Abstract
Human cytomegalovirus (HCMV) has been described to be associated with several human malignancies, though the frequency of detection remains controversial. It is unclear whether HCMV plays an active role in malignant tumor progression or becomes reactivated under pathologic conditions that result in chronic inflammation or immunosuppression. In this study, we report on the investigation of detecting HCMV in the tumors and peripheral blood of patients with newly diagnosed glioblastoma multiforme (GBM). Using immunohistochemistry, in situ hybridization, and polymerase chain reaction amplification of viral DNA, the detection of HCMV was investigated in tumor and blood specimens from patients with GBM as well as in the peripheral blood of normal volunteers and patients undergoing craniotomy for diagnoses other than GBM. We found that a high percentage (>90%) of GBM tumors, not surrounding normal brain, are associated with HCMV nucleic acids and proteins. Furthermore, a significant proportion of patients (80%) with newly diagnosed GBM have detectable HCMV DNA in their peripheral blood, while seropositive normal donors and other surgical patients did not exhibit detectable virus, suggesting either a systemic reactivation of HCMV within patients with GBM or shedding of viral DNA from infected tumor cells into the periphery. These results confirm the association of HCMV with malignant gliomas and demonstrate that subclinical HCMV viremia (presence of viral DNA in blood without clinical symptoms of infection) is a previously unrecognized disease spectrum in patients with GBM.
Keywords: human cytomegalovirus, glioblastoma, glioma, viruses
Human cytomegalovirus (HCMV) is a β-herpesvirus endemic in the human population and does not usually cause clinical disease except in immunocompromised hosts.1,2 Human herpesviruses have been implicated in a number of human malignancies including lymphoma, nasopharyngeal cancer, cervical cancer, and Kaposi’s sarcoma.3,4 Recently, HCMV antigen expression and detection of intact virus has been reported to occur in a large proportion of malignant tumors including colorectal carcinoma, prostate cancer, skin cancer, and malignant astrocytomas.5–8 The detection of HCMV in tumors, though, has been controversial, with conflicting reports in the literature regarding the detection of this virus in association with malignancies.9–14 It is not known at this time whether HCMV plays any role in the pathogenesis of malignant brain tumors and other cancers, or whether tumor growth simply provides an environment supportive of local reactivation and propagation of the virus. The presence of HCMV in tumors, whether causal or secondary to oncogenesis, may be important because of the known potential for HCMV to modulate the growth characteristics, invasiveness, and immunologic recognition of infected cells.15–20 Because of the important implications of an association between HCMV and brain tumors, we chose to investigate the detection of HCMV proteins and nucleic acids in the peripheral blood and tumors of patients with glioblastoma multiforme (GBM).
Materials and Methods
Immunohistochemistry
Human GBM, oligodendroglioma, meningioma, ependymoma, and normal brain surgical specimens were obtained in paraffin blocks (with Institutional Review Board [IRB] approval). Tumor specimens were requested based on diagnosis only from the Preston Robert Tisch Brain Tumor Center at Duke Tissue Bank. A total of 45 GBM cases confirmed by our neuropathologist (R.E.M.) were selected (36 primary GBM and 9 recurrent GBM). The group consisted of 26 males and 19 females, with a median age of 51 years.
Specimens were sectioned (6 μm) and were blocked and for endogenous peroxidase (3% H2O2, for 12 min) incubated with Fc receptor blocker (10 min at 20°C; Innovex Biosciences, Richmond, CA, USA) before the addition of a monoclonal antibody (mAb). Immunohistochemistry (IHC) was performed using three-stage horseradish peroxidase detection systems (BioGenex, San Ramon, CA, USA; Dako, Carpinteria, CA, USA; and Innovex Biosciences) with the following mAbs: anti–IE1-72 (1:25; BioGenex), anti-pp65 (1:30; Novocastra, Newcastle upon Tyne, UK), and antismooth muscle actin (1:15; BioGenex). Antibody parameters (e.g., postfixation, retrieval, and incubation time) were established for each mAb using DAB (Innovex Biosciences) as chromogen. Primary glioma cultures established for 14 to 21 days from freshly resected GBM specimens were fixed and permeabilized using cold methanol, followed by postfixation for 10 min with 10% neutral buffered formalin. Blocking of nonspecific binding was conducted using biotin block and avidin block (BioGenex) and FC receptor blockade (Innovex). Incubation with primary antibodies using isotype controls (mouse IgG1, mouse IgG2a; Invitrogen, Carlsbad, CA, USA), CD45 antibody (BD Biosciences, San Jose, CA, USA), pp28 antibody (Virusys, Sykesvile, MD, USA), glycoprotein B (gB; Virusys), and HIV p17 (Virogen, Watertown, MA, USA) was conducted for 2 h or overnight at 4°C (1 μg/ml antibody concentration) and detection conducted using BioGenex three-stage horseradish detection system.
In Situ Hybridization
For detection of HCMV nucleic acids, a biotinylated whole genomic probe specific for HCMV DNA and biotinylated positive control probe (specific for alu DNA) and negative control probe (insect genomic DNA) were obtained (BioGenex). In addition, a cocktail of six fluorescein isothiocyanate (FITC)–conjugated 40-mer probes spanning coding regions within the HCMV IE1 gene and negative (nonsense) and positive control (oligo dT) probes were obtained from GeneDetect.com. Enzyme digestion and nucleic acid denaturation of paraffin sections were performed using a Misha thermocycler (Shandon Lipshaw, Pittsburgh, PA, USA), and slides were hybridized overnight at 37°C in a humidified chamber. Probe was detected using a supersensitive detection system (chromogen NBT; BioGenex) employing anti-biotin or anti-FITC detection antibodies. Competitive in situ hybridization (ISH) experiments were done using a 50-fold molar excess of cold specific (unlabeled IE1 probe) or nonspecific probe (unlabeled nonsense probe) included during the hybridization steps (GeneDetect.com).
