Abstract
Dynamic changes in the expression of multiple genes appear to be common features that distinguish transformed cells from their normal counterparts. We compared the proteomic profiles of four glioblastoma multiforme (GBM) tissue samples and four normal brain cortex samples to examine the molecular basis of gliomagenesis. Trypsin-digested protein samples were separated by capillary isoelectric focusing with nano-reversed-phase liquid chromatography and were profiled by mass spectrometric sequencing. Wolf-Hirschhorn syndrome candidate 1 (WHSC1), along with 103 other proteins, was found only in the GBM proteomes. Western blot and immunohistochemistry verified our proteomic findings and demonstrated that 30-kDa WHSC1 expression increases with ascending tumor proliferation activity. RNA interference could suppress glioma cell growth by blocking WHSC1 expression. Our novel findings encourage the application of proteomic techniques in cancer research.
Keywords: expression, glioma, proliferation, proteomic profiling, WHSC1
Glioma, the most frequent primary brain tumor, remains a highly lethal neoplasm, refractory to current treatments. This aggressive behavior underscores the importance of understanding the molecular mechanisms of gliomagenesis. Gliomagenesis is a multitiered process with numerous, interdependent interactions among a variety of gene products. Given this diversity, molecular profiling has advantages compared with the single-candidate approach due to its ability to interrogate multiple genes and their products. Proteomic profiling can highlight protein events during cell transformation. Current proteomic profiling tools include two-dimensional polyacrylamide gel electrophoresis (2-D PAGE) and nongel/shotgun-based multidimensional liquid-chromatography protein separation, followed by tandem mass spectrometry (MS/MS) sequencing.1,2 In the present study, we used our newly developed, shotgun-based proteomic technique to distinguish the proteome of glioblastoma multiforme (GBM) from that of normal brain, by combining selective tissue-dissection-guided protein extraction with capillary isoelectric focusing (CIEF) with nano–reversed-phase liquid chromatography (nRPLC) peptide separation and MS/MS protein sequencing.3 The Wolf-Hirschhorn syndrome candidate 1 (WHSC1) protein consistently appeared in GBM proteomes but not in normal brain. Although WHSC1 possesses two conserved tumor-related domains, SET and PWWP, it has not been reported as a tumor-specific gene, and its function remains unknown.4,5 Taking WHSC1 as an indefinite gene without reported connection with gliomagenesis, we further studied its expression in gliomas and addressed its role in glioma proliferation.
Materials and Methods
Clinical Materials and Selective Tissue Dissection
All tissues were collected from the Surgical Neurology Branch at the National Institute of Neurological Disorders and Stroke or from the Brain Tumor Institute at the Cleveland Clinic Foundation. Tissues and clinical information were obtained as part of an institutional review board–approved study at these institutes. Frozen tissue included 12 gliomas: four of WHO grade II (two astrocytomas, two oligodendrogliomas), four of grade III (two astrocytomas, two oligodendrogliomas), and four GBMs. Four normal lateral temporal lobe neocortex samples were assigned for proteomic comparison with GBMs. Selective tissue dissection was performed as previously described.1
Comparison of Proteomic Profiles Generated by CIEF-nRPLC-MS/MS
An online combination of CIEF with nRPLC-MS/MS, developed by Calibrant Biosystems (Gaithersburg, MD, USA), was used for creating the proteomes from GBM and normal brain, as described previously.3 In brief, proteins prepared from microdissected cells were further digested by trypsin and filled into a CIEF capillary with ampholytes. The focused peptides were sampled into a total of 12 unique fractions. These fractions were analyzed in sequence, and the eluants from nRPLC were monitored by a quadrupole time-of-flight micro mass spectrometer (Waters, Milford, MA, USA). Peptide and protein identifications were made using MASCOT 2.0 (Matrix Science, London, UK) utilizing a reversed database search approach to determine a false-positive rate. Comparison of proteomic profiles between four GBMs and four normal brain specimens was performed using Excel software.
