Summary
Development of vaccines against severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) is crucial in the prevention of SARS reemergence. The receptor-binding domain (RBD) of SARS-CoV spike (S) protein is an important target in developing safe and effective SARS vaccines. Our previous study has demonstrated that vaccination with adeno-associated virus encoding RBD (RBD-rAAV) induces high titer of neutralizing antibodies. In this study, we further assessed the immune responses and protective effect of the immunization with RBD-rAAV prime/RBD-specific T cell peptide boost. Compared with the RBD-rAAV prime/boost vaccination, RBD-rAAV prime/RBD-peptide (RBD-Pep) boost induced similar levels of Th1 and neutralizing antibody responses that protected the vaccinated mice from subsequent SARS-CoV challenge, but stronger Th2 and CTL responses. No significant immune responses and protective effects were detected in mice vaccinated with RBD-Pep or blank AAV alone. Since T cell epitopes are highly conserved and boosting with peptides may induce the production of effector memory T cells, which may be effective against viruses with mutations in the neutralizing epitopes, our results suggest that the vaccination protocol used may be ideal for providing effective, broad and long-term protection against SARS-CoV infection.
Keywords: SARS-CoV, Receptor binding domain, Adeno-associated virus, Immune responses, Peptides to T cell epitopes
Introduction
Severe acute respiratory syndrome (SARS) is the latest-reported emerging infectious disease caused by SARS coronavirus (SARS-CoV). Since the end of the SARS outbreak in July, 2003, a number of SARS cases have been sporadically reported, raising a serious safety concern [1], [2], [3]. The presence of natural reservoir also suggests that there is a risk for reintroduction of SARS-CoV-like virus from animals into humans [4]. The development of prevention strategies, particularly the generation of effective and safe vaccines against SARS-CoV, therefore, is an important task in current SARS research.
Most existing vaccine candidates against SARS-CoV are based on the S protein. Studies have shown that vaccination of African green monkeys with an attenuated parainfluenza virus encoding SARS-CoV S protein results in the production of SARS-CoV-specific neutralizing antibodies, protecting vaccinated animals from subsequent SARS-CoV challenge [5]. It is also indicated that the S, among all structural proteins of SARS-CoV, is the only significant neutralization and protective antigen, and that a single mucosal immunization with S protein efficiently suppresses SARS-CoV infection [6]. Furthermore, the receptor-binding domain (RBD), a fragment of S protein, has been demonstrated to be a major neutralization determinant in the development of subunit vaccines against SARS [7], [8]. These reports suggest that S protein of SARS-CoV, especially RBD, is highly effective in prevention of SARS.
Recently, it has been reported that strategies in using combination of different vaccines improve humoral or cellular immune responses. Prime with SARS-CoV S-expressing DNA and boost with adenoviral vector encoding S induce optimal CD8 T cell immunity, while boost with the inactivated SARS-CoV plus adjuvant stimulate the CD4 T cell immunity and the antibody response [9]. The humoral immune response and IFN-γ secretion were also enhanced by priming with DNA-based vaccine and boosting with a recombinant adenovirus encoding N protein of SARS-CoV [10]. These reports demonstrate that an ideal SARS vaccine can be developed by rational design of appropriate prime-boost strategies.
We have previously reported that a recombinant adeno-associated virus encoding RBD of SARS-CoV S protein (RBD-rAAV) induced SARS-CoV-specific IgG antibody with neutralizing activity [11]. In the RBD region, a CD4+ T cell epitope, N60 (residues 435–444: NYNYKYRYLR) and a CD8+ T cell epitope, N50 (residues 365–374, KCYGVSATKL) have been identified, respectively [12], [13]. In this study, we primed with RBD-rAAV and boosted with RBD-peptides (RBD-Pep) specific for CD4+ and CD8+ T cell epitopes and compared the immune responses and protective effects to those vaccinated with RBD-rAAV or RBD-Pep alone in a mouse model.
Materials and methods
RBD-rAAV and RBD-Pep
RBD-rAAV was produced as previously described [11]. Briefly, RBD-rAAV plasmid was co-transfected with pHelper and pAAV-RC plasmids into HEK293T cells using a calcium phosphate transfection method (Stratagene, USA) according to the manufacturer's protocol. Transfected cells and supernatant were harvested 72 h post-transfection. After purification and titration as described previously [11], RBD-rAAV was adjusted to 1012 viral particles (VP)/ml in PBS. Peptides N50 (CD8+ T cell epitope) and N60 (CD4+ T cell epitope) derived from RBD of SARS-CoV S protein [13], plus CpG oligodeoxynucleotide (ODN) 1826 (TCC ATG ACG TTC CTG ACG TT) (RBD-Pep) were used for prime and boost immunizations in this study.
