IN the fall of 1986 my colleague, Dick Beeman, sat in my office at Kansas State University and told me a fascinating story. He worked at what was then called the U. S. Grain Marketing Research Laboratory, a U. S. Department of Agriculture facility about a mile from campus. He was trained as an insect toxicologist, but to aid in his research he had become a self-taught geneticist using the red flour beetle, Tribolium castaneum. Beeman told me about interesting data he had regarding the juxtaposition in Tribolium of the apparent orthologs of genes in the Antennapedia and bithorax complexes (Beeman 1987). He had heard me speak about the Drosophila complexes and recombinationally mapped putative orthologs in Tribolium. This interaction inspired my involvement, with many others, in the development of Tribolium as a genetic model system, and the use of this beetle for work on the evolution of developmental mechanisms (evo-devo). I will provide evidence that Tribolium now represents the third best (after Drosophila and Caenorhabditis) invertebrate for genetic and molecular studies. I will argue for the importance of genetically tractable insect systems other than Drosophila, especially given the derived nature of fly morphology. Although it is specialized in many ways, Tribolium is nevertheless relatively ancestral with regard to many morphological features and developmental events. Somewhat artificially, I am going to separate its growth as a model system from its contributions to evo-devo, where I will concentrate on studies of the Hox cluster.
EARLY TRIBOLIUM STUDIES
As interesting as I found Beeman's results, it took a bit more to convince me of Tribolium's potential as a genetic model system. Beeman made me aware of the fact that T. castaneum (and the closely related species T. confusum) had long been popular experimental organisms. Tribolium studies largely arose because of their ease of culture, relatively short generation time, and status as globally distributed pests of stored grain and grain products. Earlier work on Tribolium focused largely on population ecology (e.g., Park 1934), population genetics (e.g., Levene et al. 1965), and quantitative genetics (e.g., Englert and Bell 1970). There were extensive analyses of life history traits; inter- and intraspecific competition (including cannibalism and conditioning of media); and a number of aspects of nutrition, behavior, and physiology. Tribolium was also the object of extensive research on insecticide resistance. Much information about earlier studies is summarized in a three-volume series authored by Alexander Sokoloff (1972, 1974, 1977).
Sokoloff was also the father of Tribolium genetics. At the University of California at Berkeley and at California State University at San Bernardino, he had worked hard to advance Tribolium in the Drosophila tradition, reviewing what was known about Tribolium genetics in Sokoloff (1966) and then the trilogy mentioned above, which was designed to emulate Demerec's (1950) Biology of Drosophila. He also edited a long-standing series titled Tribolium Information Bulletin, similar to the Drosophila Information Service. I was very impressed by Sokoloff's account of the large amount known about Tribolium anatomy, development, physiology, population genetics, and population ecology. However, I had spent decades doing Drosophila genetics, and I was afflicted with an all-too-common arrogance with respect to genetic studies in other insects. In 1966 there were ∼100 variants for Tribolium and approximately another 60 by 1977. Many were incompletely penetrant, and most were poorly mapped. Further, from my Drosophila-centric view, it was hard to conceive of doing sophisticated genetics without polytene chromosomes. (Somehow I failed to note that many geneticists used other organisms effectively in their absence!)
I was mistaken in comparing Tribolium to Drosophila, rather than focusing on how far ahead of other non-drosophilid insects it was. However, Beeman quickly convinced me otherwise. Tribolium shares with flies ease of culture, a relatively rapid life cycle (albeit closer to a month than 2 weeks), and facile genetic crosses using males and virgin females separated as pupae. (Sperm precedence also results in progeny largely sired by the last male to mate with a multiply inseminated female.) The results mentioned above clearly indicated interesting existing variants, and Beeman had shown that he could perform high-resolution recombinational mapping. Moreover, Beeman had embraced the concept of crossover-suppressing balancers. Beeman et al. (1986) also had demonstrated that they could isolate numerous radiation-induced translocations by detecting pseudolinkage. These show suppression of crossing over associated with breakpoints, as predicted (e.g., Roberts 1970). At that time there was very limited usage of balancers outside of flies, although more recently balancers have made an impact on worm (Edgley et al. 1995) and mouse (Hentges and Justice 2004) genetics. Eventually, balancers were generated for about one-half of the Tribolium genome (Brown et al. 2003). Beeman et al. (1996) went on to demonstrate the efficacy of using a balancer to isolate autosomal recessive-lethal mutations. We also continued to isolate induced and spontaneous viable mutations at a steady rate, including a screen to detect mutations that failed to complement a deficiency of the Hox region (Brown et al. 2000).
