Abstract
Grade II gliomas are morphologically and clinically heterogeneous tumors for which histopathological typing remains the major tool for clinical classification. To what extent the major histological subtypes — astrocytomas, oligodendrogliomas, and oligoastrocytomas — constitute true biological entities is largely unresolved. Furthermore, morphological classification is often ambiguous and would be facilitated by specific subtype markers. In this study, 23 grade II gliomas were expression-profiled and subjected to hierarchical clustering. All six oligodendrogliomas were grouped together in one of two major clusters; a significant correlation was thus observed between gene expression and histopathological subtype. Supervised analyses were performed to identify genes differentiating oligodendrogliomas from other grade II tumors. In a leave-one-out test using 10 features for classification, 20 out of 23 tumors were correctly classified. Among the most differentially expressed genes was rPTPβ/ζ. The expression of the rPTP β/ζ protein in oligodendrogliomas and astrocytomas was further validated by immunohistochemistry in an independent set of tumors. All 11 oligodendrogliomas of this set displayed strong staining. In contrast, neoplastic astrocytes were mostly negative for rPTPβ/ζ staining. In summary, this study demonstrates a correlation between gene expression pattern and histological subtype in grade II gliomas. Furthermore, the results from the immunohistochemical analyses of rPTPβ/ζ expression should prompt further evaluation of this protein as a novel oligodendroglioma marker.
Keywords: Grade II glioma, histological marker, microarray, oligodendroglioma, rPTPβ/ζ
Grade II gliomas (astrocytomas, oligodendrogliomas, oligoastrocytomas, and ependymomas) in adults are low-malignant brain tumors, with an annual incidence of 1 – 2 per 100,000 adults.1 They are poorly circumscribed, diffusely infiltrative tumors mainly localized in the cerebral hemisphere. They are slightly more common in males, and the average age of diagnosis is about 40 years. The etiology is largely unknown. Environmental risk factors have not yet been identified.2 Stem cells as well as astrocytes, oligodendrocytes, and ependymal cells have been suggested as cell types from which these tumors originate. Development of grade II gliomas has been suggested to be a consequence of either dedifferentiation of mature glial cells or misdifferentiation of progenitor cells.
Histologically, grade II gliomas are well-differentiated tumors characterized by the absence of the criteria defining grade III – IV gliomas, such as high number of mitoses, endothelial proliferation, and necrosis.3,4 Among the most common types of grade II gliomas are astrocytic tumors called diffuse astrocytomas, which can be further divided into gemistocytic, fibrillary, and protoplastic astrocytomas. The other two most common grade II gliomas are oligodendrogliomas, and oligoastrocytomas with both astrocytic and oligodendroglial features. The occurrence of oligoastrocytomas or mixed gliomas exemplifies that a mixture of neoplastic cells with different glial phenotypes can exist within one tumor, in this case neoplastic astrocytes and presumed oligodendrocytes.
Treatment of grade II gliomas is unsatisfactory, and there is currently no cure for them.5 In most studies the median survival is 5 – 10 years, but there is a large clinical variation. For the vast majority of patients, radical surgery is not possible due to the infiltrative growth of the tumor.6 Based on data from randomized radiotherapy trials, there seems to be no overall survival advantage for immediate radiotherapy.7 Furthermore, radiotherapy is associated with a risk of side effects such as cognitive dysfunction. Therefore, the clinical management of patients has become a matter of right timing and modality of treatment for the individual patient.8
The histopathological subtypes are associated with differences in prognosis and treatment response. In general, patients with oligodendrogliomas have a better prognosis than those with astrocytomas; their 5-year survival is around 70% – 80%. The chemosensitivity of oligodendrogliomas is associated with loss of heterozygosity (LOH) on 1p and 19q.9 Most, though not all, oligodendrogliomas harbor LOH 1p, whereas only about 50% of oligoastrocytomas do so. The presence of LOH 1p and 19q is now being used as a clinical marker, complementary to the established histopathological criteria, for diagnosis of oligodendrogliomas, prediction of response to therapy, and prognosis.9 Among the grade II astrocytomas, the gemistocytic variant has a worse prognosis than the other two variants.
Gene expression profiling is becoming an established method for characterization of cancer types. It has been extensively used for analyses of glioblastomas and for comparisons of low-grade gliomas and glioblastomas. There is not yet much information about its possible use for characterization of grade II gliomas.
In this study we performed gene expression profiling of 23 cases of grade II gliomas. The results of these analyses were used in unsupervised analyses to compare correlations between the histological subtype of grade II gliomas and gene expression profiles. Also, supervised analysis was used to identify specific markers that could assist in the histopathological differential diagnostics of grade II gliomas.
