayer (FGFR1+/CDH5+ and FGFR1−/CDH5+ are included as reference measurements). Coculture experiments (Fig. 6F, mixed) where both cell populations were tracked, show that FGFR1+/CDH5− and FGFR1−/CDH5+ cell populations both lose directed movement, arguing that the loss in orientation is cell-nonautonomous. Furthermore, monolayers where all cells lack VE-cadherin show a nearly equivalent reduction in orientation compared with monolayers that have an equal mixture of VE-cadherin-positive and VE-cadherin-negative cells, consistent with a requirement for intact cell–cell junctions in the transmission of follower behavior. The same loss of follower behavior inside the sheet was observed in knockdown experiments with α-catenin (data not shown), another previously identified regulator of cell–cell coordination (Fig. 3F). Although knockdown of VE-cadherin led to a loss of cell orientation inside the monolayer, it had no significant effect on the polarity of cells at the sheet boundary (Fig. 5G).
Figure 6.
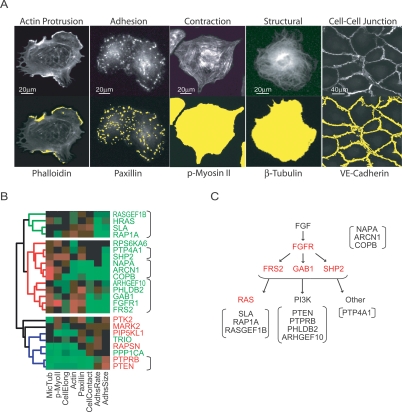
Subdivision of modules using structural and morphological characteristics. (A) All siRNAs were tested in secondary assays using molecular parameters expected to be important for sheet migration. Top panels show fluorescent images (20×) of markers. Bottom panels include the mask (in yellow, overlayed over an F-actin stain) generated by the image analysis software used to measure intensity and area of various cell structure. (B) Hierarchical clustering of directed motility genes using data from structural secondary assays. Fast and slow sheet migrators shown in red and green, respectively. (C) Pathway scheme of directed migration signaling components with their putative functional proximity to the FGFR according to their proximity in B. Identified upstream regulators are marked in red. Putative RAS-related genes, PI3K pathway components, and regulators of receptor transport are set apart with brackets.
Together, this argues that the regulatory proteins in the directed cell migration module are responsible for pioneer behavior while regulatory proteins comprising the cell–cell coordination module and the cell motility module mediate the complementary follower behavior. In combination, these modules ensure efficient sheet migration while maintaining endothelial integrity.
Subdivision of modules using structural and morphological characteristics
We tested whether it is possible to further group genes within these functional modules by subjecting transfected cells to a series of secondary assays designed to measure structural and morphological characteristics relevant for cell migration. These assays were based on immunostaining of molecular markers for cell protrusion, cell–cell adhesion, cell surface adhesion, and cell contraction. For example, we used phalloidin for F-actin levels, anti-paxillin for FA size, anti-phospho-myosin II for stress fibers, anti-tubulin for microtubules, and anti-cadherin5 for AJs (Fig. 6A). We then developed automated image analysis scripts to identify cellular structures and quantify them according to area or intensity. In addition, transfected cells were replated and fixed after 1 or 15 h to calculate the spreading rate and steady state cell area, respectively. Finally, cadherin staining was used to outline cell boundaries, allowing the determination of cell elongation (secondary analyses summarized in Supplemental Table 3; secondary measurements are provided in Supplemental Table 4).
We employed hierarchical clustering to organize genes according to these secondary assay measurements, and the results of this subclustering strategy are shown for the directed migration module in Figure 6B. Interestingly, although the hierarchical analysis does not include any kinetic parameters, it successfully separates most fast and slow migrators across the first branch point, suggesting that proximity to known regulators of sheet migration provides a useful starting point to identify mechanistic roles for the many proteins with unknown functions (Fig. 6B). In addition, the analysis can be used to generate a putative functional connection map of signaling components within the FGFR1–RAS–PI3K signaling axis (Fig. 6B,C). Identical hierarchical analyses for each of the other functional modules are included in Supplemental Figure 5.