Although I began collaborative work with Beeman on Tribolium, my main commitment was still to flies. After characterization of the Antennapedia complex (ANTC; see Denell 1994), much of my work focused on larval phenotypic analysis of Hox and other homeotic mutants. I realized that I needed to add molecular approaches to my program to remain viable. Thus, I spent a sabbatical year at the University of Washington to become a molecular biologist. Unfortunately, I showed a clear lack of talent for it at the bench. There were a couple of bright points, however. Rick Garber taught me to do immunohistochemistry on whole-mount embryos, and I met Lisa Nagy, who was a graduate student with Lynn Riddiford. Nagy was studying the expression of a Hox gene ortholog in the tobacco hornworm Manduca sexta (Nagy et al. 1991). She patiently told me about the work of the German insect embryologists Gerhard Krause (1939) and Klaus Sander (1976) on comparative studies of segmentation. Drosophila represents an extreme example of a long germ embryo, while Tribolium lies at the relatively short end of the spectrum. Together with the highly advanced nature of larval morphology in Drosophila (see below), this difference provided the rationale for comparative studies of the developmental genetics of Tribolium and Drosophila (Denell 1987). Of course, there are additional major rationales for genetic studies of Tribolium, including many physiological, nutritional, sensory, and ecological specializations as well as its importance as a model pest insect.
While I was essentially failing as a hands-on molecular biologist, an important compensating event occurred: Susan Brown joined the Manhattan beetle group. She had studied worm genetics as a graduate student but learned molecular biology as a postdoc. Brown would provide leadership in the development of Tribolium as a molecular genetic system.
Since examples of traditional model systems that turned out to have huge genomes, precluding facile molecular biology, were known, the first thing that Brown wanted to do was assess whether Tribolium would be amenable to such studies. She attacked this question using renaturation kinetics. (I remember asking Eric Davidson for advice on this approach, and he replied in astonishment that no one did that anymore and he had not thought about it in years!) Brown et al. (1990) showed that the genome is ∼2 pg in size, with >60% made up of unique sequences with a long-period repetitive dispersion pattern. Thus, its genome resembled Drosophila's and appeared to be quite tractable. This information opened the door for the construction and use of a variety of plasmid, phage, and BAC genomic libraries, as well as cDNA libraries (see Brown et al. 2003). In this pregenomic era, that allowed the cloning of dozens of genes and studies of their expression patterns (see below), as well as the construction of molecular maps of increasing resolution (Beeman and Brown 1999; Lorenzen et al. 2005).
SOPHISTICATED BEETLE GENETICS
The confluence of genetic, developmental, and molecular methods led to what I personally feel was a keystone article in showing the utility of Tribolium. Beeman et al. (1989) had described the adult phenotype and complementation behavior of 50 new dominant mutations affecting the Hox cluster. In a subsequent study of the Tribolium ortholog of abdominal-A (abd-A), Jeff Stuart et al. (1993) analyzed 25 mutations putatively affecting that gene. These included candidate recessive partial loss-of-function mutations and four classes of dominant mutations. One of these appeared to be a loss-of-function mutation and three appeared to be gain-of-function mutations that complemented the recessive lethality of the first class. Reversion mutagenesis of the latter three classes showed that they were indeed gain-of-function alleles. The interpretation of the loss-of-function mutations as haplo-insufficient was confirmed using a deficiency and a duplication of the gene. Finally, a portion of the gene was cloned and sequenced, its expression pattern described, and the phenotype of larvae homozygous for an apparent null allele ascertained. Although I will describe below the developmental significance of this work, I also maintain that it represents a landmark contribution due to its combination of genetic, developmental, and molecular studies in a new animal system, previously conceivable only in flies and worms.
While we were busy making and analyzing Hox mutants, two other groups undertook genomewide screens for new mutations in Tribolium. In Kathryn Anderson's lab at Berkeley, Ingrid Sulston isolated several mutations associated with segmental defects (Sulston and Anderson 1996), which showed that she inherited skills as a geneticist from her famous father. One mutant, jaws, proved to affect the ortholog of the Drosophila gene Krűppel (Cerny et al. 2005). Maderspacher et al. (1998) also isolated several mutations with pair-rule or gap gene phenotypes and developed protocols for the maintenance of unbalanced lethal mutations (Berghammer et al. 1999a). This work provided additional evidence of the genetic tractability of Tribolium.