Materials and methods
Patients and Tumor Samples
Frozen tumor tissue was obtained from samples collected from adult brain tumor patients (>15 years of age) at the Uppsala University Hospital. The study protocol was approved by the local ethics committee. For array analyses, a total of 23 tumor samples were collected (see Table 1). In addition, fixed embedded tissue from a subset of these cases, and from 21 additional grade II gliomas, was used (see Table 2). For the purposes of the study, all samples were reclassified by a neuropathologist according to World Health Organization (WHO) criteria.10
Table 1.
Patient characteristics
| Sample | Age at Surgery (Years)a | Gender | Histology | Tumor Localization and Size (cm) |
|---|---|---|---|---|
| O1 | 54 | F | Oligodendroglioma | T, R, >4 |
| O2 | 25 | F | Oligodendroglioma | F, L, <4 |
| O3 | 52 | F | Oligodendroglioma | F, ?, >4 |
| O4 | 47 | F | Oligodendroglioma | F, ?, ⩽4 |
| O5 | 44 | M | Oligodendroglioma | F, L, >4 |
| O6 | 50 | F | Oligodendroglioma | F, ?, ⩽4 |
| OA1 | 36 | F | Oligoastrocytoma | FT, R, >4 |
| OA2 | 31 | M | Oligoastrocytoma | F, R, >4 |
| OA3 | 33 | M | Oligoastrocytoma | T, R, >4 |
| OA4 | 37 | F | Oligoastrocytoma | FT, C, >4 |
| OA5 | 39 | M | Oligoastrocytoma | FT, L, >4 |
| OA6 | 42 | M | Oligoastrocytoma | FT, R, >4 |
| OA7 | 59 | M | Oligoastrocytoma | F, ?, ⩽4 |
| OA8 | 37 | F | Oligoastrocytoma | TP, L, >4 |
| A1 | 17 | M | Astrocytoma | F, L, <4 |
| A2 | 46 | M | Astrocytoma | F, ?, >4 |
| A3 | 53 | F | Astrocytoma | O, R, <4 |
| A4 | 46 | M | Astrocytoma | TP, L, ⩽4 |
| A5 | 27 | M | Astrocytoma | F, R, >4 |
| A6 | 71 | M | Astrocytoma | FTO, L, >4 |
| A7 | 35 | M | Astrocytoma | FT, R, ⩽4 |
| A8 | 31 | M | Astrocytoma | F, R, ⩽4 |
| A9 | 67 | F | Astrocytoma | T, ?, >4 |
Abbreviations: F, female; M, male; A, astrocytoma grade II; O, oligodendroglioma grade II; OA, oligoastrocytoma grade II; F, frontal; T, temporal; P, parietal; O, occipital; C, central; R, right; L, left; ?, data missing.
Average age = 42.6 years, SD = 13.2 years.
Table 2.
rPTPβ/ζ staining in grade II gliomas
| Sample | Histology | Fraction of Stained Tumor Cells (%) | Comments |
|---|---|---|---|
| O7 | Oligodendroglioma | 30 – 70 | A |
| O8 | Oligodendroglioma | 30 – 70 | A |
| O9 | Oligodendroglioma | 0 – 100 | A, focal pattern of staining |
| O10 | Oligodendroglioma | >70 | A |
| O11 | Oligodendroglioma | >70 | A and B |
| O12 | Oligodendroglioma | >70 | A and B |
| O13 | Oligodendroglioma | 0 – 100 | Focal pattern of staining |
| O14 | Oligodendroglioma | >70 | A and B |
| O15 | Oligodendroglioma | >70 | A and B |
| O16 | Oligodendroglioma | >70 | A |
| O17 | Oligodendroglioma | 0 – 100 | Focal pattern of staining |
| A10 | Fibrillary astrocytoma | <30 | C |
| A11 | Fibrillary astrocytoma | 30 – 70 | C |
| A12 | Fibrillary astrocytoma | 30 – 70 | Staining highest in pericellular networks |
| A13 | Fibrillary astrocytoma | <30 | C |
| A14 | Fibrillary astrocytoma | <30 | C |
| A15 | Fibrillary astrocytoma | <30 | C |
| A16 | Fibrillary astrocytoma | <30 | C |
| A17 | Gemistocytic astrocytoma | 30 – 70 | Small gemistocytes negative, C, scattered cells with pattern A |
| A18 | Gemistocytic astrocytoma | <30 | Gemistocytes negative, C |
| A19 | Gemistocytic astrocytoma | <30 | Gemistocytes negative, C |
Abbreviations: A, staining along plasma membrane and radiating processes; B, dense network between the cell bodies; C, neoplastic astrocytes negative, scattered cells with staining along plasma membrane.
For all grade II gliomas included in the array analysis, clinical information was collected from the patients’ charts regarding age (at surgery), gender, and radiological appearance of the tumor (tumor size and location on CT or MRI).