Discussion
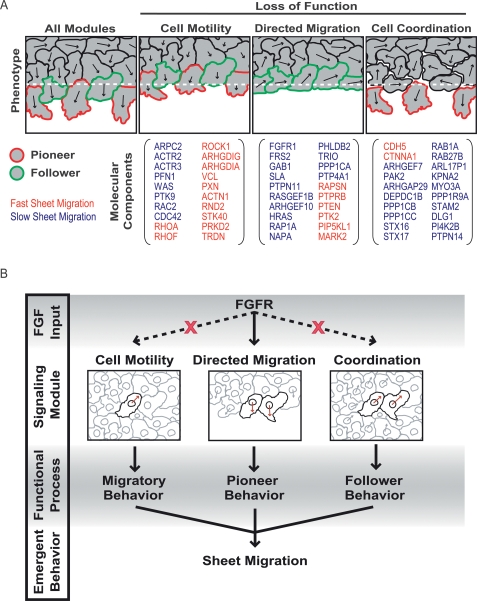
Our study combines siRNA perturbations and functional profiling to dissect growth factor-induced sheet migration. Our hierarchical decision tree analysis suggests that all regulatory proteins can be assigned into four distinct modules controlling proliferation, single cell motility, directed migration, and cell–cell coordination (Fig. 7A). Individually, each of these modules controls a key process necessary for sheet migration, and together, they generate the higher-level, emergent behavior characteristic of endothelial sheet migration.
Figure 7.
Modular control of endothelial sheet migration. (A) Schematic representation of sheet migration defects when specific functional modules are disabled by protein knockdown. A number of genes from each module are listed as examples with fast and slow sheet migration indicated as red and blue lettering, respectively. (B) Coordination of sheet migration by an FGF-dependent directed migration module and a FGF-independent cell–cell coordination module.
An FGFR pioneer cell module for directed cell migration
We found that genes associated with the FGFR–RAS–PI3K signaling pathway comprise a directed migration module that promotes sheet migration by orienting existing movement rather than enhancing cell velocity. Based on the hierarchical classification shown in Figure 6B, one can propose putative functions for genes according to their proximity to known regulators. For example, genes within the red branch contain FGFR1 and its adaptors GAB1 and FRS2, suggesting that genes within this group are likely involved in very upstream signaling activities at the receptor level (Fig. 6B). The proximity of the transport proteins COPB, NAPA, and ARCN1 to FGFR1 suggests that they may reduce the delivery of receptors or other upstream signaling components to the plasma membrane. The proximity between RAS, RASGEF1B, and RAP1A suggests that Rap and Ras may have related roles in the initiation of cell polarization. Finally, the putative function for proteins with no previously documented role in polarization and migration can be predicted based on their proximity to known regulators in the hierarchical analysis.
The importance of the FGF pathway is most apparent under serum-free conditions, where cells migrate with normal speed but fail to sense open space or respond with directed movement. By mixing FGF-responsive and FGF-unresponsive cells in the same monolayer, we demonstrate that the extension of polarized lamellipodia is an FGF-dependent and cell-autonomous process. These observations argue for a “pioneer” cell model in which cells responding to local or global FGF signals bias movements away from the cell monolayer. Mechanistically, polarized actin ruffling at the sheet boundary can be explained by a contact-mediated repression of growth factor-induced lamellipodia. In this putative AND-gate model, growth factors act as a uniform permissive signal that becomes polarized only after a local reduction in cell contact. In different physiological settings, other directional cues may also combine with permissive growth factor signaling such as chemical gradients during development or angiogenesis (Yang et al. 2002; Carmeliet 2003).
Thus, FGF signaling plays a primary role in endothelial sheet migration by acting as a costimulus for triggering directed cell migration into cell-free space. Since our studies were performed within a 15-h time window, we mostly excluded the effects of FGF signaling on cell proliferation, which likely plays a role in repopulation at later time points.