THE PATH TO TRANSGENIC BEETLES
Marjorie Hoy and Sokoloff had constructed the Berkeley synthetic strain in the 1960s with a series of crosses between many laboratory strains. The purpose of this exercise was to reconstruct the equivalent of a wild-type strain to compare to inbred lines (Sokoloff 1977). Hoy and Sokoloff noted that the strain seemed unstable and yielded a number of spontaneous mutations, including many of the homeotic variants mapped by Beeman (1987). In retrospect, it seems likely that a hybrid dysgenesis phenomenon was involved (M. Hoy, personal communication).
Beeman had a large number of lines established from beetles collected from around the world, and based on the work at Berkeley we sought to identify a currently active transposable element via observation of hybrid dysgenesis in interstrain crosses. We wanted to use such an element to perform germline transformation and transposon mutagenesis. Although we identified no active transposons, we did find and characterize an interesting and novel maternal-effect selfish element (Medea); when it is present in a mother, only the offspring receiving the element survive (Beeman et al. 1992).
Successful germline transformation and subsequent transposon mutagenesis was accomplished through vectors created and characterized in Drosophila by Horn and Wimmer (2000). As predicted by these authors, a vector based on the lepidopteran transposable element piggyBac proved effective in a number of insect species, including Tribolium (Berghammer et al. 1999b; Lorenzen et al. 2003). In addition to opening the door for manipulation of the genome, transgenesis allowed the construction of a binary system consisting of an immobilized helper providing transposase and a donor element lacking this gene and carrying an eye-specific green fluorescent protein (GFP) reporter (Lorenzen et al. 2007). Thus, closely resembling the system used so effectively in Drosophila (Rubin and Spradling 1982), the helper can mobilize the donor element, and new insertions are stable after the helper segregates away. In this case, prior to hopping, the donor element is associated with expression of GFP in the eye and (due to an enhancer trap) in the muscle. Transpositions can be identified by loss of muscle expression but retention of eye expression, indicating that the element is still in the genome but not in its previous position. In collaboration with the laboratories of Ernst Wimmer, Martin Klingler, and Gregor Bucher in Germany, we screened thousands of new insertions for associated mutant phenotypes and enhancer traps. Most work in non-drosophilid insects focuses on the roles of candidate genes predicted from Drosophila. That of course leaves a huge void with respect to genes in other insects whose roles are not shared in flies. Mutagenesis in Tribolium is a crucial approach for recognizing this class of genes, and transposon mutagenesis of course makes the genes identified easily cloned. Enhancer traps can identify genes expressed in certain contexts, allowing their role to be assessed by RNA interference (RNAi; see below). They are also useful for recombinational mapping and could be important if a GAL4-like binary ectopic expression system (Phelps and Brand 1998) can be generated.
POWERFUL REVERSE GENETICS
The use of RNAi for functional studies has proven to be especially effective in Tribolium. Soon after the injection of dsRNA into Drosophila, embryos showed the possibility of gene knockdown at that stage (Kennerdell and Carthew 1998; Misquitta and Paterson 1999). Brown et al. (1999b) showed that the same approach is efficacious in Tribolium. Bucher et al. (2002) extended that observation to show that injection of dsRNA into the female abdomen resulted in effective knockdown during early development of their offspring. RNAi is very effective in worms because the dsRNA spreads systemically throughout the organism. Anecdotal evidence from Drosophila indicated that treatment at later life stages is not effective (see Miller et al. 2008), which discouraged this approach among investigators using other insects. However, Yoshi Tomoyasu showed that RNAi is apparently systemic in Tribolium and can be used effectively at any life stage (Tomoyasu and Denell 2004). This valuable technique has proven effective in many, but far from all, insects. In one exciting extension of this technique, RNAi using ingested dsRNA appears to have potential for controlling insect pests (Gordon and Waterhouse 2007).