Western Blot
Western blot was performed following standard protocols.1 The frozen tissues were selectively dissected, and 30 μg of protein was loaded per sample. Based on the availability of antibodies and the novelty of the candidates, we applied Western blot to four candidates (see supplementary material, Table 1S, entries shown in bold) to validate our proteomic technique. Rabbit anti-SMC5 antibody (1:1,000), goat anti-BS69 antibody (1:200), and rabbit anti–prothymosin alpha antibody (1:500) were purchased from Abcam Inc. (Cambridge, MA, USA). Anti-WHSC1 polyclonal antibody (1:500; Novus Biologicals, Inc., Littleton, CO, USA) originated from goat immunized against an N-terminal common sequence (EFSIKQSPLSVQS), which was shared by the WHSC1 4a isoform and other family members. To confirm that the 30-kDa band was not from a nonspecific binding, another newly developed anti-WHSC1 antibody raised from rabbit against amino acids 219–268 of WHSC1 (1:200; Aviva Systems Biology, San Diego, CA, USA) was applied to Western blotting with identical samples. Anti–β-actin monoclonal antibody (1:500; Sigma-Aldrich, Inc., Steinheim, Germany) was applied as an internal loading control. The student t-test was applied to analyze the densitometry of immunosignal by using Proteomweaver software (Definiens, Munich, Germany).
Semiquantitative Reverse Transcriptase PCR
One microgram of total RNA was applied to reverse transcriptase (RT) PCR. To confirm that the 30-kDa WHSC1 Western blotting signal was the product of WHSC1 mRNA splicing isoform on exon 4a (here named WHSC1 4a),6 specific primers for WHSC1 4a were designed. The forward primer (TCAAAAATG-GCTCTCCAGAAAA) is located on exon 3, which is shared with other WHSC1 family members; the reverse primer (AAGTGTTCAAACTTCTTTGATTTGA), located on exon 4a, is unique to the WHSC1 4a isoform. Primers (forward, CCACGAAACTACCTTCAACTCC; reverse, TCATACTCCTGCTTGCTGATCC) were used to amplify β-actin as an internal control. Twenty-eight rounds of an amplification cycle of 94ºC for 30 s, 57ºC for 30 s, and 70ºC for 30 s were used in both PCR reactions.
Immunohistochemistry and Statistics
Paraffin-fixed tissue slides from 3 normal brains and 94 gliomas (grade II, 16 astrocytomas and 10 oligodendrogliomas; grade III, 9 astrocytomas and 9 oligodendrogliomas; grade IV, 50 GBMs) were immunostained with the anti-WHSC1 antibody (goat, 1:200) and anti–Ki-67 antibody (1:50; Dako Cytomation, Glostrup, Denmark). The staining result was reviewed by two independent observers who were unaware of specimen status. Statistical analyses were performed with SPSS for Windows software (SPSS, Inc., Chicago, IL, USA). The Spearman rank correlation test was performed to evaluate the relationship between WHSC1 expression and Ki-67 labeling index. Statistical significance was defined as p < 0.05.
RNA Interference Mediated by siRNA Transfection
WHSC1 4a-targeted short interfering RNAs (siRNAs) were designed and synthesized by Qiagen, Inc. (Valencia, CA, USA). From the four designed siRNAs, the one that achieved the highest silencing effect (CAAAGAAGTTTGAACACTTAA) was chosen for further study. Nonspecific commercial siRNA (AATTCTCCGAACGTGTCACGT) was used as a control; the fluorescence-labeled siRNA was applied to optimize the transfection efficiency. siRNA was transfected into A172, U251, and U373 human glioma cell lines (ATCC, Manassas, VA, USA), following the Qiagen RNAi Starter Kit protocol. Transfection was performed once (at the 0-h time point only) or twice (the 0-h and 24-h time points, to maximize the effect of interference) and repeated in triplicate for each cell line.
Inhibitory Effect on Cell Growth of RNA Interference Determined by Methyl Thiazole Tetrazolium Assay
The proliferation of RNA interference (RNAi)–treated cells was evaluated by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (ATCC) according to the manufacturer’s protocol. Ninety-six-well plates were used for a 96-h time-course observation (0, 4, 12, 24, 36, 48, 72, and 96 h). An absorbance value for each well was obtained by photospectrometry (Bio-tek Instruments, Inc., Winooski, VT, USA). Studies were performed in quadruplicate (12 wells each time) and reported as mean ± SD.
Results and Discussion
Combined Selective Tissue Dissection/CIEF-nRPLC-MS/MS Identified a GBM-Specific Proteome
With recent improvements in proteomic techniques, proteome profiling has been a powerful complementary approach to nucleic acid-based molecular profiling (cDNA microarray) in large-scale gene analyses implicated in tumorigenesis.1,2,7 To generate a specific proteomic profile from the nonhomogeneous sample, such as a surgical tumor specimen, selective tissue dissection is requested during sample preparation to procure pure tumor populations, in which no unwanted normal cells obscure the results.1 However, most current proteomic techniques require high sample quantities that are generally incompatible with the small amount of proteins obtained from microdissected clinical samples.8,9 We recently described an online combination of CIEF with nRPLC-MS/MS in an automated and integrated platform.3 This method provides systematic resolution of complex peptide mixtures based on their differences in isoelectric point and hydrophobicity and eliminates peptide loss and analyte dilution. Compared with 2-D PAGE and other non–gel-based profiling tools, CIEF-nRPLC permits a higher resolution and sensitivity in peptide separation with less protein loading, which allows selective tissue dissection before proteomic profiling to achieve a more specific tumor proteome.