Pioneers and followers
Migrating endothelia must not only generate bulk cell movements but also maintain sheet cohesiveness. Growth factor-dependent polarization alone does not fulfill this requirement nor can it explain oriented movements several cell diameters away from the sheet margin. Insight into this mechanism came from cell trajectories within a confluent monolayer, which showed correlated movements between neighboring cells for distances up to 200 μm. While flow patterns of cells that move into open space have been described previously (e.g., Poujade et al. 2007), our data show that coordinated migration in monolayers exists even in the absence of cell-free space. We grouped siRNA perturbations according to effects on cell–cell coordination and found that both CTNNA1 (α-catenin) and CDH5 (VE-cadherin) suppress coordination while a much larger subgroup of mostly unknown regulators enhance coordination. Markedly, siRNA perturbations that led to a highly coordinated movement also resulted in a hyper-elongated phenotype, indicating that cell–cell interaction forces are strong enough to stretch individual cells (Supplemental Fig. 6). Thus, our study argues for a model in which mechanical forces derived from cell–cell interactions have an important role in polarizing neighboring cells and cells inside the sheet during growth factor-triggered sheet migration. These cell–cell adhesion drag forces can then convert basal random migration of cells inside the sheet into a coordinated directed migration response. Such a mechanical model for coordinated migration is consistent with an observation in flow chambers experiments where flow shear forces have been shown to be sufficient to polarize endothelial cells (Simmers et al. 2007).
We further tested this mechanical model in coculture experiments with FGFR-responsive and FGFR-unresponsive cells. We found that the directional movement of boundary cells is sufficient to redirect the flow of FGF-insensitive “follower” cells toward the cell-free space. The mechanical viscous flow model is further supported by the observation that FGFR-unresponsive cells can become oriented even when FGFR-responsive cells are positioned to their sides. Finally, we confirmed that cell–cell coordination and follower behavior is generated at least in part through cell–cell adhesion forces because a reduction in VE-cadherin significantly reduces the capacity to follow deep into the sheet. This mechanical coordination of sheet movement is functionally significant because it allows a sheet to maintain cohesion when boundary cells move into open space.
We were also intrigued by our finding that cells move within intact monolayers at speeds of >10 μm/h. This suggests that cadherin and other attachments, at least in endothelial cells, are dynamic structures that dismantle and reform as cells pass by one another in a contiguous sheet. Previous work has shown that cadherin junctions form through a zippering action along cell surfaces (Adams et al. 1996). Our data add a dynamic dimension by arguing for a “slipping zipper” model where weak cadherin interactions are rapidly turned over to dynamically move cell interfaces, preserving tight and uniform contacts while allowing cells to pass one another.
A modular model for sheet migration
Together, these results argue for a simple model for endothelial sheet migration based on three modular processes: cell motility, directed migration, and cell–cell coordination. Under basal conditions, the cell motility machinery generates random cell movements within the cell monolayer. Because cells retain cell–cell contacts, the coordination module results in locally correlated cell movements (Fig. 7B). In the presence of cell-free space and growth factor stimulation, the directed migration module generates pioneer behavior in cells along the sheet margin. As pioneers begin to migrate into cell-free space, the cell–cell coordination module and cell motility modules confer follower behavior in cells within the monolayer, which then migrate directionally in response to mechanical pulling forces from the pioneer cells at the leading edge (Fig. 7B).
For clarity, the siRNA knockdown results can be used to highlight the significance of each module (Fig. 7A). In the absence of cell motility, cells can sense and respond to directional cues but move too slowly to effectively generate sheet displacement. In the absence of FGFR signaling, cells lack directed migration but continue to migrate randomly, thereby limiting monolayer extension into open space. Finally, when cell–cell coordination is reduced, cells along the sheet margin continue to generate directed motility, but cannot propagate this directional information backward, ultimately resulting in a loss of sheet integrity (Fig. 7A). Since many sheet migration processes rely on other growth factors than FGF, it will be interesting to learn whether a similar or the same control modules are also utilized for other growth factor stimuli.
According to our model, the combination of basal random migration and dynamic cell–cell adhesion are likely sufficient to close small lesions in monolayers in the absence of growth factor. In contrast, the large-scale movements necessary during development, angiogenesis or wound healing require directed motility for efficient migration and, indeed, are often characterized by an increase in growth factor secretion (Werner et al. 1992; Ortega et al. 1998; Carmeliet 2003; Nagel et al. 2004; Ghabrial and Krasnow 2006). Lastly, this pioneer and follower model may also provide a mechanistic explanation for how unchecked growth factor signaling in neoplastic cell monolayers leads to metastatic behavior by inducing directed motility.