Polymerase Chain Reaction
Thirty-four tumor specimens from patients with newly diagnosed GBM (median age 53) were obtained in accordance with the IRB and after patient consent. Twenty patients also had peripheral blood drawn intraoperatively at the time of resection (median age 52.5). Peripheral blood from 17 normal volunteers was drawn for evaluation of CMV DNA in the blood. These volunteers consisted of six patients (median age 42) undergoing surgery for a nonmalignant condition (trigeminal neuralgia) and 11 health care workers at Duke (median age 46). Volunteers were obtained through advertisement within the Duke Medical Center for volunteers over the age of 40, and the informed consent of patients over 40 was evaluated in the neurosurgery clinic by health care staff. The median and average ages of the GBM patient population evaluated in this study for detection of CMV DNA in the peripheral blood (n = 20; 52.5 years and 54 ± 11.49 years, respectively) and normal volunteers (n = 17; 42 years and 45.8 ± 10.51 years, respectively) were not significantly different (p = 2.34). Freshly resected GBM specimens and peripheral blood were collected in accordance with the IRB, and DNA was extracted from 10 mg of tissue or 20 μl of whole blood using GenScript TissueDirect Multiplex PCR (polymerase chain reaction) System (GenScript) according to the manufacturer’s instructions. To avoid contamination, no positive controls were used for PCR at the same time as clinical samples, and great care was taken to avoid cross-contamination by extracting all DNA samples in a separate room from where PCR reactions were carried out. DNA was amplified by real-time PCR using primers specific for HCMV glycoprotein B (UL55) gene21 and iQ SYBR Green SuperMix (Bio-Rad). Amplified DNA products from tumors were visualized on agarose gels with ethidium bromide, bands were cut out, and DNA was extracted and analyzed by automated sequencing (ABI 3730 and 3100 PRISM DNA Sequencers, Applied Biosystems, Foster City, CA, USA) at the Duke University DNA sequencing facility. Confirmation of HCMV sequences was performed using a National Center for Biotechnology Information BLAST search. DNA extractions, PCR amplifications, and DNA sequencing were repeated on several tumors in a blinded fashion to confirm these findings. Amplification of HCMV DNA from peripheral blood was carried out similarly using 10–20 μl of whole blood as starting material for DNA extraction.
Statistical Analysis
Detection of HCMV in the peripheral blood and tissue specimens of patients diagnosed with GBM was compared with that of normal volunteers, patients undergoing craniotomy for conditions other than GBM, and patients with nonmalignant or metastatic brain tumors using the Fisher exact test.
Results
Immunohistochemical Detection of HCMV Proteins
To determine whether HCMV proteins were expressed in malignant gliomas (MGs), we examined paraffin sections from 45 GBM specimens selected from our brain tumor bank by IHC. Detection was conducted using a mAb specific for the HCMV-encoded antigen, IE1-72. IE1-72 immunoreactivity was detected in 42 out of 45 (93%) specimens examined by IHC. Strong nuclear and cytoplasmic staining was detected in tumor cells and occasionally endothelial cells as well (Fig. 1). However, infiltrating lymphocytes and surrounding normal brain areas were devoid of immunoreactivity to the IE1-72 antibody. To further confirm specific detection of HCMV, we examined 33 of the 45 cases for reactivity to a mAb specific for the HCMV matrix protein, pp65. Thirty of the 33 cases (91%) were immunoreactive for pp65 in the tumor cells but not in areas of adjacent normal brain. pp65 reactivity was in general less ubiquitous than IE1-72 detection in tumor cells, but a majority of tumor cells in all specimens examined displayed immunoreactivity against the pp65 antibody (Fig. 1; Table 1).
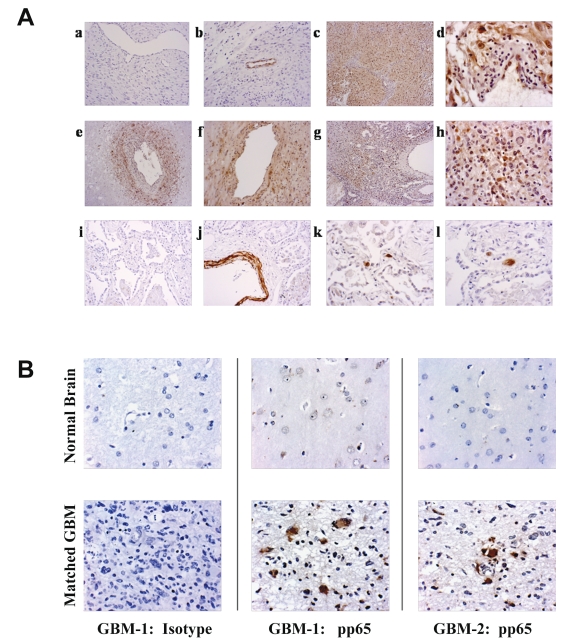
Fig. 1.