Animals and vaccinations
Female BALB/c mice at the age of 4–6 weeks were used for the vaccination. All mice for the study were purchased from Laboratory Animal Unit (LAU) in the University of Hong Kong. Animals were housed in the animal facility of the Department of Microbiology, University of Hong Kong, and maintained in accordance with the animal care protocol. All of the animal studies were approved by the Department of Health, the Government of Hong Kong Special Administration Region (SAR).
Mice were vaccinated following protocols described previously [11], [14]. Mice were separated into four groups (nine mice per group) and primed with RBD-rAAV [intramuscular (i.m.), 2 × 1011 VP/200 μl)] or RBD-pep (N50 and N60, 50 μg each) plus CpG ODN (25 μg) [subcutaneous (s.c.)] or blank AAV, and boosted with RBD-rAAV or RBD-Pep or AAV, respectively. Each group was further divided into two subgroups (five mice in subgroup 1 and four mice in subgroup 2). Forty days after the first immunization, the mice in the subgroup 1 were anaesthetized with diethyl ether. Their sera were collected by retro-orbital puncture for detection of IgG and NA by ELISA and neutralizing assay, respectively. The spleens were harvested for detection of T cell immunity by ELISPOT and flow cytometry analysis. The mice in the subgroup 2 were challenged with SARS-CoV and lung samples were collected for viral load detection 3 days post-challenge (Fig. 1 ).
Figure 1.
Immunization and detection procedures. Mice of four groups were primed with RBD-rAAV, or RBD-peptides (N50 and N60) plus CpG ODN (RBD-Pep), or blank AAV, and boosted with RBD-rAAV, RBD-Pep or AVV, respectively. Ten days post-boost, sera and spleens were collected from the mice in subgroup 1 for detection of IgG and NA titers by ELISA and neutralization assay, respectively, and for detection of Th and CTL responses by ELISPOT and flow cytometry analysis. The mice in subgroup 2 were challenged with SARS-CoV, and the viral loads in lung tissues were determined 3 days later.
Detection of specific CD4 and CD8 T cells by ELISPOT assay
Specific CD4 and CD8 T cells in vaccinated mice were detected by ELISPOT mouse kit (Mabtech, Sweden) according to the manufacturer's protocol. In brief, 96-well ELISPOT plates were coated with anti-IL-4, -IL-10 and -IFN-γ monoclonal antibodies (mAbs) overnight at 4 °C, and blocked by sterile RPMI-1640 containing 10% FBS for 2 h at room temperature. Single-cell suspensions prepared from spleens of vaccinated mice were added to the wells at the concentration of 2 × 105 cells/well. Cells were incubated for 24 h in the presence or absence of the RBD-pep N50 or N60 (1 μg/ml) plus anti-mouse-CD28 (1 μg/ml; BD Pharmingen, USA) at 37 °C with 5% CO2, followed by washes with PBS. The cells were sequentially incubated with biotinylated anti-mouse IL-4, IL-10 and IFN-γ mAbs at 1:1000 for 2 h at room temperature, streptavidin-conjugated horseradish peroxidase (HRP) for 1 h at room temperature and 3,3′,5,5′-tetramethylbenzidine (TMB) substrate solutions for around 15 min, with extensive washes between incubations. The spots of IL-4, IL-10 and IFN-γ-producing T cells were counted by ELISPOT reader system and ImmunoSpot 3 software (Cellular Technology Ltd., Ohio, USA).
Detection of specific CD8 T cells by intracellular cytokine staining and flow cytometry analysis
Specific CD8 T cells in vaccinated mice were further measured by intracellular cytokine staining and flow cytometry analysis. Single-cell suspensions (2 × 106) from spleens of vaccinated mice were stimulated with or without the RBD-pep N50 (1 μg/ml) plus anti-CD28 antibody (1 μg/ml). Phorbol myristate acetate (PMA, 5 ng/ml; Sigma, USA) and ionomycin (250 ng/ml; Sigma) were used as positive controls. Cells with stimulatory agents were incubated for 5 h at 37 °C with 5% CO2 in the presence of GolgiPlug™ containing Brefeldin A (1 μl/ml; BD Pharmingen). The cells were fixed using Cytofix/Cytoperm™ Plus kit in accordance with the manufacturer's protocol (BD Pharmingen), and stained directly with conjugated mAbs specific for cell surface antigens [anti-mouse-CD3 (PerCP) and anti-mouse-CD8 (APC)] and intracellular cytokines [anti-mouse-IL-2 (PE) and anti-mouse-IFN-γ (FITC); BD Pharmingen] for 30 min at 4 °C. Appropriate isotype-matched controls for cytokines were included in each staining. Data were acquired using a flow cytometer (FACSCaliber, BD, USA). Lymphocyte population was gated by forward light scatter versus side light scatter, and 20,000 events for CD3+CD8+ lymphocyte subpopulation were acquired to determine the percentage of CD8+ T cells positive for specific cytokines. Data were analyzed by CellQuest software (BD Biosciences, USA).