Conclusion
Our study argues for a modular model of growth factor-triggered sheet migration based on “pioneers and followers.” Experimental evidence for this model was derived from our ability to group 100 identified sheet migration regulators into four independent functional modules controlling proliferation, cell motility, directed migration and cell–cell coordination. We found that growth factor signals feed primarily into the directed migration module with little effect on cell motility or cell–cell coordination. This argues that growth factor is important primarily at boundary positions, where it elicits directed migration, but is less relevant for random, diffusive migration inside the sheet. Our mixed cell experiments, where cells in the monolayer varied in their capacity to activate specific control modules, demonstrate that boundary cells become pioneers in the presence of growth factor. Additionally, cells near these pioneers follow through growth factor-independent, coordinated, random migration. Thus, endothelial sheet migration is a modular process that couples growth factor-triggered formation of pioneer cells at the sheet boundary with growth factor-independent follower behavior, the latter requiring the activity of cell motility and cell–cell coordination modules within the monolayer.
Materials and methods
Diced siRNA library
Genes from the NCBI RefSeq database were selected according to the presence of known signaling domains such as PH, SAM, C1, C2, EF, kinase, and phosphatase. Pools of siRNA were generated with ∼500-bp PCR products that were transcribed, diced, and purified in vitro according to Liou et al. (2005).
Antibodies and reagents
Antibodies were purchased from BD Transduction Laboratories (VE-cadherin, Paxillin), Cell Signaling Technologies (p-Myo), and Sigma (Tubulin-Cy3). Diced pools of siRNA targeting GL3 luciferase was used as a control. Chemically synthesized siRNAs targeting FGFR1, FRS2, GAB1, and PTEN were purchased from Dharmacon.
Cell culture and transfection
HUVEC were cultured in EGM Bullet Kit (Clonetics) and plated at 10,000 cells per well onto tissue culture treated plates (COSTAR) coated with 300 μg/mL Collagen I (PureCol) for 1 h at 37°C. Cells were plated 16 h before transfecting 40 nM siRNA with Lipofectin (Invitrogen) according to manufacturer’s protocol. Assays were performed 48–72 h after siRNA transfection. Serum starved cells were placed in Endothelial SFM (Gibco) supplemented with 0.1% BSA (Sigma) or 0.1% BSA with 2 ng/mL bFGF (Invitrogen) at least 6 h prior to experimentation.
Screening and bioinformatics
Cells were stained with 10 μg/mL wheat germ agglutinin conjugated to AlexaFluor594 (Invitrogen) for 10 min before scraping. Cells were washed three times with PBS before addition of SFM + FGF. After 15 h, cell monolayers were fixed with 4% formaldehyde and stained with fluorescein-phalloidin (Invitrogen). All images were taken with a 4× objective on an automated fluorescent microscope (ImageXpress 5000A, Molecular Devices). Cell-free areas were determined by thresholding for dark objects using IxConsole software. Sheet migration rates from the primary screen represent the average of duplicate wells normalized according to the median and standard deviation of each plate. For hit validation and secondary screening, all plates were normalized to the median and standard deviation of control wells containing siRNA pools against GL3 (firefly luciferase). Once confirmed, subsequent knockdowns were performed with the most potent siRNA pool (first versus second coding sequence). Principal components analysis was performed with normalized secondary assay measurements with outliers restricted within three standard deviations. For decision tree hierarchical clustering, best fit correlation lines were approximated based on highest point density, with outliers defined as points further than two standard deviation units from the correlation line.
Secondary assays
For most secondary assays, cells were transfected at confluence and replated at 25% of the original density before staining with phalloidin, Hoescht, and one of several antibodies against various cell markers including microtubules, p-myosin, and paxillin. For VE-cadherin, cells were stained within a confluent monolayer. Additionally, an adhesion assay was performed in which cells were allowed to attach for 1 or 12 h before fixation and actin staining to determine spreading rates. Cell number and nuclear areas were determined using Hoescht staining. For all intensity measurements, images were first background subtracted. Each cell was identified using the nuclear stain, and the cell boundaries were determined with the actin staining. The average intensity within the third channel was then calculated for each cell using this actin mask (analysis performed in IXConsole).