ARGUMENTS FOR GENOMIC SEQUENCING
Brown, Denell, Beeman, and Richard Gibbs wrote a successful white paper proposing sequencing of the Tribolium genome to the National Human Genome Research Institute (see Brown et al. 2003). In addition to Tribolium's efficacy as a genetic and molecular model, they argued for sequencing based on its status as a major global pest of stored grain and cereal products, usefulness in evo-devo studies, importance as a genetic model for many medically and agriculturally important coleopteran species, and the likelihood that it would provide useful information linking other sequenced insect and vertebrate genomes. We worried that the fact that relatively few laboratories (largely in Manhattan, Kansas, and Germany) were taking genetic and molecular approaches to Tribolium research would doom our proposal. Thus, to paraphrase the famous line from the movie Field of Dreams, our strategy was “Build it and they will come.” The Tribolium genome was sequenced at the Baylor Human Genome Sequencing Center under the leadership of Stephen (“fringy”) Richards. In the end, the publication in Nature describing the genome was authored by >100 members of the Tribolium Genome Sequencing Consortium (Richards et al. 2008) and was accompanied by dozens of companion articles from many sources, demonstrating that they did indeed come.
TRIBOLIUM AND THE HOX CLUSTER
As noted above, Beeman (1987) mapped probable orthologs of genes in the Drosophila Antennapedia and bithorax complexes and showed that they form a single cluster in Tribolium, which he called the homeotic complex (HOM-C). He did not explicitly hypothesize that the single complex was ancestral, although in a companion article Akam (1987) pointed out that this conclusion was implied. Molecular analysis of orthologs in humans and the mouse further supported the idea of a single ancestral complex (Akam 1989). Intact complexes in many animal species were called HOM-C until the term was largely superseded by Hox cluster.
There were several arguments for examining the genetic regulation of developmental decisions in insect species other than Drosophila. First, fly larval morphology is highly specialized. Although I have seldom heard a Drosophilist use the word maggot, the legless and (through head involution) headless larva is greatly modified from ancestral larval morphology. The adult Drosophila also shows specialized morphological features, such as the gnathal appendages, hind wings, and terminalia. Our working hypothesis was that changes in Hox gene function were important to insect morphological evolution. In addition to Tribolium's technical advantages compared to other non-drosophilid insects, the larva (complete with all head segments and thoracic legs) is much more ancestral in morphology (Figure 1A). (Another impediment for the Drosophila-centric scientists of that era, which I personally had to overcome, was the tendency to think about how other insects were evolutionarily modified with respect to Drosophila, rather than how Drosophila evolved differences with respect to ancestral insects.) The adult beetle is fairly specialized morphologically, but in some ways different from Drosophila (Figure 1B). Thus, a major focus of our work for several decades has been a molecular and functional analysis of Tribolium Hox genes and comparisons to Drosophila and to other insects. Until the advent of RNAi in other insects used for comparative studies, Tribolium dominated functional studies, owing to the possibility of using mutations.
Figure 1.—
A late instar larva (A) and an adult (B) T. castaneum.
I argued above that the work of Stuart et al. (1993) was groundbreaking in its demonstration of the sophisticated genetic analysis possible for Tribolium. I further maintain that it made important and novel contributions to demonstrating the correlation between changes in Hox gene function and morphological evolution. In insects, the posterior body comprises the abdomen proper and the post-abdomen, which includes the genitalia and analia. In Drosophila larvae, abdominal-A is expressed throughout the abdomen, and Abdominal-B in the posterior abdomen and post-abdomen. Null mutations of abdominal-A show anteriorly directed transformations of anterior abdominal parasegments, and more posterior abdominal parasegments have only subtle anterior transformations. Abdominal-B mutants show transformations of both the posterior abdomen and the post-abdomen. In Tribolium, the abdominal-A ortholog is also expressed throughout the abdomen (Stuart et al. 1993), while Abdominal-B expression is restricted to the post-abdomen (He 1996). In contrast to Drosophila, Tribolium abdominal-A null mutations show strong anterior transformations of the entire abdomen. This observation led to several conclusions. First, although the transformations in the anterior abdomen are parallel in the two insects, the segment-specific morphological features are quite different, implying (not surprisingly) that Hox gene activity is conserved but transcriptionally regulates nonconserved downstream genes. Second, the transformations in Tribolium were clearly parasegmental, representing the first demonstration of the functional significance of parasegmental Hox gene expression outside of Drosophila. Finally, these observations suggested that the ancestral domain of functional significance of abdominal-A and Abdominal-B were in the abdomen and post-abdomen, respectively, and that abdominal-A function in the posterior abdomen was largely assumed by Abdominal-B during the evolution of Drosophila. Studies in other insects (Kelsh et al. 1993) support the hypothesis that the Abdominal-B ancestral function is restricted to the post-abdomen. Thus, this study indicated that, without a notable change in the expression domain, the functional domain of abd-A changed significantly in the Drosophila lineage, the first such observation made. During a period of comparative studies in which conservation of candidate gene expression patterns was usually interpreted to reflect conservation of function, the Tribolium abdominal-A ortholog (and fushi tarazu discussed below) provided worrisome counterexamples. This work was not often cited in that context.