Detection of SARS-CoV-specific IgG antibodies by ELISA
Specific IgG antibody against SARS-CoV in sera of vaccinated mice was tested by ELISA using the protocol described previously with some modification [11]. Briefly, serially diluted mouse sera were added to 96-well microtiter plates pre-coated with the protein mixture from SARS-CoV viral lysates. The plates were incubated at 37 °C for 30 min, followed by four washes with PBS containing 0.1% Tween 20 (PBS-T). Bound antibodies were then reacted with HRP-conjugated goat anti-mouse IgG (DAKO, Denmark) at 37 °C for 20 min. After four washes, TMB substrate solutions were added to the plates and the reaction was stopped by adding 1N H2SO4. The absorbance at 450 nm was measured by an ELISA plate reader (Victor 1420 Multilabel Counter, PerkinElmer, USA).
Detection of neutralizing antibodies by neutralization assay
Titers of NA against SARS-CoV in sera of vaccinated mice were measured by neutralization assay in Vero E6 cells as previously described [11], [15], [16]. Briefly, cells were seeded at 104/well in 96-well culture plates and cultured at 37 °C to form a monolayer. Serial two-fold dilutions of serum samples were separately mixed with 100 TCID50 of SARS-CoV GZ50 strain (GenBank accession no. AY304495), incubated at 37 °C for 1 h, and added to the monolayer of Vero E6 cells in tetraplicate. Cells infected with 100 TCID50 SARS-CoV and without the virus were applied as positive and negative controls, respectively. Cytopathic effect (CPE) in each well was observed daily and recorded on day 3 post-infection. The neutralizing titers of mouse antisera that completely prevented CPE in 50% of the wells were calculated by Reed–Muench method.
SARS-CoV challenge in the mouse model
Forty days post-vaccination, mice were anaesthetized with isoflurane and i.n. inoculated with 50 μl of SARS-CoV strain GZ50 (5 × 105 TCID50) according to the national animal care and use guidelines in an approved animal BSL-3 laboratory. The mice were sacrificed 3 days after virus challenge and mouse lungs were removed. The lung tissues were stored at −80 °C for virological tests.
Q-RT-PCR
The viral RNA copies in lung tissues of challenged mice were determined by quantitative reverse-transcriptase polymerase chain reaction (Q-RT-PCR) according to the protocol described previously with some modification [15], [17], [18]. Briefly, total RNA was extracted from 20 mg of lung tissues using RNeasy Mini kit (Qiagen Inc, USA). cDNA was synthesized using random primers and SuperScript II RT kit (Invitrogen, USA). Extracted RNA (10 μl) was reverse transcribed in a 20 μl reaction mixture containing 1× first strand buffer, 100 mM DTT, 10 mM each dNTP, 50 ng random primers, 40 U RNaseOUT, and 200 U SSIIRT at 42 °C for 50 min, followed by 15 min at 70 °C. The solution was incubated with RNase H (Invitrogen, USA) at 37 °C for 20 min. Synthesized cDNA was quantified using Power SYBR Green PCR Master Mix (Applied Biosystems, USA) in 20 μl mixture containing 5 μl cDNA (1:10), 10 μl 2× Power SYBR Green PCR Master Mix, 3 μl RNase-free H2O, 10 μM forward primer F (5′-GCTTAGGCCCTTTGAGAGAGACA-3′) and reverse primer R (5′-GCCAATGCCAGTAGTGGTGTAA-3′) in Mx3000 QPCR System (Stratagene, USA).
Statistical analysis
Values were presented as mean with standard error (SE). Statistical significance among different vaccination groups was calculated by Student's t test using Stata statistical software. The P values less than 0.05 were considered significant.
Results
RBD-rAAV prime/RBD-Pep boost induced both Th1 and Th2 responses
Th1 and Th2 responses induced by vaccinations were evaluated by detection of IFN-γ-producing cells (Th1) and IL-4-/IL-10-secreting cells (Th2) in splenocyte cultures of vaccinated mice 24 h post-stimulation with N60, a peptide corresponding to the CD4+ T cell epitope in RBD. As shown in Fig. 2 , RBD-rAAV prime/RBD-Pep boost vaccination induced similar frequencies of IFN-γ-producing cells (Th1) as those of RBD-rAAV prime/RBD-rAAV boost (P > 0.05), which were significantly higher than those of other groups (P < 0.05) (Fig. 2A). RBD-rAAV prime/RBD-Pep boost immunization elicited a higher level (P < 0.05) of IL-4-producting Th2 cells which are responsible for up-regulation of immune responses, but a lower level (P < 0.05) of IL-10-secreting Th2 cells that play roles in down-regulation of immune responses, as compared to those of RBD-rAAV prime/RBD-rAAV boost vaccination (Fig. 2B, 2C). However, boost doses of RBD-Pep or blank AAV did not induce significant generation of IL-4- and IL-10-producing Th2 cells in the vaccinated mice (Fig. 2B and C). The above results suggested that RBD-rAAV prime/RBD-pep boost vaccination was able to induce similar Th1 and stronger Th2 responses than RBD-rAAV prime/RBD-rAAV boost vaccination.