Cell tracking and analysis
HUVEC were stained with 500 ng/mL Hoescht 33342 (Invitrogen) for 1 h at 37°C. Cells were imaged every 20 min in 37°C, 5% CO2 chamber for at least 4 h. Cells were tracked using a MatLab particle tracking algorithm originally developed by John Crocker and David Grier but adapted for MatLab by Daniel Blair and Eric Dufresne. In brief, the routine identifies bright objects and links them in consecutive frames based on minimum Euclidean distance (see http://physics.georgetown.edu/matlab/tutorial.html for more details). Individual cell velocities were calculated as the average displacement over all frames, and velocity for a particular treatment was the average of all cell velocities within the movie. Directional correlation was determined using a pair-wise comparison of cell trajectories throughout an entire time course. Only cells that moved at least 7.5 μm were included in the analysis, which calculates the average angular displacement between cells as a function of distance between cells. For screening purposes, the average angular displacement for cells separated by 75–150 μm were used as a metric for proximal correlation.
Acknowledgments
We thank Mark Hammer for discussions and sharing his directed motility analysis and colored tracking scripts, Onn Brandman for discussions and his protein–protein interaction script for the bioinformatics analysis, and Annette Salmeen for providing scientific feedback and her development of an automated primer design program. We appreciate the consultation from Gabriel Fenteany concerning the high-throughput scratch tool. We also extend our gratitude to James Nelson and Julie Theriot for their careful reading of the manuscript. This work was supported by National Institutes of Health grant R01-GM063702 and the National Science Foundation Predoctoral Fellowship Program.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1725808.
References
- Adams C.L., Nelson W.J., Smith S.J. Quantitative analysis of cadherin-catenin-actin reorganization during development of cell–cell adhesion. J. Cell Biol. 1996;135:1899–1911. doi: 10.1083/jcb.135.6.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikfalvi A., Klein S., Pintucci G., Rifkin D.B. Biological roles of fibroblast growth factor-2. Endocr. Rev. 1997;18:26–45. doi: 10.1210/edrv.18.1.0292. [DOI] [PubMed] [Google Scholar]
- Bindschadler M., McGrath J.L. Sheet migration by wounded monolayers as an emergent property of single-cell dynamics. J. Cell Sci. 2007;120:876–884. doi: 10.1242/jcs.03395. [DOI] [PubMed] [Google Scholar]
- Brandman O., Liou J., Park W.S., Meyer T. STIM2 is a feedback regulator that stabilizes basal cytosolic and endoplasmic reticulum Ca2+ levels. Cell. 2007;131:1327–1339. doi: 10.1016/j.cell.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P. Angiogenesis in health and disease. Nat. Med. 2003;9:653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- Chaffer C.L., Thompson E.W., Williams E.D. Mesenchymal to epithelial transition in development and disease. Cells Tissues Organs. 2007;185:7–19. doi: 10.1159/000101298. [DOI] [PubMed] [Google Scholar]
- Ciruna B., Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev. Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- Collins C.S., Hong J., Sapinoso L., Zhou Y., Liu Z., Micklash K., Schultz P.G., Hampton G.M. A small interfering RNA screen for modulators of tumor cell motility identifies MAP4K4 as a promigratory kinase. Proc. Natl. Acad. Sci. 2006;103:3775–3780. doi: 10.1073/pnas.0600040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker J.C., Grier D.G. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 1996;179:298–310. [Google Scholar]
- D’Amore P.A., Smith S.R. Growth effects on cells of the vascular wall: A survey. Growth Factors. 1993;8:61–75. doi: 10.3109/08977199309029135. [DOI] [PubMed] [Google Scholar]