In addition to abdominal-A, sequences and expression patterns were described for the orthologs of labial (Nie et al. 2001), proboscipedia (Shippy et al. 2000a,b), Deformed (Brown et al. 1999a, 2000), fushi tarazu (Brown et al. 1994), Sex combs reduced (Curtis et al. 2001), Antennapedia (Brown et al. 2002b and our unpublished results), Ultrabithorax (Bennett et al. 1999), and Abdominal-B (He 1996). Except for the latter gene and some other minor exceptions, expression patterns are well conserved in the two insects, showing that the basis of the effect of Hox genes on morphological differences lies in changes in target and other downstream genes (see below).
We went on to complete the analysis of the significance of the Hox genes for embryonic development and larval morphology in Tribolium. Genetic variants were available for the orthologs of proboscipedia, Sex combs reduced, Antennapedia, Ultrabithorax, abdominal-A, and (most probably) Abdominal-B, but not for labial or Deformed. We undertook a search for new EMS-induced variants failing to complement two overlapping deficiencies that deleted the entire complex except the Abdominal-B ortholog. This strategy directly paralleled that used to define genetically the Drosophila Antennapedia complex (see Denell 1994). This effort generated a Deformed loss-of-function allele (Brown et al. 2000) as well as new alleles at some of the other genes. No labial variants were found, and we know now that knockdown of the gene by RNAi also does not result in a detectable phenotype (S. Brown, personal communication).
ANTENNAPEDIA COMPLEX AND FUSHI TARAZU
With respect to Antennapedia complex genes, Tribolium orthologs showed conserved expression domains, and mutations in orthologous genes affect similar body regions. However, the mutant phenotypes observed were in every case different from those in Drosophila (Denell et al. 1996; Brown et al. 2002b). Stuart et al. (1991) had shown earlier that homozygotes for a deletion of most of the HOM-C display a spectacular phenotype in which antennae form on all gnathal, thoracic, and abdominal segments. Brown et al. (2002a) used double-mutant combinations and deficiencies to show that the role of most of the beetle's Antennapedia complex Hox genes includes the suppression of antennal development, which represents a default state in the absence of Hox gene expression. This was presumably an ancestral function of these genes. Together, these studies provided strong evidence for the hypothesis that the evolution of the highly derived anterior larval morphology in Drosophila involved considerable modification of Antennapedia complex gene function and support the general concept of the importance of Hox gene changes to morphological evolution.
Studies of the HOM-C also gave some new insights into the evolution of some non-Hox genes in the Drosophila Antennapedia complex: fushi tarazu, bicoid, zerknüllt (zen), and its paralog zen2. (Other non-Hox genes, including a cluster of cuticle genes, are also present.) Surprisingly, it was thought that Drosophila had no ortholog of the class 3 Hox genes, although one was clearly present in the ancestral cluster. Studies of Tribolium and the grasshopper showed that zen and zen2 are indeed derived members of the class 3 Hox family that had lost a role in anterior-posterior patterning (Falciani et al. 1996). The extraembryonic expression of the Tribolium zen paralogs resembled that of the Drosophila genes, but we did not recover any variants in our mutant screen.
The Drosophila Antennapedia complex also includes fushi tarazu, a derived Hox paralog with a pair-rule function important for the segmentation process. When studying the deficiency of the Hox cluster described above, Stuart et al. (1991) noted that, if a fushi tarazu ortholog is present at the same location as in the Drosophila complex, it would be deleted by this rearrangement. Nevertheless, embryos homozygous for this deficiency do not show a pair-rule phenotype. Thereafter, Brown et al. (1994) showed that a fushi tarazu ortholog is indeed present in the Hox cluster and is expressed in a pattern very much like that in Drosophila, albeit with a shift in register and altered temporal dynamics. In the absence of evidence from the deficiency, one would conclude that Tribolium fushi tarazu has a pair-rule function, but somewhat different from that in Drosophila. The deficiency phenotype instead argues against a function in segmentation, a conclusion supported by RNAi studies (S. Brown, personal communication).