Gene Expression Profiling
RNA was isolated by the TRIZOL (Invitrogen, Paisley, UK) method. RNA quality was confirmed with a Bio-analyzer (Agilent Technologies, Santa Clara, CA, USA) and by photospectrometry. For each sample, 10 – 30 μg of total RNA was used to generate Cy-dye – labeled cDNA by direct incorporation of Cy-dye – coupled nucleotides in a reverse transcription reaction as described by Wellcome Trust Sanger Institute protocol 5 (“Generation of Fluorescently-Labelled Single-Stranded cDNA Target Using Direct Incorporation of Cye Dyes”; http://www.sanger.ac.uk/Projects/Microarrays/arraylab/protocol5.pdf). As a reference for the competitive hybridization, a mixture of RNA from normal cerebrum from an autopsy case and RNA from four of the tumors of the study was used.
Samples were hybridized overnight to Sanger Institute Hver 1.2.1 gene chips, containing about 10,000 elements (about 6,000 genes), as described by Wellcome Trust Sanger Institute protocol 6 “Competitive Hybridisation of Labelled ss cDNAs onto Microarrays”; http://www.sanger.ac.uk/Projects/Microarrays/arraylab/protocol6.pdf). Each tumor was hybridized together with the reference sample in duplicate or triplicate with dye-swap (17 cases) or in a single experiment (6 cases), depending on the amount of RNA available.
Shortly thereafter, for each hybridization, labeled sample and labeled reference in a volume of 66 μl were mixed with 4 μl cot-1 DNA (1 mg/ml; Gibco-BRL, Carlsbad, CA, USA), 4 μl polyadenylic acid (2 μg/ml; Sigma- Aldrich Sweden AB, Stockholm, Sweden), 218 μl 70% ethanol, and 7 μl 3 M sodium acetate pH 5.2. After precipitation by incubation for 30 min at – 70oC, samples were centrifuged at 15,000 × g for 20 min at 4oC. Pellets were washed with 70% ethanol and air dried for 60 min, dissolved in 8 μl water and 40 μl hybridization solution (5× saline sodium citrate [SSC], 6× Denhardt’s solution, 60 mM Tris-HCl pH 7.6, 0.12% sarkosyl, 48% formamide), heated at 100oC for 5 min, and cooled to room temperature. Samples were placed on a precooled microarray chip, covered by a cover-glass, and incubated for 16 h at 47oC in a Corning hybridization chamber containing 40 μl 40% formamide and 2× SSC. The chips were washed once in 2× SSC for 5 min, three times in 0.1× SSC with 0.1% sodium dodecyl sulfate for 20 min, and once in 0.1× SSC for 10 min. The chips were finally dried by centrifugation at 100 × g for 2 minutes. Chips were scanned with a ScanArray 5000 (GSI Lumonics, Rugby, UK), using ScanArray software version 3.1 (Packard BioChip Technologies, Billerica, MA, USA). Expression intensity values were quantified using QuantArray software version 3.0.0.0 (Packard BioChip Technologies). Unreliable spots were flagged, and signals were quantified by the histogram method. All raw data files for the microarray analysis have been deposited to ArrayExpress (http://www.ebi.ac.uk/arrayexpress) at the European Bioinformatics Institute (Hinxton, UK), and are publicly available under accession number E-MEXP-938.
Analyses of Array Data
Data were loaded into GeneSpring GX (Agilent Technologies) and then LOWESS (locally weighted sum of squares) normalized. A list of genes with varying expression between individual tumors was generated by analysis of variance (ANOVA) between the replicates (p < 0.001; samples were not assumed to have equal variance, and the significance calculations were corrected for multiple testing according to Bonferroni). The list was subsequently used to cluster the tumors hierarchically. Shortly thereafter, the expression data were loaded into BRB ArrayTools version 3.5.0 beta 1 (Biometric Research Branch, NCI, Bethesda, MD, USA), expression values were log2-transformed, genes were median centered, and the samples were subjected to uncentered complete linkage clustering using the cluster function.11 Following clustering, reproducibility and discrimination indexes were calculated as previously described.12
Genes with predicative potential between oligodendrogliomas, as compared to astrocytomas and mixed tumors, were identified by weighted voting in a leave-one-out validation using GeneCluster 2.0.13–15
To evaluate expression differerences in gene ontology categories between the histological groups, functional class scoring analysis was performed, as previously described,16 by using the function included in BRB ArrayTools version 3.5.0 beta 1.
Immunohistochemical Analyses
For the diagnostic characterization, sections were immunostained with either rabbit polyclonal or mouse monoclonal antibody against glial fibrillary acidic protein (GFAP; DAKO, Glostrup, Denmark) and mouse monoclonal antibody against Ki-67 (clone MIB-1; DAKO). Immunostaining for the protein product of the selected up-regulated gene rPTPβ/ζ was performed on sections from fixed paraffin-embedded tissue with commercially available mouse monoclonal antibody against rPTPβ/ζ clone 12 BD (Transduction Laboratories, San Diego, CA, USA). Antibody was used at a final concentration of 2.5 μg/ml. Stainings were performed in a Ventana Discovery system (Ventana Medical Systems, Illkirch, France) with heat-induced epitope retrieval for 48 min in a Tris/ borate/EDTA buffer pH 8.0. Primary antibody was incubated for 1 h at 37°C. Bound antibody was visualized with the DABMap kit (Ventana Medical Systems) by incubating with biotin-conjugated secondary antibody for 30 min and developing with diaminobenzidine.