(A) Immunohistochemical detection of human cytomegalovirus (HCMV) proteins: (a) negative control (no primary antibody; objective lens ×10); (b) antismooth-muscle actin (mouse IgG2a mAb; ×10); (c) glioblastoma multiforme (GBM) specimen 1 stained with anti-HCMV IE1 (mouse IgG2a mAb, ×10); (d) higher magnification of anti-IE1 staining shows positive tumor cells and endothelial cells but negative lymphocytes and vascular intima (×20); (e) GBM specimen 2 stained with anti-HCMV IE1 showing staining of perivascular tumor cells but lack of detection in necrotic areas (×10); (f) perivascular tumor cells stained with anti-IE1 mAb (×20); (g and h) GBM specimen 3 stained with anti-HCMV pp65 mAb showing nuclear and perinuclear staining of tumor cells scattered throughout the GBM specimen (×10 and ×20, respectively); (i and j) CMV-infected lung stained with antismooth-muscle actin mAb (×10); (k) CMV-infected lung stained with anti-HCMV IE1 (×20); (l) CMV-infected lung stained with anti-HCMV pp65 mAb (×20). (B) HCMV detection in matched GBM and normal brain. Representative histochemical sections from two GBM specimens containing areas of normal brain and tumor were stained for detection using isotype control antibodies (patient 1, left column), or anti-HCMV pp65 (patient 1 tumor, middle column; patient 2, right column). Focal areas of reactivity against the HCMV pp65 antibody was observed throughout the tumor-involved areas, but normal brain was devoid of immunoreactivity to the HCMV-specific antibodies (IE1 staining showed identical findings with more ubiquitous detection of IE1 in the tumor, not shown). All photographs taken at ×40 objective magnification.
Table 1.
Summary of HCMV detection in GBM specimens
| HCMV | GBM Tissue Specimen | Primary GBM Cultures |
|---|---|---|
| IE1 IHC | 42/45 (93%)a | 4/4 (100%) |
| pp65 IHC | 30/33 (91%)a | 12/12 (100%) |
| HCMV DNA ISH | 16/16 (selected cases ) | not tested |
| gB PCR | 21/34 (61.7%)b | 13/17 (70.6%) |
| IE1 PCR | 8/34 (24%) b | 9/17 (53%) |
Abbreviations: HCMV, human cytomegalovirus; GBM, glioblastoma multiforme; IHC, immunohistochemistry; ISH, in situ hybridization; gB, glycoprotein B; PCR, polymerase chain reaction.
Other tumors tested by IHC were negative for HCMV: oligodendroglioma (n = 5; one case exhibited focal detection of HCMV IE1 in endothelial cells but no reactivity within tumor parenchyma); meningioma (n = 5); ependymoma (n = 5).
PCR products were isolated from 21 gB PCR reactions and 6 IE1 PCR and confirmed by DNA sequencing to be specific for HCMV.
Tumor cells have been described as having a higher propensity to display false-positive immunoreactivity, either due to nonspecific binding to mAbs or higher levels of endogenous peroxidases that react with detection substrate. To rule out the possibility of nonspecific detection in tumor cells, we performed IHC on tumor sections with isotype- and concentration-matched control mAbs. Isotype-matched, control antibodies used at identical concentration to the HCMV-specific mAbs showed no immunoreactivity within tumor cells, and an isotype-matched mAb to smooth muscle actin demonstrated reactivity to blood vessels within tumor and normal brain sections (Fig. 1A) but no reactivity with tumor cells. Examination results of meningiomas (n = 5), ependymomas (n = 5), and oligodendrogliomas (n = 5) were negative for detection of IE1 and pp65, except for focal endothelial staining observed in a single case of oligodendroglioma with the IE1 monoclonal antibody (Table 1).
Detection of HCMV Nucleic Acids Using ISH
To demonstrate the localization of HCMV nucleic acids in the same areas as detection by IHC within GBM specimens, we performed ISH using a cocktail of six overlapping 40-mer HCMV IE1 antisense DNA probes as well as using a biotinylated whole genomic HCMV DNA probe on selected GBM cases shown to be positive by IHC. Specific hybridization to tumor cells but not to blood vessels or normal brain was observed in all examined cases (n = 16), while a guanine/cytosine content–matched control cocktail probe consisting of six nonsense 40-base-pair oligonucleotide probes or negative control DNA exhibited no hybridization (Fig. 2; Table 1). A positive-control probe specific for polyadenylated mRNA and alu DNA sequences hybridized with all specimens (Fig. 2). To further confirm the specificity of this detection, cold competition experiments were performed using a 50-fold excess of unlabeled specific or nonspecific competitor DNA. Hybridization signal could be competed effectively with specific cold competitor, while nonspecific DNA exhibited no effect on hybridization of the IE1 probes (Fig. 2).
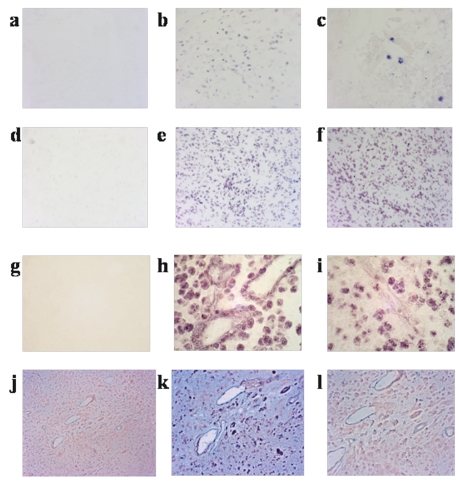
Fig. 2.
In situ hybridization. Detection of human cytomegalovirus (HCMV) nucleic acids using HCMV IE1 and whole genomic probe. First row: CMV-infected lung hybridized with (a) a nonsense DNA probe (negative control) and (b) poly-AAA probe (positive control); (c) HCMV IE1 probe demonstrates hybridization to large cytomegalic cells throughout the lung specimen. Second row: (d) GBM specimen hybridized with nonsense DNA probe and (e) poly-AAA antisense probe; (f) detection of HCMV IE1 in a GBM specimen shows hybridization of tumor cells throughout the specimen. Third row: (g) GBM specimen stained with nonsense whole genomic probe (negative control); (h) alu DNA probe (positive control) and (i) pan HCMV genomic DNA probe, demonstrating HCMV detection within tumor cells but not vasculature within the specimen. Fourth row: Competition experiment showing ability to block detection of IE1 hybridization using a 50-fold excess of unlabeled specific competitor DNA probe but not using an excess of the nonsense probe; (j) lack of detection with nonsense DNA probe; (k) HCMV IE1 antisense probe with 50-fold excess nonsense competitor probe; (l) HCMV IE1 antisense probe with 50-molar excess of IE1 competitor probe. Results show localization of HCMV DNA within GBM tumor cells and capacity to specifically compete hybridization with HCMV-specific competitor probe.