ANTERIOR EMBRYONIC POSITIONAL INFORMATION
In Drosophila, the maternal-effect gene bicoid encodes the anterior determinant during early embryogenesis. Although a bicoid ortholog that originated as a Hox 3 duplication is recognizable in lower diptera (Stauber et al. 1999; Brown et al. 2001), as described below there is no such gene within the Tribolium Hox (Brown et al. 2001, 2002a; Richards et al. 2008). Additional studies of Tribolium and other insects support the role of orthodenticle as the ancestral anterior determinant (see Schroder et al. 2008).
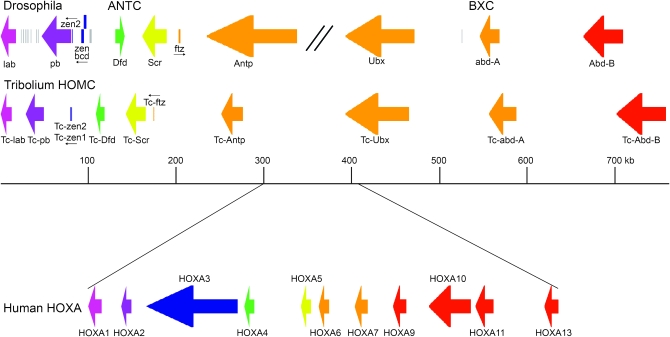
Tribolium studies have provided a series of insights into the evolution of Hox clusters. I have discussed the evidence that an intact ancestral complex was divided in Drosophila melanogaster, and additional cluster rearrangements have been demonstrated in other dipteran species (see Shippy et al. 2008). Brown et al. (2002a) sequenced three BAC clones that contained the Antennapedia-complex-like portion of the Hox cluster. Although complete clusters had been sequenced in a number of deuterostomes, among protostomes such information was then available only for Drosophila and the highly degenerate cluster of Caenorhabditis elegans. For many protostomes, Hox genes themselves had been sequenced (e.g., Grenier et al. 1997), but organization of the cluster as a whole was unknown. The Drosophila Antennapedia complex is very large (∼400 kb) compared to the vertebrate clusters (∼100 kb). The ANTC-like portion of the Tribolium complex (∼280 kb) is smaller than the ANTC itself, but still considerably larger than the vertebrate clusters (Figure 2). Each of the Hox genes predicted is present in one copy and in the same order as in flies, although (like vertebrates and unlike flies) they are all transcribed off of the same strand. This provided strong evidence that the ancestral cluster included genes of consistent transcriptional orientation. fushi tarazu and two zerknüllt paralogs are present, but orthologs of bicoid and the nonhomeobox genes found in the Drosophila clusters are not present. This work has been extended by Shippy et al. (2008), based on the newly available full sequence of the Hox cluster. The results are consistent with earlier studies of the Antennapedia-complex-like portion: the cluster is intact, all of the genes in the cluster are transcribed off of the same strand, and Hox genes are the only protein-coding transcription units present. Thus, in contrast with Drosophila, ancestral Hox cluster organization has been conserved. These observations are consistent with the idea that the constraints that have maintained Hox cluster integrity over vast evolutionary distances were relaxed during dipteran evolution, allowing rearrangements and the invasion of non-Hox genes. These observations argue for the potential utility of Tribolium in understanding further the basis of the conservation of Hox cluster organization.
Figure 2.—
A comparison of the size and organization of the Hox clusters of Drosophila and Tribolium, as well as one of the human clusters. The sizes of the primary transcripts are shown, and for most the direction of transcription is indicted. For Hox genes, more closely related genes are indicated by common colors. Additional non-Hox genes in the Drosophila cluster are shown in gray.
I cannot do justice here to the contributions of Tribolium studies to an understanding of the evolution of arthropod segmentation or, more generally, to the flood of new information on the genetic control of embryogenesis in Tribolium and its evolutionary implications that preceded and accompanied the sequencing of its genome. Ongoing and future application of such methodologies as mutagenesis, microarray analysis, DNA tiling arrays, and RNAi (including high-throughput screens) will allow Tribolium evo-devo studies to move beyond the study of candidate genes to discover those with ancestral functions no longer found in Drosophila. This should be a fruitful field of study for some time.
Acknowledgments
I thank Teresa Shippy for making the figures and for comments on a draft of this essay as well as Sue Brown and and Dick Beeman for comments on a draft of this essay.