Statistical Analyses
To assess whether discrete variables were associated, the chi-square test was used. To illustrate the expression range of features in different histological entities, MINITAB 14 (Minitab Incorporation, Coventry, UK) was used.
Results
Subsets of Grade II Gliomas Defined by Gene Expression Profiles Significantly Overlap with Histological Subsets
Microarray analyses were performed on 23 grade II gliomas (Table 1). These included nine astrocytomas, six oligodendrogliomas, and eight oligoastrocytomas. The mean age at surgery was 42.6 years (range, 17 – 71). The sex distribution was 13 males and 10 females (Table 1).
Array analyses were performed on cDNA chips containing approximately 10,000 spots representing more than 6,000 unique genes. Seventeen of the tumor samples were hybridized in triplicate or duplicate including dye-swap; six were hybridized only once due to limiting amounts of RNA. Hierarchical clustering analysis performed based on 82 cDNAs showed differential expression between individual tumors with p-values less than 0.001 in an ANOVA among the 23 tumor samples.
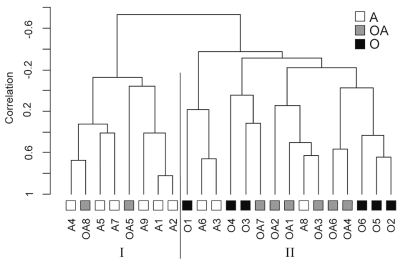
As shown in Fig. 1, this analysis yielded two major clusters (I – II) of tumors. Of interest, the three histopathological subtypes showed a significant distribution among the gene expression – based clusters (p ⩽ 0.025, chi-square). Six of six oligodendrogliomas were included in cluster II, whereas six of nine astrocytomas were grouped in cluster I. Among the oligoastrocytomas, two were grouped in cluster I and six in cluster II. The robustness of this clustering was tested as previously described,12 and yielded an overall robustness index value of 0.653 and discrimination index value of 4.96. Cluster-specific robustness values for clusters I and II were 0.802 and 0.613, respectively.
Fig. 1.
Subsets of grade II gliomas defined by gene expression overlap with histopathological subtypes. Twenty-three grade II gliomas were clustered based on 82 genes showing differential expression between the tumors. Tumors of different histopathological types were distributed nonrandomly between clusters I and II (p = 0.025, chi-square). The different histopathological subsets are color-coded with astrocytomas (A) shown in white, oligoastrocytomas (OA) in gray, and oligodendrogliomas (O) in black.
It was concluded from these analyses that a significant correlation exists between histopathologically defined subtypes and subsets identified based on gene expression.
Identification of Genes Differentially Expressed in Oligodendrogliomas
The data sets from the microarray analyses of the grade II tumors were subsequently used in a supervised analysis to identify putative novel marker genes for the different histopathological subsets.
Supervised analyses (see Materials and Methods for details) were used to investigate the possibility of generating a “gene classifier” for distinguishing oligodendrogliomas and identifying novel oligodendroglioma markers. When these analyses were performed in a leave-one-out format with gene lists varying in length between 1 and 10 genes, 20 or 21 of the 23 tumors were correctly classified.
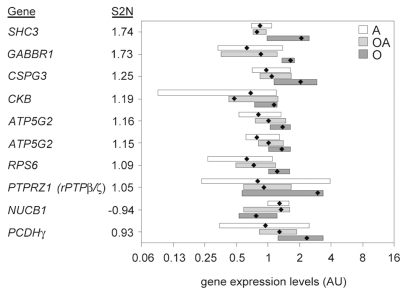
The 10 genes that most commonly occurred in the 23 leave-one-out tests using 10 classifying features are listed in Fig. 2. All genes displayed a fairly broad expression range within the oligodendroglioma group and within the other tumor subsets. The two genes that showed the largest signal-to-noise ratio were the intracellular signaling protein SHC3 and GABA B receptor 1 (GABBR1). It is noteworthy that, for all features except SHC3, CKB, and NUCB1, the mean expression in the oligoastrocytomas falls between the median expression in oligodendrogliomas and astrocytomas (Fig. 2).
Fig. 2.
Supervised gene analysis identifies a set of genes differentially expressed in oligodendrogliomas (O). Weighted voting classification (Golub et al.15) was used for identification of genes able to separate grade II oligodendrogliomas from other grade II gliomas (astrocytomas [A]; oligoastrocytomas [OA]). The expression range of the top 10 features most frequently occurring in a leave-one-out validation are illustrated for the different histopathological subtypes. The bars indicates minimum and maximum expression in each histopathological subtype, and median expression in each subtype is indicated by a black diamond. The features are arranged according to absolute signal-to-noise (S2N) values.