In the present study, CIEF-nRPLC-MS/MS identified approximately 7,000 fully tryptic peptides and led to the identification of about 1,800 distinct proteins from each of the GBM samples as well as the control samples from normal brain tissues. Identifications were generated from three runs of a single tissue sample (consuming 10 μg of protein per run) and were based on high-mass-accuracy (60 ppm) and high-confidence (5% false-positive) hits to fully tryptic peptides. Proteins found exclusively in all four samples of either the GBM group or the normal brain group were defined as “group-steady proteins,” and the differences between two groups of proteins identified tumor-specific and normal-specific proteins. In this study, 104 proteins were found to be GBM specific (see supplementary material, Table 1S). The high output of protein identifications from our selective tissue dissection/CIEF-nRPLC-MS/MS technique is attributed to its high resolving power, high concentration, narrow analyte bands, and effective usage of electrospray ionization-tandem MS in peptide identifications. Compared with our previous 2-D PAGE profiling studies, which required approximately 50 μg of protein loading and revealed no more than 1,200 visible silver-stained spots (only about half of these could be sequenced) over a limited pH (4–7) range, CIEF-nRPLC-MS/MS produced at least a 15-fold increase in protein identification.1
Comparative Study of GBM Proteome Revealed Tumor-Specific Expression of a Novel Protein, WHSC1 4a, in Human Gliomas
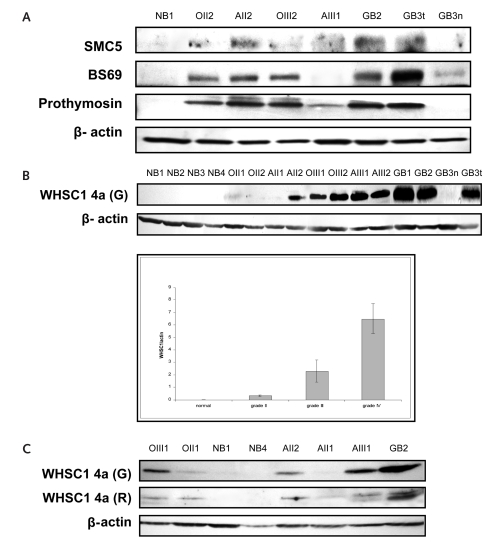
Direct comparison of the GBM proteome with that of normal brain tissue may reveal proteins that underlie events important in gliomagenesis. We identified 104 GBM-specific proteins. Most of these proteins, such as Forkhead Transcription Factor, Ets Transcription Factor, KDR, and MAX, have been found to be coupled with gliomagenesis.10–13 According to the availability of antibodies and the novelty of identified proteins, we selected the SMC5, BS69, prothymosin alpha, and WHSC1 proteins for Western blot analysis. We found these proteins to be expressed in gliomas but not in normal brain tissue (Fig. 1A). In the present study, we focus on the WHSC1 protein because the strength of expression is also correlated with glioma grades (Fig. 1B).
Fig. 1.
Proteomic findings are validated by Western blot. The specimens included four normal brain tissues (NB1–NB4); tissues from oligodendroglioma grade II (OII1, OII2) and grade III (OIII1, OIII2), astrocytoma grade II (AII1, AII2) and grade III (AIII1, AIII2), and glioblastoma multiforme (GBM; GB1–GB4); and tissues from a microdissected normal region (GB3n) and tumor (GB3t), both from the same GBM patient. (A) SMC5, BS69, and prothymosin are absent in normal brain tissue and are expressed at varied levels in gliomas. (B) Wolf-Hirschhorn syndrome candidate 1 (WHSC1) isoform 4a protein expression is correlated with the malignancy of gliomas. Polyclonal anti-WHSC1 antibody raised from goat recognized a single immunoblotting signal at 30 kDa from all brain tumor tissues (upper panel). The immunosignal was completely absent in normal brain and in normal brain tissue from the microdissected GBM. The blotting signal density was higher in GBMs than in grade III tumors and was higher in grade III than in grade II tumors. As a loading control, 42-kDa β-actin protein was detected from the same blotting membrane after stripping out WHSC1 4a immunosignals. The lower panel summarizes the densitometry results of the WHSC1 4a expression. (C) The WHSC1 4a protein expression was reconfirmed by Western blotting by using another polyclonal anti-WHSC1 antibody that was recently developed from rabbit. Both antibodies identified a consistent pattern of WHSC1 expression.