Figure 2.
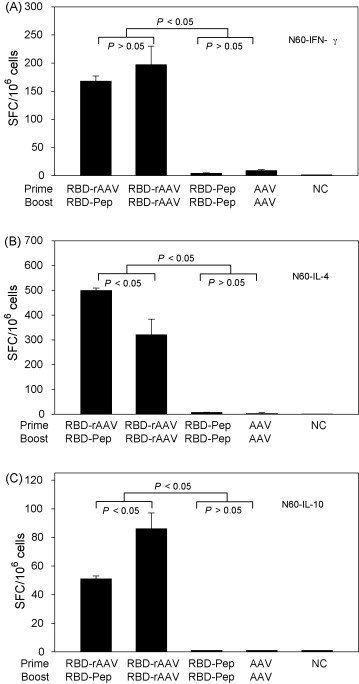
Detection of SARS-CoV RBD-specific Th responses by ELISPOT. Splenocytes from vaccinated mice were stimulated with RBD-specific CD4+ T cell peptide N60 plus anti-CD28 for 24 h. Anti-CD28 alone was applied as the negative control (NC). Frequencies of cytokine-producing cells are expressed as mean ± SE of cytokine spot-forming cells (SFC)/106 cells of 5 independent experiments. (A) Detection of IFN-γ-producing CD4+ (Th1) cells. (B) Detection of IL-4-producing CD4+ (Th2) cells. (C) Detection of IL-10-producing CD4+ (Th2) cells.
RBD-rAAV prime/RBD-Pep boost vaccination elicited strong CTL response
To examine the induction of CTL responses by different vaccination protocols, cells from spleens of vaccinated mice were stimulated with or without RBD-pep N50 (CD8+ T cell epitope) and detected for IFN-γ- and IL-2-producing cells by ELISPOT. As shown in Fig. 3A, RBD-rAAV prime/RBD-Pep boost vaccination induced the highest level of IFN-γ-secreting cells (197 SFC/106 splenocytes), seconded by RBD-rAAV prime/RBD-rAAV boost vaccination (123 SFC/106 splenocytes). Both RBD-Pep and RBD-rAAV prime/RBD-rAAV boost vaccinations elicited similar levels of IL-2-secreting cells, which were significantly higher than other vaccinations (P < 0.05) (Fig. 3B). The CTL responses were further confirmed by detection of IFN-γ- and IL-2-producing CD8+ T lymphocytes using cell surface markers and intracellular cytokine staining followed by flow cytometry analysis (Fig. 4 ). The results demonstrated that RBD-rAAV prime/RBD-Pep boost vaccination induced higher level of IFN-γ-producing CD8+ T cells (0.51%) than that of RBD-rAAV prime/RBD-rAAV boost (0.28%), while IL-2-producing CD8+ T lymphocytes induced by these two vaccinations were similar. Nevertheless, no or very low levels of IFN-γ- and IL-2-secreting CD8+ T cells were detected in splenocytes from mice with RBD-Pep prime/RBD-Pep boost, and AAV prime/AAV boost vaccinations. These results demonstrated that RBD-rAAV prime/RBD-pep boost induced stronger CTL response, especially IFN-γ-producing CD8+ T lymphocytes, than other vaccinations.
Figure 3.
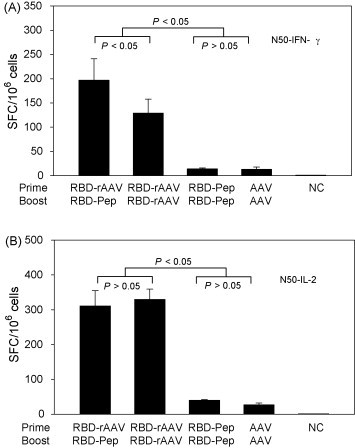
Detection of SARS-CoV RBD-specific CTL response by ELISPOT. Splenocytes from vaccinated mice were stimulated with SARS-CoV RBD-specific CD8+ T cell peptide N50 plus anti-CD28 for 24 h. Anti-CD28 alone was applied as the negative control (NC). Frequencies of IFN-γ-producing cells are expressed as mean ± SE of cytokine spot-forming cells (SFC)/106 cells of 5 independent experiments. (A) Detection of IFN-γ-producing CD8+ (CTL) cells. (B) Detection of IL-2-producing CD8+ (CTL) cells.
Figure 4.