To gain further understanding of the potential biological differences between oligodendrogliomas and other grade II tumors in gene expression, gene set expression comparison was performed. This analysis indicated that the two gene ontology categories displaying the most significant differences between oligodendrogliomas and other grade II gliomas were 14 “epidermal growth factor receptor signaling pathway components” and 76 “transmembrane receptor protein tyrosine kinase signaling pathway components,” with permutation p-values < 0.001 for both groups.
We also noted that the micro-array-based expression analyses were able to identify a set of genes that, when analyzed together, could successfully classify the great majority of tumors according to histopathological subtype. However, the array analyses failed to identify single genes, even in this relatively small set of tumors, that showed a nonoverlapping expression pattern between histopathological subsets.
Immunohistochemical Analyses of rPTPβ/ζ in Grade II Oligodendrogliomas and Astrocytomas
Among the genes identified as up-regulated in oligodendrogliomas, a commercial antibody for immunohistochemical validation could be identified for rPTPβ/ζ.
For initial analyses, protein expression was analyzed on tissue sections of two grade II astrocytomas and two oligodendrogliomas included in the array analyses. Expression was clearly higher in tumor cells than in the surrounding normal brain (data not shown).
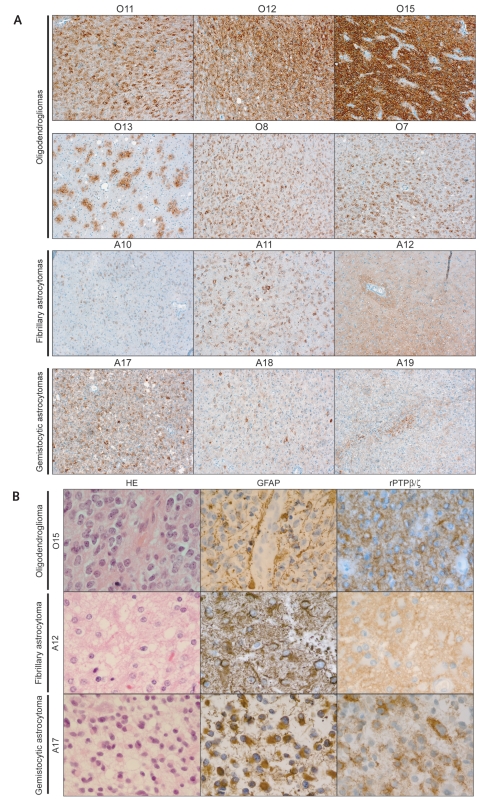
In the complementary analyses, a novel set of 11 grade II oligodendrogliomas and 10 grade II astrocytomas were analyzed. The latter included seven fibrillary astrocytomas and three gemistocytic variants (Table 2, Fig. 3A). Nonneoplastic oligodendrocytes in the white matter were immunonegative or at most showed very faint cytoplasmic staining for rPTPβζ/ (data not shown). All 11 oligodendrogliomas showed strong staining (Table 2, Fig. 3A). The intensity of staining as well as the fraction of positive cells showed some variability, as illustrated, for example, by tumors O7 and O15 in Fig. 3A. Also, some tumors displayed a focal staining pattern with clusters of cells showing strong immunoreactivity, as exemplified by tumor O13 in Fig. 3A. More detailed analyses of rPTPβ/ζ staining in the oligodendrogliomas revealed that the characteristic “fried-egg” oligodendroglioma cells were outlined by a rim of immunopositivity consistent with plasma membrane localization (Fig. 3B). The immunostaining also extended into short processes radiating from the cell body, and in regions with high cell density the rPTPβ/ζ-immunopositive cell processes appeared to merge into a strongly stained continuous network between the tumor-cell bodies (Fig. 3B).
Fig. 3.
Consistent up-regulation of rPTPβ/ζ in oligodendrogliomas. The immunohistochemical analyses of rPTPβ/ζ were extended to an independent set of tumors composed of 11 grade II oligodendrogliomas and 10 grade II astrocytomas (see Table 2 for details): (A) is composed of representative pictures showing the overall pattern of rPTPβ/ζ expression, whereas (B) shows close-ups and also includes glioma tissue stained with glial fibrillary acidic protein (GFAP) antibodies and hematoxylin- and eosin-stained tissue sections. All oligodendrogliomas showed prominent expression of rPTPβ/ζ in tumor cells, which occurred in either a homogeneous or a focal pattern. In general the staining in astrocytomas was weaker and predominantly localized to networks surrounding cell bodies. Gemistocytic and other astrocytic tumor cells were mostly negative for rPTPβ/ζ staining.