This Perspectives is dedicated to Judith Plesset and the late DeLill Nasser, program officers at the National Science Foundation who supported our transition to Tribolium genetic studies and in general fostered the growth of evo-devo.
References
- Akam, M., 1987. Insect evolution: molecules and morphology. Nature 327 184–185. [DOI] [PubMed] [Google Scholar]
- Akam, M., 1989. Hox and HOM: homologous gene clusters in insects and vertebrates. Cell 57 347–349. [DOI] [PubMed] [Google Scholar]
- Beeman, R. W., 1987. A homeotic gene cluster in the red flour beetle. Nature 327 247–249. [Google Scholar]
- Beeman, R. W., and S. J. Brown, 1999. RAPD-based genetic linkage maps of Tribolium castaneum. Genetics 153 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeman, R. W., T. R. Johnson and S. M. Nanis, 1986. Chromosome rearrangements in Tribolium castaneum. J. Hered. 77 451–456. [Google Scholar]
- Beeman, R. W., J. J. Stuart, M. S. Haas and R. E. Denell, 1989. Genetic analysis of the homeotic gene complex (HOM-C) in the beetle Tribolium castaneum. Dev. Biol. 133 196–209. [DOI] [PubMed] [Google Scholar]
- Beeman, R. W., K. S. Friesen and R. E. Denell, 1992. Maternal-effect selfish genes in flour beetles. Science 256 89–92. [DOI] [PubMed] [Google Scholar]
- Beeman, R. W., J. J. Stuart, M. S. Haas and K. S. Friesen, 1996. Chromosome extraction and revision of linkage group 2 in Tribolium castaneum. J. Hered. 87 224–232. [DOI] [PubMed] [Google Scholar]
- Bennett, R. L., S. J. Brown and R. E. Denell, 1999. Molecular and genetic analysis of the Tribolium Ultrabithorax ortholog, Ultrathorax. Dev. Genes Evol. 209 608–619. [DOI] [PubMed] [Google Scholar]
- Berghammer, A., G. Bucher, F. Maderspacher and M. Klingler, 1999. a A system to efficiently maintain embryonic lethal mutations in the flour beetle Tribolium castaneum. Dev. Genes Evol. 209 382–389. [DOI] [PubMed] [Google Scholar]
- Berghammer, A. J., M. Klingler and E. A. Wimmer, 1999. b A universal marker for transgenic insects. Nature 402 370–371. [DOI] [PubMed] [Google Scholar]
- Brown, S. J., C. W. Black, IV, J. Henry and R. E. Denell, 1990. Molecular genetic manipulation of the red flour beetle: genomic organization and cloning of a ribosomal protein gene. Insect Biochem. 20 185–193. [Google Scholar]
- Brown, S. J., R. B. Hilgenfeld and R. E. Denell, 1994. The beetle Tribolium castaneum has a fushi tarazu homolog expressed in stripes during segmentation. Proc. Natl. Acad. Sci. USA 91 12922–12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, S., S. Holtzman, T. Kaufman and R. Denell, 1999. a Characterization of the Tribolium Deformed ortholog and its ability to directly regulate Deformed target genes in the rescue of a Drosophila Deformed null mutant. Dev. Genes Evol. 209 389–398. [DOI] [PubMed] [Google Scholar]
- Brown, S. J., J. P. Mahaffey, M. D. Lorenzen, R. E. Denell and J. W. Mahaffey, 1999. b Using RNAi to investigate orthologous homeotic gene function during development of distantly related insects. Evol. Dev. 1 11–15. [DOI] [PubMed] [Google Scholar]
- Brown, S., M. DeCamillis, K. Gonzalez-Charneco, M. Denell, R. Beeman et al., 2000. Implications of the Tribolium Deformed mutant phenotype for the evolution of Hox gene function. Proc. Natl. Acad. Sci. USA 97 4510–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, S., J. Fellers, T. Shippy, R. Denell, M. Stauber et al., 2001. A strategy for mapping bicoid on the phylogenetic tree. Curr. Biol. 11 R43–R44. [DOI] [PubMed] [Google Scholar]
- Brown, S. J., J. P. Fellers, T. D. Shippy, E. A. Richardson, M. Maxwell et al., 2002. a Sequence of the Tribolium castaneum homeotic complex: the region corresponding to the Drosophila melanogaster antennapedia complex. Genetics 160 1067–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, S. J., T. D. Shippy, R. W. Beeman and R. E. Denell, 2002. b Tribolium Hox genes repress antennal development in the gnathos and trunk. Mol. Phylogenet. Evol. 24 384–387. [DOI] [PubMed] [Google Scholar]
- Brown, S. J., R. E. Denell and R. W. Beeman, 2003. Beetling around the genome. Genet. Res. 82 155–161. [DOI] [PubMed] [Google Scholar]
- Bucher, G., J. Scholten and M. Klingler, 2002. Parental RNAi in Tribolium (Coleoptera). Curr. Biol. 12 R85–R86. [DOI] [PubMed] [Google Scholar]
- Cerny, A. C., G. Bucher, R. Schroder and M. Klingler, 2005. Breakdown of abdominal patterning in the Tribolium Kruppel mutant jaws. Development 132 5353–5363. [DOI] [PubMed] [Google Scholar]
- Curtis, C. D., J. A. Brisson, M. A. DeCamillis, T. D. Shippy, S. J. Brown et al., 2001. Molecular characterization of Cephalothorax, the Tribolium ortholog of Sex combs reduced. Genesis 30 12–20. [DOI] [PubMed] [Google Scholar]
- Demerec, M., 1950. The Biology of Drosophila. John Wiley & Sons, New York.