- Deng C.X., Wynshaw-Boris A., Shen M.M., Daugherty C., Ornitz D.M., Leder P. Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes & Dev. 1994;8:3045–3057. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- Drees F., Pokutta S., Yamada S., Nelson W.J., Weis W.I. α-Catenin is a molecular switch that binds E-cadherin–β-catenin and regulates actin-filament assembly. Cell. 2005;123:903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernig D.G., Gallagher J.T. Fibroblast growth factors and their receptors: An information network controlling tissue growth, morphogenesis and repair. Prog. Growth Factor Res. 1994;5:353–377. doi: 10.1016/0955-2235(94)00007-8. [DOI] [PubMed] [Google Scholar]
- Folkman J. Angiogenesis: An organizing principle for drug discovery? Nat. Rev. Drug Discov. 2007;6:273–286. doi: 10.1038/nrd2115. [DOI] [PubMed] [Google Scholar]
- Galvez T., Teruel M.N., Heo W.D., Jones J.T., Kim M.L., Liou J., Myers J.W., Meyer T. siRNA screen of the human signaling proteome identifies the PtdIns(3,4,5)P3–mTOR signaling pathway as a primary regulator of transferrin uptake. Genome Biol. 2007;8:R142. doi: 10.1186/gb-2007-8-7-r142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial A.S., Krasnow M.A. Social interactions among epithelial cells during tracheal branching morphogenesis. Nature. 2006;441:746–749. doi: 10.1038/nature04829. [DOI] [PubMed] [Google Scholar]
- Goshima G., Vale R.D. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol. Biol. Cell. 2005;16:3896–3907. doi: 10.1091/mbc.E05-02-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton S.L., Waterman-Storer C.M. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 2006;125:1361–1374. doi: 10.1016/j.cell.2006.05.029. [DOI] [PubMed] [Google Scholar]
- Heath J.P. Epithelial cell migration in the intestine. Cell Biol. Int. 1996;20:139–146. doi: 10.1006/cbir.1996.0018. [DOI] [PubMed] [Google Scholar]
- Hegerfeldt Y., Tusch M., Bröcker E.B., Friedl P. Collective cell movement in primary melanoma explants: Plasticity of cell–cell interaction, β1-integrin function, and migration strategies. Cancer Res. 2002;62:2125–2130. [PubMed] [Google Scholar]
- Kametani Y., Takeichi M. Basal-to-apical cadherin flow at cell junctions. Nat. Cell Biol. 2007;9:92–98. doi: 10.1038/ncb1520. [DOI] [PubMed] [Google Scholar]
- Konturek S.J., Brzozowski T., Majka J., Szlachcic A., Bielanski W., Stachura J., Otto W. Fibroblast growth factor in gastroprotection and ulcer healing: Interaction with sucralfate. Gut. 1993;34:881–887. doi: 10.1136/gut.34.7.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger D.A., Horwitz A.F. Cell migration: A physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- Liou J., Kim M.L., Heo W.D., Jones J.T., Myers J.W., Ferrell J.E., Meyer T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y., Ebisuya M., Honjoh S., Nishida E. ERK activation propagates in epithelial cell sheets and regulates their migration during wound healing. Curr. Biol. 2004;14:731–735. doi: 10.1016/j.cub.2004.03.060. [DOI] [PubMed] [Google Scholar]
- Meili R., Firtel R.A. Follow the leader. Dev. Cell. 2003;4:291–293. doi: 10.1016/s1534-5807(03)00064-9. [DOI] [PubMed] [Google Scholar]
- Montell D.J. Border-cell migration: The race is on. Nat. Rev. Mol. Cell Biol. 2003;4:13–24. doi: 10.1038/nrm1006. [DOI] [PubMed] [Google Scholar]
- Myers J.W., Chi J.-T., Gong D., Schaner M.E., Brown P.O., Ferrell J.E., Jr. Minimizing off-target effects by using diced siRNAs for RNA interference. J RNAi Gene Silencing. 2006;2:181–194. [PMC free article] [PubMed] [Google Scholar]
- Nagel M., Tahinci E., Symes K., Winklbauer R. Guidance of mesoderm cell migration in the Xenopus gastrula requires PDGF signaling. Development. 2004;131:2727–2736. doi: 10.1242/dev.01141. [DOI] [PubMed] [Google Scholar]
- Ogita H., Takai Y. Cross-talk among integrin, cadherin, and growth factor receptor: Roles of nectin and nectin-like molecule. Int. Rev. Cytol. 2008;265:1–54. doi: 10.1016/S0074-7696(07)65001-3. [DOI] [PubMed] [Google Scholar]