Immunohistochemical Detection of HCMV Proteins in Primary GBM Culture
To determine whether freshly resected glioma specimens from newly diagnosed patients retained virus after propagation in vitro, we established short-term cultures (7–14 days) from 17 GBM specimens cultured directly after removal from the patient. Immunohistochemical detection of HCMV proteins was confirmed in 16 out of 17 GFAP-positive, astrocytic tumor cell lines using mAbs to HCMV IE1, pp65 gB, and pp28 (Fig. 3; summarized in Table 1). Isotype and concentration-matched mAbs specific for hematopoietic cells (CD45), human immunodeficiency virus p17, and isotypic IgG controls showed no immunoreactivity against tumor cell lines established in vitro.
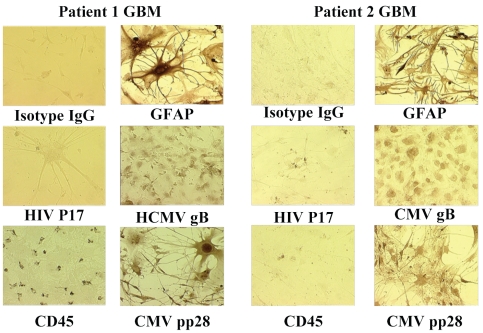
Fig. 3.
Detection of human cytomegalovirus (HCMV) in primary glioblastoma multiforme (GBM) cultures. Short-term GBM cultures from two patients were stained with a variety of monoclonal antibodies to detect HCMV proteins or astrocyte markers (GFAP). Cells were fixed, permeabilized, and stained with equimolar concentrations of isotype control antibodies (mouse IgG), anti-GFAP antibody, anti-HIV p17, anti-HCMV glycoprotein B (gB), anti-CD45, and anti-HCMV pp28. Photos demonstrate strong nuclear reactivity of astrocytic tumor cells to anti-HCMV gB and pp28 with scant cytoplasmic staining in two primary GBM cultures. Isotype controls and HIV p17 staining were negative in all samples examined. CD45 immunoreactivity was observed in scattered hematopoietic cells in one culture (patient 1 GBM, bottom left panel), but did not stain astrocytic tumor cells.
PCR Amplification of HCMV DNA
To further confirm the presence of HCMV in MGs, we amplified DNA extracted directly from freshly resected, newly diagnosed GBM specimens using real-time polymerase chain reaction (RT-PCR). All DNA extractions were performed in a separate tissue culture room, and PCR amplification was conducted in a dedicated molecular biology laboratory. Positive controls were validated once and then eliminated from all test PCR amplifications, while negative controls were included on each run to ensure that no DNA contamination occurred. To optimize detection of HCMV, we evaluated primers and PCR amplification conditions for six different HCMV genes (a total of 15 different primer sets) in their capacity to amplify limiting dilutions of an HCMV genomic control template. We found primers to glycoprotein B to be the most sensitive and consistent in detection of limiting quantity of HCMV DNA and used this set of primers to screen GBM specimens for viral DNA. We detected HCMV DNA in 21 out of 34 examined specimens (62%) by PCR. Amplification of the correct size product (121 base pairs) was observed, and these bands were isolated from agarose gels, DNA extracted, and analyzed by direct DNA sequencing. Sequencing confirmed the products as specific for HCMV glycoprotein B in all 21 cases. Variation in nucleotide sequences across the 141 base pair products (4%–31% variation in nucleotide usage) demonstrated unique viral isolates in patient specimens, indicating that viral DNA detection was not due to a contaminating source of HCMV. Primary GBM cultures demonstrated the presence of HCMV DNA in 13 out of 17 cultures examined (70%). HCMV DNA was not detected in template-negative samples or from metastatic breast or colon cancer (n = 3), meningiomas (n = 3), acoustic neuromas (n = 3), or epidermoid tumors (n = 3; Table 2).
Table 2.
HCMV gB PCR detection in matched blood and tumor samples
| Blood vs. Tumor Detection of gB | GBMs | Other Brain Tumorsa | Metastaticb Tumors | Normal Volunteers |
|---|---|---|---|---|
| Blood(+)/tumor(+) | 10/20 (50%) | 0/3 (0%) | 0/3 (0%) | NA |
| Blood(+)/tumor(−) | 6/20 (30%) | 0/3 (0%) | 0/3 (0%) | NA |
| Blood(−)/tumor(+) | 1/20 (5%) | 0/3 (0%) | 0/3 (0%) | NA |
| Blood(−)/tumor(−) | 3/20 (15%) | 3/3 (100%) | 3/3 (100%) | NA |
| Blood(+) overall | 16/20 (80%) | 0/3 (0%) | 0/3 (0%) | 0/17 (0%) |
| Blood(−) overall | 4/20 (20%) | 3/3 (100%) | 3/3 (100%) | 17/17 (100%) |
Abbreviations: HCMV, human cytomegalovirus; gB, glycoprotein B; PCR, polymerase chain reaction; GBM, glioblastoma multiforme.
Other brain tumors: acoustic neuroma (1), epidermoid (1), meningioma (1).
Metastatic tumors: breast cancer (2), colon cancer (1).