WHSC1 wasidentified in 1998 as a 90-kb gene family that maps to a 165-kb area on chromosome 4p16.3. Hemizygous deletion of this region has been identified in most patients with Wolf-Hirschhorn syndrome (WHS, a multiple malformation syndrome), and a t(4;14)(p16, q32) gene translocation that includes WHSC1 appears to be associated with poor outcome of patients with multiple myelomas.4,6 There has been no direct evidence proving that WHSC1 participates in developing either WHS or myelomas. Based on our proteomic evidence and the fact that it contains two conserved, tumor-related domains, we hypothesize that WHSC1 plays a role in gliomagenesis. It contains a SET domain that appears to be a transcriptional mediator that influences chromatin- mediated regulating mechanisms in tumor cells,4 and a PWWP domain that functions as a DNA binding domain by directing protein transport. Hepatoma-Derived Growth Factor, an extracellular heparin-binding, acidic, nuclear polypeptide with mitogenic activity, contains a functional PWWP domain. The DNA mismatch repair gene MSH6, alterations of which contribute to nonpolyposis colorectal cancers, also contains a PWWP domain.5,6
Previous studies of WHSC1 gene expression focused exclusively on its transcriptional level, indicating that WHSC1 mRNA is expressed ubiquitously in early development and undergoes complex alternative splicing processes to create a WHSC1 family with eight individual members with predicted protein sizes from 30 kDa to 157 kDa.6,14 Protein expression of WHSC1 has not been previously addressed. Our Western blotting detected a glioma-specific, single immunosignal at 30 kDa, which was expected to represent the WHSC1 splicing 4a isoform (Fig. 1B; see also supplementary data, Fig. 1S). The specificity of immunoblotting has been reconfirmed by applying another independent, newly commercialized anti-WHSC1 antibody in Western blotting (Fig. 1C). Our data indicate that WHSC1 4a expression increases with the ascending glioma grade and that it is tumor specific (t-test, p < 0.01 between any conjunctive groups). Using microdissection on a large GBM specimen, we separated the tumor cells from their adjacent normal tissue and applied Western blotting on these two cell populations. The result clearly indicated that WHSC1 was expressed only in the tumor cells (lane GB3t, Fig. 1B) and not in the normal cells (lane GB3n, Fig. 1B). From the protein structure analysis, WHSC1 4a lacks one tumor-related domain (SET) during RNA splicing; it still conserves a modified PWWP domain. The modification of PWWP domain may thus affect DNA binding and promote transformation toward gliomagenesis (see supplementary data, Fig. 1S).6
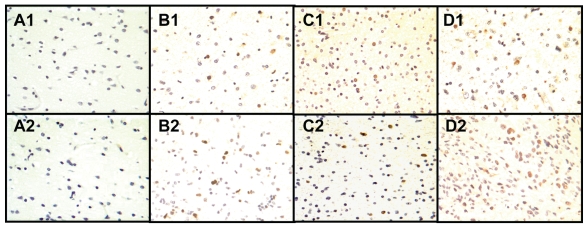
WHSC1 4a Is Localized in Tumor Nuclei, and Its Expression Level Correlates with Glioma Proliferative Activity
Immunohistochemistry was performed on 94 gliomas and 3 normal brain tissues to gain insight into WHSC1 4a’s subcellular distribution (Fig. 2). More intense staining, consistent with the result from Western blotting, occurred in higher grade specimens. Staining was completely absent in normal brain cells (Fig. 2, A1). Recently, Keats et al.6 investigated the intracellular distribution of the WHSC1 4a isoform (also known as MMSET III). In their study, WHSC1 4a was fused with a fluorescent tag in living cells and the protein was localized wholly within the nuclei. Consistent with these results, our data indicate that endogenous WHSC1 4a protein is expressed in the nuclei of tumor cells.
Fig. 2.