The astrocytomas in general showed less staining with regard to both intensity and fraction of stained cells (Table 2, Fig. 3A). This was noted in both the fibrillary and gemistocytic variants. Although some astrocytomas showed relatively strong staining, as illustrated by tumors A12 and A17 (Fig. 3A), the pattern of staining was different from that in the oligodendrogliomas. More specifically, in the gemistocytic variants, gemistocytic cells consistently remained negative for rPTPβ/ζ, although a weaker irregular network of staining could be observed between gemistocytes (tumor A17, Table 2, Fig. 3A, B). Similarly, in the fibrillary astrocytomas, cell bodies of neoplastic GFAP-positive astrocytes remained largely negative (Table 2, Fig. 3B). However, in most astrocytomas (Table 2, Fig. 3A, B) there were scattered positive cells between the astrocytic cells.
In summary, these immunohistochemical analyses confirmed the indications of the array analyses that rPTPβ/ζ should be further validated as a novel protein with characteristic staining pattern in grade II oligodendrogliomas.
Discussion
This study reports an array-based comparison of the three major histological subtypes of grade II gliomas. The gene expression analyses created clusters of tumors that significantly overlapped with histopathological subtypes (Fig. 1). Supervised gene analyses identified a set of candidate oligodendroglioma markers. SHC3 and GABBR1 showed the largest differential mRNA expression (Fig. 2). Finally, the immunohistochemical analyses of rPTPβ/ζ indicate that this protein should be further evaluated as a marker for distinguishing oligodendrogliomas from astrocytomas (Table 2, Fig. 3).
Gene expression profiling has been extensively used to characterize brain tumors.17 Most of these studies have focused on defining new biological subsets of glioblastomas, in combination with identification of markers predicting survival.18–22 Less attention has been paid to grade II gliomas. One previous study, which included only five grade II tumors, indicated that tumors of the same histopathological subtype cluster together.23 A second study analyzed the expression profiles of 17 oligodendrogliomas and 15 grade II astrocytomas.24 In this study, unsupervised analyses did not clearly indicate relationships between gene expression pattern and histological subtype. However, supervised analyses using 79 differentially expressed features perfectly separated oligodendrogliomas and astrocytomas. In comparison with the present study it should be noted that the study of Huang et al.24 used a smaller array of 1,176 cancer-associated genes.
In our study we used supervised analyses to identify genes showing differential expression in the histopathological subsets. Among the nine differentially expressed genes in Fig. 2 were three cell surface proteins, protocadherin-γ, GABA B receptor 1 (GABBR1), and RPTPβ/ζ (PTPRZ1). The list also included intracellular signaling molecules (SHC3); secreted proteins with potential roles in growth regulation (CSPG3); and two proteins involved in energy homeostasis, ATP synthase (ATP5G2) and creatine kinase, brain (CKB).
Of interest, all three cell surface proteins have previously been implicated in oligodendroglioma biology. Protocadherin-γ and GABBR1 were both included in the list of oligodendroglioma genes in Huang et al.24 The observation from the analyses of differentially expressed gene ontology categories, that growth factor signaling appears to differ between histological types of grade II gliomas, also merits further study.
A series of observations also link rPTPβ/ζ to gliomas and physiological neurobiology. The protein has also attracted interest as a cancer drug target.25 Many studies suggest roles of rPTPβ/ζ in neuronal migration, neurite outgrowth, and gliogenesis.26–28 A stimulatory role of rPTPβ/ζ in migration of both glioblastoma and neuronal cells has been demonstrated.29,30 Of interest, this protein is also important for function of oligodendrocytes, since in rPTPβ/ζ knockout mice oligodendrocyte recovery after demyelinating lesions is reduced.31 Most recently, antitumor activities of native and saporin-coupled rPTPβ/ζ-directed antibodies have been demonstrated.32 Our study does not address the functional significance of rPTPβ/ζ expression in oligodendrogliomas. However, the consistently high expression levels suggest that this tumor subset might be particularly interesting to consider for rPTPβ/ζ-targeted therapies.
The histopathological distinction between oligodendrogliomas and astrocytomas is still hampered by the lack of reliable immunohistochemical markers for oligodendroglioma cells. The distinction between oligodendrogliomas and astrocytomas has become increasingly important due to the divergent developments in their therapies, which also have significant effects on their respective prognoses. The characteristic immunohistochemical staining pattern for rPTPβ/ζ may provide an auxiliary, confirmatory method for definite histopathological diagnosis of oligodendrogliomas. Even though rPTPβ/ζ antibody does not exclusively stain oligodendroglioma cells, the rPTPβ/ζ staining pattern could be used as a confirmatory marker similar to that suggested for jun-D in the differential diagnosis between oligodendrogliomas (nuclear jun-D staining) and astrocytomas (cytoplasmic jun-D staining).24
In conclusion, these analyses of grade II gliomas show an overlap between subsets defined by unsupervised analyses of expression data and histological subtypes of these tumors. Our study thus gives further support for the idea that the different histological subgroups of grade II gliomas indeed reflect separate biological entities with different pathogenic pathways. The identification of the tyrosine phosphatase rPTPβ/ζ as a new putative marker for oligodendrogliomas also opens up new possibilities for more refined histopathological diagnosis and suggests continued exploration of targeting of rPTPβ/ζ in oligodendrogliomas.