Detection of HCMV in Peripheral Blood
Detection of HCMV in GBM tumors could represent a local reactivation of virus or a systemic HCMV reactivation with specific localization of virus in astrocytomas in the brain. To determine if virus could be detected in the periphery, we analyzed the peripheral blood of patients with newly diagnosed GBMs and normal volunteers for detection of HCMV DNA using PCR. We were able to obtain matched GBM tissue and intraoperative blood samples from 20 patients undergoing primary tumor resection. We found that 16 out of 20 (80%) patients exhibited detectable viral DNA in their whole blood, while none of the 17 normal volunteers, including 11 seropositive donors, demonstrated any detectable viral DNA. The results indicate that HCMV is frequently present in patients with GBM compared with normal, age-matched volunteers (p < 0.001). Intraoperative blood samples taken from patients diagnosed with benign tumors or undergoing craniotomy for metastatic tumors also did not demonstrate detectable CMV DNA in their blood, ruling out transient viremia induced by surgical intervention (p < 0.01; Table 2).
In summary, the results of our studies confirm the association of HCMV infection with malignant astrocytomas initially reported by Cobbs et al.5 and demonstrate a global, subclinical state of HCMV reactivation in many patients with GBM. Further studies are warranted to determine whether HCMV plays a role in gliomagenesis and tumor progression, or is a secondary event resulting from global immunosuppression known to exist in patients with MGs.
Discussion
A role for HCMV in malignant disease initiation has long been proposed since the findings of the oncogenic potential of the HCMV viral particles or gene particles in in vitro cultured cells.9,22–25 Furthermore, the establishment of lifelong latency of HCMV within progenitor cells in the bone marrow, brain, and possibly other tissues seems consistent with a predisposing risk factor for viral transformation of immortalized and pluripotent cell types.26–30 However, a simpler and more plausible explanation for virus detection in cancer patients is a secondary reactivation of virus after cancer-related, and possibly treatment-related, immunosuppression. GBMs are known to exert a variety of local and systemic immunosuppressive effects in patients, all of which could contribute to the establishment of an environment permissive of HCMV reactivation.31 Among other immunosuppressive factors, we have found that newly diagnosed patients with GBM are often profoundly lymphopenic, with particular deficits in their CD4+ T-cell compartment that result in impaired cell-mediated immunity.32,33 Given the known role of the cellular immune system in maintaining viral latency, cell-mediated immunologic defects in patients with GBM and other cancers may cause viral reactivation and propagation. In addition to being susceptible to tumor-related immunosuppression, patients with GBM are often placed on corticosteroids at the time of diagnosis, which has a known capacity to elicit further immunosuppression that could lead to HCMV reactivation.34 Thus, our findings and those reported by Cobbs et al.5 could simply be attributed to a subclinical reactivation of virus secondary to tumor-related or treatment-related immunosuppression. However, viral reactivation secondary to treatment-related immunosuppression is an unlikely mechanism because newly diagnosed patients with GBM at our institution have usually undergone less than 2 weeks of corticosteroid therapy prior to resection, and the detection of virus in the blood and throughout the tumor specimens at the time of initial resection seems indicative of viral reactivation and a more prolonged course of viral replication.35 Studies in patients undergoing bone marrow transplantation and organ transplantation who are placed on much more rigorous immunosuppressive regimens demonstrate a time course of HCMV reactivation, typically on the order of several weeks to months posttherapy.36,37
A few lines of evidence, however, support a more closely linked association of HCMV with GBM. The frequency of detection of virus in tumor samples in our studies is higher than the expected frequency of latently infected individuals in the population (50%–70%), so one would expect that if HCMV were simply secondarily reactivated by immunosuppression in these patients that a frequency more closely linked to the general population would be found. The seropositive status of patients whose tumors were examined from our tumor bank is unknown, however, and much larger epidemiologic studies beyond the scope of this report would be needed to provide any meaningful investigation of this type of analysis.
As reported by Cobbs et al.,5 we have also detected HCMV proteins and nucleic acids in the tumors—but not surrounding normal brain—of patients with MGs. Preferential viral replication within astrocytomas may be explained by the relative permissiveness of astrocytes and neural progenitors to HCMV infection compared with other brain-cell types.38–40 Of interest, astrocytoma cell lines have been used for years to propagate HCMV in vitro because they are one of the few permissive cell lines that allow for culture of the virus.41,42 Another plausible explanation for preferential viral tropism in brain tumors is recent identification of the epidermal growth factor receptor (EGFR) as a cellular binding and incorporation site for the entry of HCMV into cells.43 GBMs almost uniformly demonstrate amplified EGFR expression, while normal brain is largely negative.44–46
We noted that IHC was more sensitive in our hands in the detection of CMV than PCR (Table 1). We attribute this difference to the fact that normal brain and necrotic tissue, which may be included in the gross tumor specimens provided during resection, are devoid of CMV; thus, sampling error may result in missing CMV-infected viable tumor when directly extracting DNA from small quantities of tissues for PCR. This sampling error is avoided during IHC evaluation since viable tumor tissue is selected by a trained neuropathologist prior to immunohistochemical evaluation. We have also observed that IE1 is generally more ubiquitously expressed in GBM tissue than pp65, and in a minority of tumor samples, focal reactivity could be observed (Fig. 1B). While evaluation of the demographics and prognosis of the few cases where a focal pattern was observed did not reveal any distinguishing characteristics, a more extensive evaluation of the levels of CMV in GBM tissue—based on quantitative PCR analysis, immunohistochemical evaluation of large areas of tumor, or intracellular FACS analysis of CMV proteins in dissociated tumor tissue—may reveal whether the levels of CMV or staining patterns have any prognostic or predictive value. Such analysis is the focus of future research.