- Omelchenko T., Vasiliev J.M., Gelfand I.M., Feder H.H., Bonder E.M. Rho-dependent formation of epithelial ‘leader’ cells during wound healing. Proc. Natl. Acad. Sci. 2003;100:10788–10793. doi: 10.1073/pnas.1834401100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega S., Ittmann M., Tsang S.H., Ehrlich M., Basilico C. Neuronal defects and delayed wound healing in mice lacking fibroblast growth factor 2. Proc. Natl. Acad. Sci. 1998;95:5672–5677. doi: 10.1073/pnas.95.10.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons-Wingerter P., Elliott K.E., Clark J.I., Farr A.G. Fibroblast growth factor-2 selectively stimulates angiogenesis of small vessels in arterial tree. Arterioscler. Thromb. Vasc. Biol. 2000;20:1250–1256. doi: 10.1161/01.atv.20.5.1250. [DOI] [PubMed] [Google Scholar]
- Perlin J.R., Talbot W.S. Signals on the move: Chemokine receptors and organogenesis in zebrafish. Sci. STKE. 2007;2007:pe45. doi: 10.1126/stke.4002007pe45. [DOI] [PubMed] [Google Scholar]
- Pignatelli M. Integrins, cadherins, and catenins: Molecular cross-talk in cancer cells. J. Pathol. 1998;186:1–2. doi: 10.1002/(SICI)1096-9896(199809)186:1<1::AID-PATH135>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Poujade M., Grasland-Mongrain E., Hertzog A., Jouanneau J., Chavrier P., Ladoux B., Buquin A., Silberzan P. Collective migration of an epithelial monolayer in response to a model wound. Proc. Natl. Acad. Sci. 2007;104:15988–15993. doi: 10.1073/pnas.0705062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., Horwitz A.R. Cell migration: Integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Rohde L.A., Heisenberg C.P. Zebrafish gastrulation: Cell movements, signals, and mechanisms. Int. Rev. Cytol. 2007;261:159–192. doi: 10.1016/S0074-7696(07)61004-3. [DOI] [PubMed] [Google Scholar]
- Sawin K.E., LeGuellec K., Philippe M., Mitchison T.J. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- Sharp D.J., McDonald K.L., Brown H.M., Matthies H.J., Walczak C., Vale R.D., Mitchison T.J., Scholey J.M. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J. Cell Biol. 1999;144:125–138. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmers M.B., Pryor A.W., Blackman B.R. Arterial shear stress regulates endothelial cell-directed migration, polarity, and morphology in confluent monolayers. Am. J. Physiol. Heart Circ. Physiol. 2007;293:H1937–H1946. doi: 10.1152/ajpheart.00534.2007. [DOI] [PubMed] [Google Scholar]
- Slavin J. Fibroblast growth factors: At the heart of angiogenesis. Cell Biol. Int. 1995;19:431–444. doi: 10.1006/cbir.1995.1087. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V., Fuchs E. Actin dynamics and cell–cell adhesion in epithelia. Curr. Opin. Cell Biol. 2001;13:76–84. doi: 10.1016/s0955-0674(00)00177-0. [DOI] [PubMed] [Google Scholar]
- Werner S., Peters K.G., Longaker M.T., Fuller-Pace F., Banda M.J., Williams L.T. Large induction of keratinocyte growth factor expression in the dermis during wound healing. Proc. Natl. Acad. Sci. 1992;89:6896–6900. doi: 10.1073/pnas.89.15.6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T.P., Harpal K., Henkemeyer M., Rossant J. fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes & Dev. 1994;8:3032–3044. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- Yang X., Dormann D., Münsterberg A.E., Weijer C.J. Cell movement patterns during gastrulation in the chick are controlled by positive and negative chemotaxis mediated by FGF4 and FGF8. Dev. Cell. 2002;3:425–437. doi: 10.1016/s1534-5807(02)00256-3. [DOI] [PubMed] [Google Scholar]
- Zhao W.M., Fang G. MgcRacGAP controls the assembly of the contractile ring and the initiation of cytokinesis. Proc. Natl. Acad. Sci. 2005;102:13158–13163. doi: 10.1073/pnas.0504145102. [DOI] [PMC free article] [PubMed] [Google Scholar]