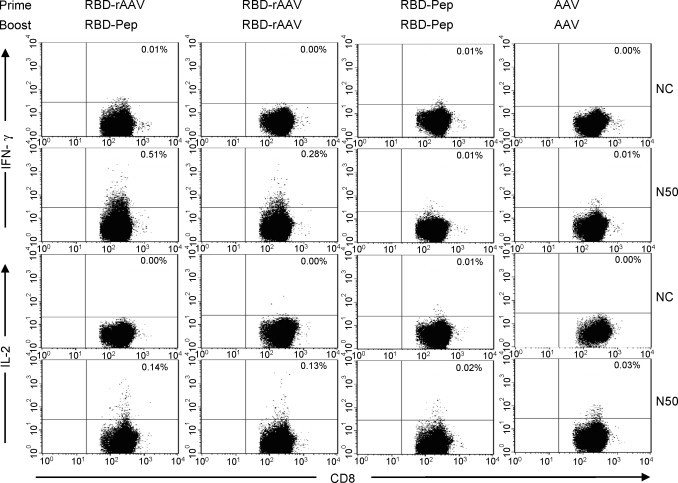
Detection of CTL responses by flow cytometry analysis. Specific CTL responses in vaccinated mice were further detected for frequencies of IL-2 and IFN-γ-producing CD8+ T cells in splenocytes by cell surface marker and intracellular cytokine triple-staining using flow cytometry analysis. Cells were incubated with RBD-peptide N50 plus anti-CD28. Anti-CD28 alone was used as the negative control (NC). At least 20,000 CD3+/CD8+ lymphocytes were first gated, and frequencies of IFN-γ+ and IL-2+ cells were then analyzed and indicated as percentages of CD3+/CD8+ T cells. The graphs are presented as mean value of five independent experiments. Numbers in the upper right corner of each graph represent the frequencies of IFN-γ- or IL-2-producing CD8+ T cells.
RBD-rAAV prime/RBD-Pep boost induced SARS-CoV-specific IgG and neutralizing antibody responses
Humoral immune responses in vaccinated mice were evaluated by detection of SARS-CoV-specific serum IgG titers using ELISA. As shown in Fig. 5A, RBD-rAAV prime/RBD-Pep boost was able to induce high level of SARS-CoV-specific IgG production, in consistent with RBD-rAAV prime/RBD-rAAV boost vaccination. However, RBD-Pep prime/RBD-Pep boost only induced low levels of SARS-CoV-specific antibodies. RBD-rAAV prime/RBD-Pep boost could also induce similar level of neutralizing antibody against SARS-CoV as that of the RBD-rAAV prime/RBD-rAAV boost vaccination (Fig. 5B). In contrast, the titer of neutralizing antibody induced by RBD-Pep prime/RBD-Pep boost vaccination was significantly lower than the above two vaccination strategies (P < 0.05). Similar to serum IgG, no significant neutralizing antibody was detected in mice immunized with AAV prime/AAV boost (Fig. 5A and B). The above results indicated that RBD-rAAV prime with RBD-Pep boost vaccination could induce strong neutralizing antibody response.
Figure 5.
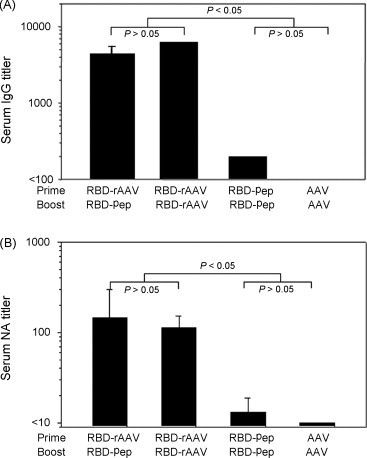
Detection of antibody responses in vaccinated mice. Vaccinated mice were detected for humoral immune responses 40 days after the first vaccination. (A) SARS-CoV-specific IgG titer in serum samples was detected by ELISA. The data are presented as mean ± SE of five mice per group. (B) Titer of serum neutralizing antibody was measured by neutralization assay. The titers were determined as the highest dilutions of sera that could completely prevent CPE in at least 50% of the wells, and presented as mean ± SE of five mice each group.
RBD-rAAV prime/RBD-Pep boost provided strong protection against SARS-CoV challenge
Protective efficacies of vaccinations with different protocols were further investigated in mice (4 mice/group) intranasally challenged with SARS-CoV strain GZ50 40 days post-vaccination. Mice were sacrificed 3 days after the challenge, and virus replication in lung tissues was detected by Q-RT-PCR. It was shown in Fig. 6 that SARS-CoV viral load in lung tissues was significantly reduced in mice vaccinated with RBD-Pep. Very low level of viral load was detected in lung tissues of RBD-rAAV prime/RBD-Pep boost group, similar to that in lung tissues of RBD-rAAV prime/RBD-rAAV boost group. Notably, boost doses of RBD-Pep and blank AAV did not provide protection to SARS-CoV challenge, with significantly higher level of viral load (P < 0.05, Fig. 6). These results demonstrated that like boost doses of RBD-rAAV, vaccination of RBD-rAAV prime/RBD-pep boost was able to significantly inhibit SARS-CoV infection.