Acknowledgments
A.Ö. and M.N. were supported by grants from the Swedish Cancer Society, Karolinska Institutet, the Cancer Society in Stockholm, and the Swedish Medical Research Council. Members of the laboratory of A.Ö. are acknowledged for contributing with critical discussions throughout the project, and Aris Moustakas and Marcin Kowanetz are acknowledged for sharing their experience in microarray analysis. The Sanger microarray consortium is funded by the Wellcome Trust, Cancer Research UK, and the Ludwig Institute for Cancer Research. We thank the staff of the Sanger Institute Microarray Facility TU for supplying arrays, lab protocols, and technical advice (David Vetrie, Cordelia Langford, Adam Whittaker, Neil Sutton), and Quantarray/GeneSpring data files and databases relating to array elements (Kate Rice, Rob Andrews, Adam Butler, Harish Chudasama). The human I.M.A.G.E. (Integrated Molecular Analysis of Genomes and Their Expression) cDNA clone collection was from the MRC HGMP Resource Centre (Hinxton, UK). cDNA clone resequencing was performed by Team 56 at the Sanger Institute. Immunohistochemical staining for rPTPβ/ζ was performed by the MTA (Mouse Tissue Analysis) Core Facility at Karolinska Institutet/Karolinska University Hospital.
References
- 1.Bampoe J, Bernstein M. The role of surgery in low grade gliomas. J Neurooncol. 1999;42:259–269. doi: 10.1023/a:1006103010109. [DOI] [PubMed] [Google Scholar]
- 2.Davis FG, McCarthy BJ. Epidemiology of brain tumors. Curr Opin Neurol. 2000;13:635–640. doi: 10.1097/00019052-200012000-00004. [DOI] [PubMed] [Google Scholar]
- 3.McLendon RE, Cleveland L, Pegram C, Bigner SH, Bigner DD, Friedman HS. Immunohistochemical detection of the DNA repair enzyme O6–methylguanine-DNA methyltransferase in formalin-fixed, paraffin-embedded astrocytomas. Lab Invest. 1998;78:643–644. [PubMed] [Google Scholar]
- 4.Kleihues P, Louis DN, Scheithauer BW, et al. The WHO classification of tumors of the nervous system. J Neuropathol Exp Neurol. 2002;61:215–225. doi: 10.1093/jnen/61.3.215. discussion 226–229. [DOI] [PubMed] [Google Scholar]
- 5.Leighton C, Fisher B, Bauman G, et al. Supratentorial low-grade glioma in adults: an analysis of prognostic factors and timing of radiation. J Clin Oncol. 1997;15:1294–1301. doi: 10.1200/JCO.1997.15.4.1294. [DOI] [PubMed] [Google Scholar]
- 6.Keles GE, Lamborn KR, Berger MS. Low-grade hemispheric gliomas in adults: a critical review of extent of resection as a factor influencing outcome. J Neurosurg. 2001;95:735–745. doi: 10.3171/jns.2001.95.5.0735. [DOI] [PubMed] [Google Scholar]
- 7.Karim AB, Afra D, Cornu P, et al. Randomized trial on the efficacy of radiotherapy for cerebral low-grade glioma in the adult: European Organization for Research and Treatment of Cancer Study 22845 with the Medical Research Council study B RO4: an interim analysis. Int J Radiat Oncol Biol Phys. 2002;52:316–324. doi: 10.1016/s0360-3016(01)02692-x. [DOI] [PubMed] [Google Scholar]
- 8.Pignatti F, Van Den Bent M, Curran D, et al. Prognostic factors for survival in adult patients with cerebral low-grade glioma. J Clin Oncol. 2002;20:2076–2084. doi: 10.1200/JCO.2002.08.121. [DOI] [PubMed] [Google Scholar]
- 9.Reifenberger G, Louis DN. Oligodendroglioma: toward molecular definitions in diagnostic neuro-oncology. J Neuropathol Exp Neurol. 2003;62:111–126. doi: 10.1093/jnen/62.2.111. [DOI] [PubMed] [Google Scholar]
- 10.Kleihues P, Cavenee WK. World Health Organization Classification of Tumours. New York: Oxford University Press; 2000. Pathology and Genetics: Tumours of the Nervous System. [Google Scholar]
- 11.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McShane LM, Radmacher MD, Freidlin B, Yu R, Li MC, Simon R. Methods for assessing reproducibility of clustering patterns observed in analyses of microarray data. Bioinformatics. 2002;18:1462–1469. doi: 10.1093/bioinformatics/18.11.1462. [DOI] [PubMed] [Google Scholar]