Wolf-Hirschhorn syndrome candidate 1 (WHSC1) isoform 4a protein expression correlates with glioma proliferative activity by immunohistochemical staining. Tissue labeling: normal cortex (A), astrocytoma grade II (B), oligodendroglioma grade III (C), and glioblastoma multiforme (GBM) grade IV (D). Anti-WHSC1 antibody (labeled 1 in each tissue) and anti–Ki-67 antibody (labeled 2 in each tissue) were applied. Both antibodies picked up their specific immunosignals with dark brown color in the nuclei of positive cells. Consistent with an increased glioma proliferative activity demonstrated by elevated Ki-67 staining, anti-WHSC1 antibody stained a few scattered cells in low-grade gliomas (B) and was more robust in grade III tumors (C) and highest in GBMs (D). Neither antibody stained normal brain tissue. (Original magnification, ×400.)
We also performed immunohistochemistry with anti–Ki-67 antibody on the adjacent slides of WHSC1 staining, from the same group of gliomas to correlate the WHSC1 4a expression level with tumor proliferation (Fig. 2, Table 1). The percentage of WHSC1-positive cells increased with increasing glioma grade. WHSC1 4a expression was statistically correlated with the Ki-67 labeling index, a marker of tumor proliferative activity (p = 0.03).
Table 1.
Association of glioma grades with Wolf-Hirschhorn syndrome candidate 1 (WHSC1) isoform 4a expression and Ki-67 labeling index
| Tumor Grade | Number of Cases (n) | WHSC1 Expression (%,Mean ± SD) | Ki-67 Labeling Index (%,Mean ± SD) |
|---|---|---|---|
| II | 26 | 3.13 ± 1.77 | 2.33 ± 1.51 |
| III | 18 | 9.42 ± 5.60 | 12.1 ± 7.87 |
| IV | 50 | 15.77 ± 11.92 | 30.5 ± 13.2 |
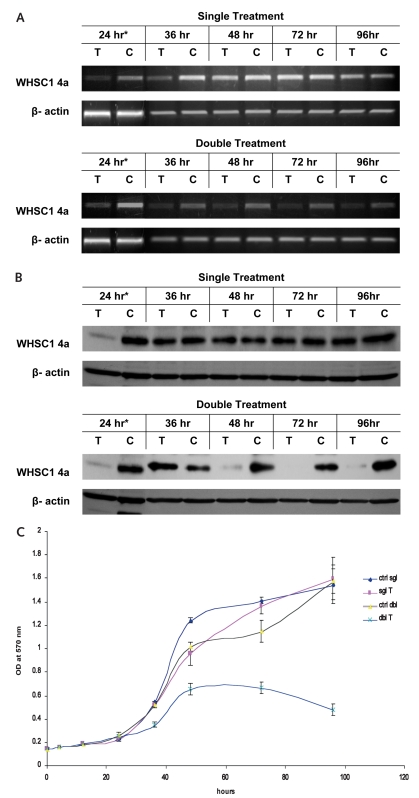
Inhibition of WHSC1 4a Expression by RNAi Suppresses Viability of GBM Cells In Vitro
We used RNAi to assess the role WHSC1 4a may play in proliferation of glioma cells in vitro. Endogenous WHSC1 4a expression was blocked efficiently at 24 h after siRNA transfection and slowly recovered afterward (Fig. 3A,B). The maximal inhibitory effect on glioma proliferation appeared around 48 h after transfection, achieving about 23% suppression (Fig. 3C). Growth suppression abated after 48 h and disappeared by 72 h. This is perhaps because either the growth of the control (non-specific siRNA transfected cells) was restricted within the 96-well plate after a prolonged culture or the RNAi effect was diminished and the treated cells resumed their growth with normal expression levels of WHSC1 4a. Thus, we repeated siRNA transfection at 24 h for sustained suppression of WHSC1 4a expression. As shown by the blue curve in Fig. 3C, the double-treatment group achieved about a 36% decrease of tumor proliferation at 48 h, which persisted to 72 h and maximized to about 70% at 96 h (longer observation was not practical because of the saturation of cell density for the control group). These RNAi studies, although preliminary, suggest that WHSC1 4a may play a heretofore unsuspected role in glioma proliferation.
Fig. 3.