Figure 6.
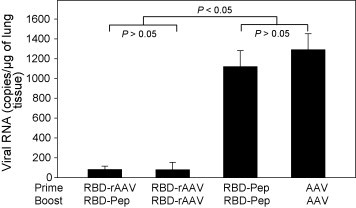
Detection of viral replication in lung tissues of SARS-CoV-challenged mice. Vaccinated mice in subgroup 2 were challenged with SARS-CoV. Viral loads in the mouse lung tissues were detected by Q-RT-PCR 3 days after the challenge. The data are expressed as mean ± SE of viral RNA copies/μg of lung tissues of four mice each group.
Discussion
Development of vaccines is needed for prevention of SARS in case of the disease reemergence. Most reported vaccines against SARS-CoV are based on the full-length S protein or its fragments [19], [20], but vaccination with MVA-based vaccine containing the full-length S protein may cause enhanced hepatitis in the vaccinated animals [21]. In this regard, fragments of SARS-CoV S protein are more practical and safer for developing vaccines. It has been shown that the S1 domain, especially the RBD region, is the main fragment and major determinant in eliciting NA against SARS-CoV [7], [8]. In previous studies, we have also demonstrated that candidate vaccines based on the RBD induced high titers of neutralizing antibody and prevented subsequent SARS-CoV challenge in vaccinated mice [11], [15]. These suggest that RBD of SARS S protein is an important target in developing safe and effective SARS vaccines [22], [23], [24].
Prime-boost vaccinations have been used as a strategy for development of SARS vaccines. DNA vaccines boosted with inactivated virus or viral protein fragments have been shown to improve immune responses, including neutralizing antibody response, against SARS-CoV [9], [25], [26]. It was also reported that the immunization by priming with a DNA plasmid encoding SARS-CoV N protein (pcDNA3.1-N) and boosting with an adenoviral vector expressing the same protein (rAd-N) effectively elicited strong SARS-CoV-N-specific humoral and cellular immune responses in vaccinated mice [27]. In this study, we further demonstrated that the immune responses could be enhanced by priming with RBD-rAAV and boosting with RBD-pep corresponding to the T cell epitopes in the RBD region of SARS-CoV S protein. Compared with the RBD-rAAV prime/boost vaccination, RBD-rAAV prime/RBD-Pep boost induced similar levels of Th1 and neutralizing antibody responses but stronger Th2 and CTL responses (Figure 2, Figure 3, Figure 4, Figure 5). Like the RBD-rAAV prime/boost vaccination, the RBD-rAAV prime/RBD-pep boost vaccination could also induce potent protective activity in mice against SARS-CoV challenge (Fig. 6). However, no significant immune responses and protective effect were detected in mice vaccinated with RBD-pep or blank AAV alone. These results indicated that RBD-rAAV prime/RBD-Pep boost vaccination can greatly promote cellular immune responses and neutralizing antibody production in the vaccinated mice, which may play important roles in the suppression of SARS-CoV infection in the challenged mice.
Recombinant AAV (rAAV)-based vaccine has been shown to be a promising vaccine candidate due to its ability in inducing antigen-specific humoral and/or cellular immune responses against other pathogens [28], [29], [30]. However, i.m. vaccination with rAAV, especially repeated doses, might induce a humoral immune response against viral AAV capsid proteins [31], affecting the immune efficacy of rAAV-based vaccines. In this respect, RBD-rAAV prime with specific peptide boost may prevent the disadvantages induced by repeated i.m. RBD-rAAV vaccinations, while eliciting similar or even higher immune responses and protective effects.
One key issue for vaccine development is how to provide cross-protection against infection by rapidly mutated viruses, because the mutants may escape from protective effects of the vaccination. Generally, T cell epitopes are much more conserved than neutralizing antibody epitopes. We have compared amino acid sequences of N50 and N60 peptides, which were used for boost vaccination, between 25 human SARS-CoV strains and 5 civet cat SARS-CoV strains deposited in Genebank. N50 is completely conserved in these strains, and only one strain (Sino1-11) shows a single amino acid change in N60. Thus, the strategy of boosting with T cell peptide may plausibly provide cross-protection against quickly mutated viruses. In this study, both RBD-rAAV prime/RBD-pep boost and RBD-rAAV prime/boost vaccinations provided similar protection against subsequent SARS-CoV challenge, which may be attributed to the neutralizing antibody responses against the challenged virus induced by both strategies. However, the former may probably induce more effective protection against infection by viruses with mutations in the neutralizing epitopes than the latter because the former can elicit significantly stronger specific CTL response than the latter.