Regardless of whether HCMV is an early or late event associated with gliomagenesis, the presence of the virus within tumor cells holds significance for several reasons: (1) HCMV is known to down-regulate the immunogenecity of infected cells through inhibition of antigen presentation, down-regulation of surface MHC expression, elaboration of TGF-β from infected cells (particularly astrocytes), and secretion of a viral interleukin 10 homologue (vIL-10).47–49 All of these factors may contribute to the immunologic evasion of infiltrative tumor cells and make MGs more difficult for the immune system to eradicate. (2) HCMV could modulate other properties that could contribute to a more malignant phenotype in tumor cells, including increasing angiogenesis, invasiveness, and cell proliferation, as well as decreasing susceptibility of infected tumor cells to cell death through blockade of apoptopic pathways.17 (3) The presence of viral antigens specifically in tumor cells lends the potential for targeting HCMV as a tumor-associated antigen in gliomas, lending the vast array of reagents and extensive experience in immunotherapeutic targeting of HCMV as tools to leverage against malignant brain tumors.50
Three other groups recently investigating the presence of HCMV in gliomas have failed to confirm the findings published by Cobbs et al.5 and reported by us in this manuscript.12–14 While the reasons for these discrepancies are unclear, one possibility is differences in the sensitivities of the assays employed by the different investigators’ laboratories. We have found, for instance, that detection of HCMV by IHC in brain tumors requires optimal antigen retrieval as well as blockade of nonspecific binding of isotype controls; IHC protocols using less optimized processes revealed negative detection in tumors. Nonoptimized staining protocols, however, were sufficient for detection of HCMV in cases of HCMV pneumonia used as positive controls in this study. While quantification of viral load was not examined in this study, these results, and the fact that GBM patients do not exhibit clinical signs of HCMV infection, suggest that very low levels of virus may propagate within these patients and require more sensitive detection methods than are necessary for detection in cases of symptomatic viral infection. Also, extensive comparison of primer sets and PCR detection methods revealed variability among the sensitivities of various primer sets and PCR conditions using known concentrations of viral standards. Our efforts at optimizing the recovery of low levels of viral DNA and PCR amplification of HCMV DNA have demonstrated that detection of HCMV levels present in patients with MG is not a trivial issue; therefore, controls having limiting quantities of virus should be used to ensure retention and detection of small numbers of viral copies per sample. Ultrasensitive detection techniques run the risk of detecting latent viral genomes persistent in a very small fraction of cells present in normal hosts. However, our inability to detect HCMV viral DNA in the whole blood of normal hosts or patients with nonmalignant brain tumors indicates that our detection methods are likely not sensitive enough to pick up latent virus and demonstrates a specific association of HCMV with GBM tumors.
We found that direct isolation of DNA from tumor samples and blood was consistently more reliable in detection than using DNA purification prior to PCR amplification, where detection of low-copy viral DNA required considerably larger sample size in order to purify sufficient viral DNA (Mitchell et al., unpublished data). Because we have confirmed the amplification of HCMV by DNA sequencing in more than 21 GBM samples, we are confident that our detection is due neither to artifact nor contamination by laboratory viral DNA. Consistent negative results obtained in samples from normal patients further support this conclusion. Finally, it is possible, based on the demographic profile of patients examined within various laboratories reporting on HCMV detection, that wide variation in HCMV association may exist, although we do not think this is likely to explain the inability of some laboratories to detect HCMV in association with GBM.
These findings warrant further study to determine whether the presence of viral DNA in the blood or tumors of patients, or quantification of viral load among HCMV-positive patients with GBM, holds any prognostic or predictive significance. Studies are underway in our laboratory to determine the clinical significance of HCMV detection in patients with GBM, along with efforts aimed at targeting these tumors through use of HCMV-targeted immunotherapy.
Acknowledgments
We thank Deb Smith, R.N., for assistance with specimen collection and Michael J. Ellis (University of Manitoba Medical School) for technical assistance with immunohistochemistry. This work was supported by grants from the NIH/NINDS SPORE in Brain Cancer 1P50 CA108786 01 (J.H.S.), The Brain Tumor Society Billy Grey Chair of Research Award (D.A.M.), and Accelerate Brain Cancer Cure Foundation Young Investigator’s Award (D.A.M.). None of the contributing authors reports any conflicts of interest related to this work.