Down-regulation of Wolf-Hirschhorn syndrome candidate 1 (WHSC1) isoform 4a gene expression suppresses cell proliferation of glioblastoma multiforme (GBM) in vitro. Reverse transcriptase PCR (A) and Western blot (B) show that successful silencing of WHSC1 4a occurred 24 h after transfection (T, transfected with short interfering RNA (siRNA) targeted against WHSC1 4a; C, nonspecific siRNA control; *, identical samples); after double treatment, the suppression of gene expression remained through 96 h. Methyl thiazole tetrazolium bromide assay (C) demonstrates a 23% maximal drop of cell proliferation in the experiment with one siRNA transfection at time 0 h (sgl T, purple curve) and a 70% drop in the twice-transfected experiment (dbl T, blue curve), compared with the nonspecific siRNA control groups (sgl ctrl and dbl ctrl). Experiments using U251 and U373 cell lines produced similar results (data not shown).
Conclusion
To examine the molecular basis of gliomagenesis, we used a newly developed proteomic technique to identify the glioma-specific proteome. In a representative follow-up of our proteomic findings, WHSC1 (as one of the 104 GBM-specific proteins) was examined in further investigations. WHSC1 expression level is correlated with glioma grade, and it appears to be directly involved in the proliferative capacity of GBM cells in vitro. These findings suggest that the novel combination of selective tissue dissection/CIEF-nRPLC-MS/MS is a reliable technique to identify promising protein candidates that may serve for the diagnosis, prognosis, and therapy of clinical diseases.
Supplementary Material
Acknowledgment
This research was supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke, NIH.
References
- 1.Furuta M, Weil RJ, Vortmeyer AO, et al. Protein patterns and proteins that identify subtypes of glioblastoma multiforme. Oncogene. 2004;23:6806–6814. doi: 10.1038/sj.onc.1207770. [DOI] [PubMed] [Google Scholar]
- 2.Washburn MP, Ulaszek R, Deciu C, Schieltz DM, Yates JR., III Analysis of quantitative proteomic data generated via multidimensional protein identification technology. Anal Chem. 2002;74:1650–1657. doi: 10.1021/ac015704l. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Rudnick PA, Evans EL, et al. Proteome analysis of microdissected tumor tissue using a capillary isoelectric focusing-based multidimensional separation platform coupled with ESI-tandem MS. Anal Chem. 2005;77:6549–6556. doi: 10.1021/ac050491b. [DOI] [PubMed] [Google Scholar]
- 4.Stec I, Wright TJ, Van Ommen GJ, et al. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet. 1998;7:1071–1082. doi: 10.1093/hmg/7.7.1071. [DOI] [PubMed] [Google Scholar]
- 5.Miyaki M, Konishi M, Tanaka K, et al. Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet. 1997;17:271–272. doi: 10.1038/ng1197-271. [DOI] [PubMed] [Google Scholar]
- 6.Keats JJ, Maxwell CA, Taylor BJ, et al. Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients. Blood. 2005;105:4060–4069. doi: 10.1182/blood-2004-09-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeRisi J, Penland L, Brown PO, et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 8.Shen Y, Jacobs JM, Camp DG, II, et al. Ultra-high-efficiency strong cation exchange LC/RPLC/MS/MS for high dynamic range characterization of the human plasma proteome. Anal Chem. 2004;76:1134–1144. doi: 10.1021/ac034869m. [DOI] [PubMed] [Google Scholar]
- 9.Wu SL, Hancock WS, Goodrich GG, Kunitake ST. An approach to the proteomic analysis of a breast cancer cell line (SKBR-3) Proteomics. 2003;3:1037–1046. doi: 10.1002/pmic.200300382. [DOI] [PubMed] [Google Scholar]
- 10.Seoane J, Le HV, Shen L, Anderson SA, Massague J. Integration of Smad and Forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 11.Hirvonen HE, Salonen R, Sandberg MM, et al. Differential expression of myc, max and RB1 genes in human gliomas and glioma cell lines. Br J Cancer. 1994;69:16–25. doi: 10.1038/bjc.1994.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll RS, Zhang J, Bello L, Melnick MB, Maruyama T, Black PM. KDR activation in astrocytic neoplasms. Cancer. 1999;86:1335–1341. doi: 10.1002/(sici)1097-0142(19991001)86:7<1335::aid-cncr32>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 13.Valter MM, Hugel A, Huang HJ, et al. Expression of the Ets-1 transcription factor in human astrocytomas is associated with Fms-like tyrosine kinase-1 (Flt-1)/vascular endothelial growth factor receptor-1 synthesis and neoangiogenesis. Cancer Res. 1999;59:5608–5614. [PubMed] [Google Scholar]
- 14.Wright TJ, Ricke DO, Denison K, et al. A transcript map of the newly defined 165 kb Wolf-Hirschhorn syndrome critical region. Hum Mol Genet. 1997;6:317–324. doi: 10.1093/hmg/6.2.317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.