Another important issue for vaccine development is whether the vaccination can provide long-term protection. Our previous results showed that majority of the N50- and N60-specific IFN-γ-producing T cells were IL-7Rα+ (CD127) and CD62L− in both CD4+ and CD8+ T cell populations [13]. Our present work also demonstrated that the peptide N50 boost vaccination induced much higher level of CD8+ IFN-γ-producing T cells. It has been found that IL-7Rα+ antigen-specific T cells preferentially develop into long-term (effector) memory T cells [32] and effector memory T cells predominantly persist in peripheral sites, the key portals of entry for pathogens [33]. Thus, vaccination with RBD-rAAV prim/RBD-pep boost may provide long-term protection against SARS-CoV.
In conclusion, prime with RBD-rAAV and boost with T cell epitope peptides may be an ideal vaccine strategy to provide effective, cross- and long-term protective effects against infection by SARS-CoV, including those bearing mutations in the neutralizing epitopes.
Acknowledgements
This study was supported by the Research Fund for the Control of Infectious Diseases, the Food and Health Bureau of the Hong Kong SAR Government, by the National 973 Basic Research Program of China (2005CB523001), and by the National Institutes of Health (NIH) of the United States (RO1 AI68002).
Contributor Information
Yusen Zhou, Email: yszhou@nic.bmi.ac.cn.
Bo-Jian Zheng, Email: bzheng@hkucc.hku.hk.
References
- 1.Orellana C. Laboratory-acquired SARS raises worries on biosafety. Lancet Infect Dis. 2004;4(2):64. doi: 10.1016/S1473-3099(04)00911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Normile D. Infectious diseases Mounting lab accidents raise SARS fears. Science. 2004;304(5671):659–661. doi: 10.1126/science.304.5671.659. [DOI] [PubMed] [Google Scholar]
- 3.Normile D. Infectious diseases SARS experts want labs to improve safety practices. Science. 2003;302(5642):31. doi: 10.1126/science.302.5642.31a. [DOI] [PubMed] [Google Scholar]
- 4.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 5.Bukreyev A., Lamirande E.W., Buchholz U.J., Vogel L.N., Elkins W.R., St Claire M. Mucosal immunisation of african green monkeys (cercopithecus aethiops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS. Lancet. 2004;363(9427):2122–2127. doi: 10.1016/S0140-6736(04)16501-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchholz U.J., Bukreyev A., Yang L., Lamirande E.W., Murphy B.R., Subbarao K. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc Natl Acad Sci USA. 2004;101(26):9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He Y., Lu H., Siddiqui P., Zhou Y., Jiang S. Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies. J Immunol. 2005;174(8):4908–4915. doi: 10.4049/jimmunol.174.8.4908. [DOI] [PubMed] [Google Scholar]
- 8.He Y., Zhou Y., Liu S., Kou Z., Li W., Farzan M. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem Biophys Res Commun. 2004;324(2):773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong W.P., Xu L., Stadler K., Ulmer J.B., Abrignani S., Rappuoli R. Modulation of the immune response to the severe acute respiratory syndrome spike glycoprotein by gene-based and inactivated virus immunization. J Virol. 2005;79(22):13915–13923. doi: 10.1128/JVI.79.22.13915-13923.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma C., Yao K., Zhou F., Zhu M. Comparative immunization in BALB/c mice with recombinant replication-defective adenovirus vector and DNA plasmid expressing a SARS-CoV nucleocapsid protein gene. Cell Mol Immunol. 2006;3(6):459–465. [PubMed] [Google Scholar]
- 11.Du L., He Y., Wang Y., Zhang H., Ma S., Wong C.K. Recombinant adeno-associated virus expressing the receptor-binding domain of severe acute respiratory syndrome coronavirus S protein elicits neutralizing antibodies: implication for developing SARS vaccines. Virology. 2006;353(1):6–16. doi: 10.1016/j.virol.2006.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhi Y., Kobinger G.P., Jordan H., Suchma K., Weiss S.R., Shen H. Identification of murine CD8 T cell epitopes in codon-optimized SARS-associated coronavirus spike protein. Virology. 2005;335(1):34–45. doi: 10.1016/j.virol.2005.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J., Cao Y.N., Du J.L., Bu X.Z., Ma R., Wu C.Y. Priming with SARS CoV S DNA and boosting with SARS CoV S epitopes specific for CD4 + and CD8 + T cells promote cellular immune responses. Vaccine. 2007;25(39–40):6981–6991. doi: 10.1016/j.vaccine.2007.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mancheno-Corvo P., Martin-Duque P. Viral gene therapy. Clin Transl Oncol. 2006;8(12):858–867. doi: 10.1007/s12094-006-0149-y. [DOI] [PubMed] [Google Scholar]