References
- 1.Michelson-Fiske S. Human cytomegalovirus: a review of developments between 1970 and 1976. Part II. Experimental developments. Biomedicine. 1977;26:86–97. [PubMed] [Google Scholar]
- 2.Sissons JG, Carmichael AJ. Clinical aspects and management of cytomegalovirus infection. J Infect. 2002;44:78–83. doi: 10.1053/jinf.2001.0949. [DOI] [PubMed] [Google Scholar]
- 3.Rafferty KA., Jr Herpes viruses and cancer. Sci Am. 1973;229:26–33. doi: 10.1038/scientificamerican1073-26. [DOI] [PubMed] [Google Scholar]
- 4.Kadow JF, Regueiro-Ren A, Weinheimer SP. The role of viruses in human cancer development and antiviral approaches for intervention. Curr Opin Investig Drugs. 2002;3:1574–1579. [PubMed] [Google Scholar]
- 5.Cobbs CS, Harkins L, Samanta M, et al. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 2002;62:3347–3350. [PubMed] [Google Scholar]
- 6.Harkins L, Volk AL, Samanta I, et al. Specific localisation of human cytomegalovirus nucleic acids and proteins in human colorectal cancer. Lancet. 2002;360:1557–1563. doi: 10.1016/S0140-6736(02)11524-8. [DOI] [PubMed] [Google Scholar]
- 7.Samanta M, Harkins L, Klem K, Britt WJ, Cobbs CS. High prevalence of human cytomegalovirus in prostatic intraepithelial neoplasia and prostatic carcinoma. J Urol. 2003;170:998–1002. doi: 10.1097/01.ju.0000080263.46164.97. [DOI] [PubMed] [Google Scholar]
- 8.Zafiropoulos A, Tsentelierou E, Billirik, Spandidos DA. Human herpes viruses in non-melanoma skin cancers. Cancer Lett. 2003;198:77–81. doi: 10.1016/s0304-3835(03)00269-6. [DOI] [PubMed] [Google Scholar]
- 9.Roche JK, Cheung KS, Boldogh I, Huang ES, Lang DJ. Cytomegalovirus: detection in human colonic and circulating mononuclear cells in association with gastrointestinal disease. Int J Cancer. 1981;27:659–667. doi: 10.1002/ijc.2910270513. [DOI] [PubMed] [Google Scholar]
- 10.Hart H, Neill WA, Norval M. Lack of association of cytomegalovirus with adenocarcinoma of the colon. Gut. 1982;23:21–30. doi: 10.1136/gut.23.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grail A, Norval M. Elution of cytomegalovirus antibodies from adenocarcinoma of the colon. Gut. 1985;26:1053–1058. doi: 10.1136/gut.26.10.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau SK, Chen YY, Chen WG, et al. Lack of association of cytomegalovirus with human brain tumors. Mod Pathol. 2005;18:838–843. doi: 10.1038/modpathol.3800352. [DOI] [PubMed] [Google Scholar]
- 13.Sabatier J, Uro-Coste E, Pommepuy I, et al. Detection of human cytomegalovirus genome and gene products in central nervous system tumours. Br J Cancer. 2005;92:747–750. doi: 10.1038/sj.bjc.6602339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poltermann S, Schlehofer B, Steindorf K, et al. Lack of association of herpesviruses with brain tumors. J Neurovirol. 2006;12:90–99. doi: 10.1080/13550280600654573. [DOI] [PubMed] [Google Scholar]
- 15.Cinatl J, Jr, Kotchetkov R, Scholz M, et al. Human cytomegalovirus infection decreases expression of thrombospondin-1 independent of the tumor suppressor protein p53. Am J Pathol. 1999;155:285–292. doi: 10.1016/S0002-9440(10)65122-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basta S, Bennink JR. A survival game of hide and seek: cytomegaloviruses and MHC class I antigen presentation pathways. Viral Immunol. 2003;16:231–242. doi: 10.1089/088282403322396064. [DOI] [PubMed] [Google Scholar]
- 17.Cinatl J, Jr, Vogel JU, Kotchetkov R, et al. Oncomodulatory signals by regulatory proteins encoded by human cytomegalovirus: a novel role for viral infection in tumor progression. FEMS Microbiol Rev. 2004;28:59–77. doi: 10.1016/j.femsre.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Hoever G, Vogel JU, Lukashenko P, et al. Impact of persistent cytomegalovirus infection on human neuroblastoma cell gene expression. Biochem Biophys Res Commun. 2005;326:395–401. doi: 10.1016/j.bbrc.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 19.Bego MG, St Jeor S. Human cytomegalovirus infection of cells of hematopoietic origin: HCMV-induced immunosuppression, immune evasion, and latency. Exp Hematol. 2006;34:555–570. doi: 10.1016/j.exphem.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 20.Soderberg-Naucler C. Does cytomegalovirus play a causative role in the development of various inflammatory diseases and cancer? J Intern Med. 2006;259:219–246. doi: 10.1111/j.1365-2796.2006.01618.x. [DOI] [PubMed] [Google Scholar]
- 21.Kuhn JE, Wendland T, Eggers HJ, et al. Quantitation of human cytomegalovirus genomes in the brain of AIDS patients. J Med Virol. 1995;47:70–82. doi: 10.1002/jmv.1890470114. [DOI] [PubMed] [Google Scholar]
- 22.Rapp F, Li JL. Demonstration of the oncogenic potential of herpes simplex viruses and human cytomegalovirus. Cold Spring Harb Symp Quant Biol. 1975;39:747–763. doi: 10.1101/sqb.1974.039.01.087. [DOI] [PubMed] [Google Scholar]
- 23.Geder KM, Lausch R, O’Neill F, Rapp F. Oncogenic transformation of human embryo lung cells by human cytomegalovirus. Science. 1976;192:1134–1137. doi: 10.1126/science.179143. [DOI] [PubMed] [Google Scholar]
- 24.Geder L, Rapp F. Evidence for nuclear antigens in cytomegalovirus-transformed human cells. Nature. 1977;265:184–186. doi: 10.1038/265184a0. [DOI] [PubMed] [Google Scholar]
- 25.Buhren J, Christoph AH, Buslei R, et al. Expression of the neurotrophin receptor p75NTR in medulloblastomas is correlated with distinct histological and clinical features: evidence for a medulloblastoma subtype derived from the external granule cell layer. J Neuropathol Exp Neurol. 2000;59:229–240. doi: 10.1093/jnen/59.3.229. [DOI] [PubMed] [Google Scholar]