- 13.Tamayo P, Slonim D, Mesirov J, et al. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reich M, Ohm K, Angelo M, Tamayo P, Mesirov JP. GeneCluster 2.0: an advanced toolset for bioarray analysis. Bioinformatics. 2004;20:1797–1798. doi: 10.1093/bioinformatics/bth138. [DOI] [PubMed] [Google Scholar]
- 15.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 16.Pavlidis P, Qin J, Arango JJ, Sibille E. Using the gene ontology for microarray data mining: a comparison of methods and application to age effects in human prefrontal cortex. Neurochem Res. 2004;29:1213–1222. doi: 10.1023/b:nere.0000023608.29741.45. [DOI] [PubMed] [Google Scholar]
- 17.Mischel PS, Cloughesy TF, Nelson SF. DNA-microarray analysis of brain cancer: molecular classification for therapy. Nat Rev Neurosci. 2004;5:782–792. doi: 10.1038/nrn1518. [DOI] [PubMed] [Google Scholar]
- 18.Rich JN, Hans C, Jones B, et al. Gene expression profiling and genetic markers in glioblastoma survival. Cancer Res. 2005;65:4051–4058. doi: 10.1158/0008-5472.CAN-04-3936. [DOI] [PubMed] [Google Scholar]
- 19.Freije WA, Castro-Vargas FE, Fang Z, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 20.Nutt CL, Mani DR, Betensky RA, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63:1602–1607. [PubMed] [Google Scholar]
- 21.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 22.Tso CL, Freije WA, Day A, et al. Distinct transcription profiles of primary and secondary glioblastoma subgroups. Cancer Res. 2006;66:159–167. doi: 10.1158/0008-5472.CAN-05-0077. [DOI] [PubMed] [Google Scholar]
- 23.Shai R, Shi T, Kremen TJ, et al. Gene expression profiling identifies molecular subtypes of gliomas. Oncogene. 2003;22:4918–4923. doi: 10.1038/sj.onc.1206753. [DOI] [PubMed] [Google Scholar]
- 24.Huang H, Okamoto Y, Yokoo H, et al. Gene expression profiling and subgroup identification of oligodendrogliomas. Oncogene. 2004;23:6012–6022. doi: 10.1038/sj.onc.1207781. [DOI] [PubMed] [Google Scholar]
- 25.Muller S, Lamszus K, Nikolich K, Westphal M. Receptor protein tyrosine phosphatase zeta as a therapeutic target for glioblastoma therapy. Expert Opin Ther Targets. 2004;8:211–220. doi: 10.1517/14728222.8.3.211. [DOI] [PubMed] [Google Scholar]
- 26.Holland SJ, Peles E, Pawson T, Schlessinger J. Cell-contact-dependent signalling in axon growth and guidance: Eph receptor tyrosine kinases and receptor protein tyrosine phosphatase beta. Curr Opin Neurobiol. 1998;8:117–127. doi: 10.1016/s0959-4388(98)80015-9. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Tullai JW, Yu WH, Salton SR. Regulated expression during development and following sciatic nerve injury of mRNAs encoding the receptor tyrosine phosphatase HPTPzeta/RPTPbeta. Brain Res Mol Brain Res. 1998;60:77–88. doi: 10.1016/s0169-328x(98)00175-2. [DOI] [PubMed] [Google Scholar]
- 28.Stoker A, Dutta R. Protein tyrosine phosphatases and neural development. Bioessays. 1998;20:463–472. doi: 10.1002/(SICI)1521-1878(199806)20:6<463::AID-BIES4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 29.Muller S, Kunkel P, Lamszus K, et al. A role for receptor tyrosine phosphatase zeta in glioma cell migration. Oncogene. 2003;22:6661–6668. doi: 10.1038/sj.onc.1206763. [DOI] [PubMed] [Google Scholar]
- 30.Maeda N, Noda M. Involvement of receptor-like protein tyrosine phosphatase zeta/RPTPbeta and its ligand pleiotrophin/heparin-binding growth-associated molecule (HB-GAM) in neuronal migration. J Cell Biol. 1998;142:203–216. doi: 10.1083/jcb.142.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harroch S, Furtado GC, Brueck W, et al. A critical role for the protein tyrosine phosphatase receptor type Z in functional recovery from demyelinating lesions. Nat Genet. 2000;32:411–414. doi: 10.1038/ng1004. [DOI] [PubMed] [Google Scholar]
- 32.Foehr ED, Lorente G, Kuo J, Ram R, Nikolich K, Urfer R. Targeting of the receptor protein tyrosine phosphatase beta with a monoclonal antibody delays tumor growth in a glioblastoma model. Cancer Res. 2006;66:2271–2278. doi: 10.1158/0008-5472.CAN-05-1221. [DOI] [PubMed] [Google Scholar]