- 15.Du L., Zhao G., He Y., Guo Y., Zheng B.J., Jiang S. Receptor-binding domain of SARS-CoV spike protein induces long-term protective immunity in an animal model. Vaccine. 2007;25(15):2832–2838. doi: 10.1016/j.vaccine.2006.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng B.J., Wong K.H., Zhou J., Wong K.L., Young B.W., Lu L.W. SARS-related virus predating SARS outbreak, Hong Kong. Emerg Infect Dis. 2004;10(2):176–178. doi: 10.3201/eid1002.030533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng B.J., Guan Y., Tang Q., Du C., Xie F.Y., He M.L. Prophylactic and therapeutic effects of small interfering RNA targeting SARS-coronavirus. Antivir Ther. 2004;9(3):365–374. [PubMed] [Google Scholar]
- 18.Chan V.S., Chan K.Y., Chen Y., Poon L.L., Cheung A.N., Zheng B. Homozygous L-SIGN (CLEC4M) plays a protective role in SARS coronavirus infection. Nat Genet. 2006;38(1):38–46. doi: 10.1038/ng1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keng C.T., Zhang A., Shen S., Lip K.M., Fielding B.C., Tan T.H. Amino acids 1055 to 1192 in the S2 region of severe acute respiratory syndrome coronavirus S protein induce neutralizing antibodies: implications for the development of vaccines and antiviral agents. J Virol. 2005;79(6):3289–3296. doi: 10.1128/JVI.79.6.3289-3296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428(6982):561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weingartl H., Czub M., Czub S., Neufeld J., Marszal P., Gren J. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J Virol. 2004;78(22):12672–12676. doi: 10.1128/JVI.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He Y., Li J., Heck S., Lustigman S., Jiang S. Antigenic and immunogenic characterization of recombinant baculovirus-expressed severe acute respiratory syndrome coronavirus spike protein: implication for vaccine design. J Virol. 2006;80(12):5757–5767. doi: 10.1128/JVI.00083-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang S., He Y., Liu S. SARS vaccine development. Emerg Infect Dis. 2005;11(7):1016–1020. doi: 10.3201/eid1107.050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He Y., Jiang S. Vaccine design for severe acute respiratory syndrome coronavirus. Viral Immunol. 2005;18(2):327–332. doi: 10.1089/vim.2005.18.327. [DOI] [PubMed] [Google Scholar]
- 25.Woo P.C., Lau S.K., Tsoi H.W., Chen Z.W., Wong B.H., Zhang L. SARS coronavirus spike polypeptide DNA vaccine priming with recombinant spike polypeptide from Escherichia coli as booster induces high titer of neutralizing antibody against SARS coronavirus. Vaccine. 2005;23(42):4959–4968. doi: 10.1016/j.vaccine.2005.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zakhartchouk A.N., Liu Q., Petric M., Babiuk L.A. Augmentation of immune responses to SARS coronavirus by a combination of DNA and whole killed virus vaccines. Vaccine. 2005;23(35):4385–4391. doi: 10.1016/j.vaccine.2005.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chunling M., Kun Y., Jian X., Jian Q., Hua S., Minsheng Z. Enhanced induction of SARS-CoV nucleocapsid protein-specific immune response using DNA vaccination followed by adenovirus boosting in BALB/c mice. Intervirology. 2006;49(5):307–318. doi: 10.1159/000094247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xin K.Q., Ooki T., Mizukami H., Hamajima K., Okudela K., Hashimoto K. Oral administration of recombinant adeno-associated virus elicits human immunodeficiency virus-specific immune responses. Hum Gene Ther. 2002;13(13):1571–1581. doi: 10.1089/10430340260201662. [DOI] [PubMed] [Google Scholar]
- 29.Kuck D., Lau T., Leuchs B., Kern A., Muller M., Gissmann L. Intranasal vaccination with recombinant adeno-associated virus type 5 against human papillomavirus type 16 L1. J Virol. 2006;80(6):2621–2630. doi: 10.1128/JVI.80.6.2621-2630.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z., Allen J.M., Riddell S.R., Gregorevic P., Storb R., Tapscott S.J. Immunity to adeno-associated virus-mediated gene transfer in a random-bred canine model of duchenne muscular dystrophy. Hum Gene Ther. 2007;18(1):18–26. doi: 10.1089/hum.2006.093. [DOI] [PubMed] [Google Scholar]
- 31.Riviere C., Danos O., Douar A.M. Long-term expression and repeated administration of AAV type 1, 2 and 5 vectors in skeletal muscle of immunocompetent adult mice. Gene Ther. 2006;13(17):1300–1308. doi: 10.1038/sj.gt.3302766. [DOI] [PubMed] [Google Scholar]
- 32.Kaech S.M., Tan J.T., Wherry E.J., Konieczny B.T., Surh C.D., Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4(12):1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 33.Roberts A.D., Ely K.H., Woodland D.L. Differential contributions of central and effector memory T cells to recall responses. J Exp Med. 2005;202(1):123–133. doi: 10.1084/jem.20050137. [DOI] [PMC free article] [PubMed] [Google Scholar]