- 26.Hahn G, Jores R, Mocarski ES. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc Natl Acad Sci U S A. 1998;95:3937–3942. doi: 10.1073/pnas.95.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slobedman B, Mocarski ES. Quantitative analysis of latent human cytomegalovirus. J Virol. 1999;73:4806–4812. doi: 10.1128/jvi.73.6.4806-4812.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagasse N, Dhooge I, Govaert P. Congenital CMV-infection and hearing loss. Acta Otorhinolaryngol Belg. 2000;54:431–436. [PubMed] [Google Scholar]
- 29.Cohrs RJ, Gilden DH. Human herpesvirus latency. Brain Pathol. 2001;11:465–474. doi: 10.1111/j.1750-3639.2001.tb00415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reeves MB, Coleman H, Chadderton J, et al. Vascular endothelial and smooth muscle cells are unlikely to be major sites of latency of human cytomegalovirus in vivo. J Gen Virol. 2004;85:3337–3341. doi: 10.1099/vir.0.80285-0. [DOI] [PubMed] [Google Scholar]
- 31.Gomez GG, Kruse CA. Mechanisms of malignant glioma immune resistance and sources of immunosuppression. Gene Ther Mol Biol. 2006;10(a):133–146. [PMC free article] [PubMed] [Google Scholar]
- 32.Dix AR, Brooks WH, Roszman TL, Morford LA. Immune defects observed in patients with primary malignant brain tumors. J Neuroimmunol. 1999;100:216–232. doi: 10.1016/s0165-5728(99)00203-9. [DOI] [PubMed] [Google Scholar]
- 33.Fecci PE, Mitchell DA, Whitesides JF, et al. Increased regulatory T-cell fraction amidst a diminished CD4 compartment explains cellular immune defects in patients with malignant glioma. Cancer Res. 2006;66:3294–302. doi: 10.1158/0008-5472.CAN-05-3773. [DOI] [PubMed] [Google Scholar]
- 34.Dalessandro L, Bottaro E. Reactivation of CMV retinitis after treatment with subtenon corticosteroids for immune recovery uveitis in a patient with AIDS. Scand J Infect Dis. 2002;34:780–782. doi: 10.1080/00365540260348644. [DOI] [PubMed] [Google Scholar]
- 35.Ashby LS, Ryken TC. Management of malignant glioma: steady progress with multimodal approaches. Neurosurg Focus. 2006;20:E3. [PubMed] [Google Scholar]
- 36.Preiser W, Bräuninger S, Schwerdtfeger R, et al. Evaluation of diagnostic methods for the detection of cytomegalovirus in recipients of allogeneic stem cell transplants. J Clin Virol. 2001;20:59–70. doi: 10.1016/s1386-6532(00)00156-6. [DOI] [PubMed] [Google Scholar]
- 37.Zekri AR, Mohamed WS, Samra MA, et al. Risk factors for cytomegalovirus, hepatitis B, and C virus reactivation after bone marrow transplantation. Transpl Immunol. 2004;13:305–311. doi: 10.1016/j.trim.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 38.van Den Pol AN, Mocarski E, Saederup N, Vieira J, Meier TJ. Cytomegalovirus cell tropism, replication, and gene transfer in brain. J Neurosci. 1999;19:10948–10965. doi: 10.1523/JNEUROSCI.19-24-10948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Den Pol AN, Vieira J, Spencer DD, Santarelli JG. Mouse cytomegalovirus in developing brain tissue: analysis of 11 species with GFP-expressing recombinant virus. J Comp Neurol. 2000;427:559–580. doi: 10.1002/1096-9861(20001127)427:4<559::aid-cne5>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 40.Lecointe D, Dugas N, Leclerc P, Hery C, Delfraissy JF, Tardieu M. Human cytomegalovirus infection reduces surface CCR5 expression in human microglial cells, astrocytes, and monocyte-derived macrophages. Microbes Infect. 2002;4:1401–1408. doi: 10.1016/s1286-4579(02)00022-9. [DOI] [PubMed] [Google Scholar]
- 41.Reis B, Bogner E, Reschke M, et al. Stable constitutive expression of glycoprotein B (gpUL55) of human cytomegalovirus in permissive astrocytoma cells. J Gen Virol. 1993;74:1371–1379. doi: 10.1099/0022-1317-74-7-1371. [DOI] [PubMed] [Google Scholar]
- 42.Marr KA, Carter RA, Boeckh M, Martin P, Corey L. Invasive aspergillosis in allogeneic stem cell transplant recipients: changes in epidemiology and risk factors. Blood. 2002;100:4358–4366. doi: 10.1182/blood-2002-05-1496. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Huong SM, Chin ML, Raab-Traub N, Huang ES. Epidermal growth factor receptor is a cellular receptor for human cytomegalovirus. Nature. 2003;424:456–461. doi: 10.1038/nature01818. [DOI] [PubMed] [Google Scholar]
- 44.Wong AJ, Bigner SH, Bigner DD, et al. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci U S A. 1987;84:6899–6903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Humphrey PA, Wong AJ, Vogelstein B, et al. Amplification and expression of the epidermal growth factor receptor gene in human glioma xenografts. Cancer Res. 1988;48:2231–2238. [PubMed] [Google Scholar]
- 46.Liu TF, Tatter SB, Willingham MC, et al. Growth factor receptor expression varies among high-grade gliomas and normal brain: epidermal growth factor receptor has excellent properties for interstitial fusion protein therapy. Mol Cancer Ther. 2003;2:783–787. [PubMed] [Google Scholar]
- 47.Hengel H, Brune W, Koszinowski UH. Immune evasion by cytomegalovirus—survival strategies of a highly adapted opportunist. Trends Microbiol. 1998;6:190–197. doi: 10.1016/s0966-842x(98)01255-4. [DOI] [PubMed] [Google Scholar]
- 48.Reddehase MJ. The immunogenicity of human and murine cytomegaloviruses. Curr Opin Immunol. 2000;12:390–396. doi: 10.1016/s0952-7915(00)00106-0. [DOI] [PubMed] [Google Scholar]
- 49.Kossmann T, Morganti-Kossmann MC, Orenstein JM, et al. Cytomegalovirus production by infected astrocytes correlates with transforming growth factor-beta release. J Infect Dis. 2003;187:534–541. doi: 10.1086/373995. [DOI] [PubMed] [Google Scholar]
- 50.Riddell SR, Greenberg PD. Principles for adoptive T cell therapy of human viral diseases. Annu Rev Immunol. 1995;13:545–586. doi: 10.1146/annurev.iy.13.040195.002553. [DOI] [PubMed] [Google Scholar]