- Denell, R., 1987. Insect developmental genetics: moving beyond Drosophila. BioEssays 6 77–79. [Google Scholar]
- Denell, R., 1994. Discovery and genetic definition of the Drosophila Antennapedia complex. Genetics 138 549–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denell, R. E., S. J. Brown and R. W. Beeman, 1996. Evolution of the organization and function of insect homeotic complexes. Semin. Dev. Biol. 7 527–538. [Google Scholar]
- Edgley, M. L., D. L. Baillie, D. L. Riddle and A. M. Rose, 1995. Genetic balancers. Methods Cell Biol. 48 147–184. [PubMed] [Google Scholar]
- Englert, D. C., and A. E. Bell, 1970. Selection for time of pupation in Tribolium castaneum. Genetics 64 541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falciani, F., B. Hausdorf, R. Schroder, M. Akam, D. Tautz et al., 1996. Class 3 Hox genes in insects and the origin of zen. Proc. Natl. Acad. Sci. USA 93 8479–8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, K. H., and P. M. Waterhouse, 2007. RNAi for insect-proof plants. Nat. Biotechnol. 25 1231–1232. [DOI] [PubMed] [Google Scholar]
- Grenier, J. K., T. L. Garber, R. Warren, P. M. Whitington and S. Carroll, 1997. Evolution of the entire arthropod Hox gene set predated the origin and radiation of the onychophoran/arthropod clade. Curr. Biol. 7 547–553. [DOI] [PubMed] [Google Scholar]
- He, J., 1996. Molecular and genetic analysis of the Abdominal-B homolog of the beetle Tribolium castaneum. Ph.D. Thesis, Kansas State University, Manhattan, KS.
- Hentges, K. E., and M. J. Justice, 2004. Checks and balancers: balancer chromosomes to facilitate genome annotation. Trends Genet. 20 252–259. [DOI] [PubMed] [Google Scholar]
- Horn, C., and E. A. Wimmer, 2000. A versatile vector set for animal transgenesis. Dev. Genes Evol. 210 630–637. [DOI] [PubMed] [Google Scholar]
- Kelsh, R., I. Dawson and M. Akam, 1993. An analysis of abdominal-B expression in the locust Schistocerca gregaria. Development 117 293–305. [DOI] [PubMed] [Google Scholar]
- Kennerdell, J. R., and R. W. Carthew, 1998. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95 1017–1026. [DOI] [PubMed] [Google Scholar]
- Krause, G., 1939. Die Eitypen der Insekten. Biol. Zbl. 59 495–536. [Google Scholar]
- Levene, H., I. M. Lerner, A. Sokoloff, F. K. Ho and I. R. Franklin, 1965. Genetic load in Tribolium. Proc. Natl. Acad. Sci. USA 53 1042–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen, M. D., A. J. Berghammer, S. J. Brown, R. E. Denell, M. Klingler et al., 2003. piggyBac-mediated germline transformation in the beetle Tribolium castaneum. Insect Mol. Biol. 12 433–440. [DOI] [PubMed] [Google Scholar]
- Lorenzen, M. D., Z. Doyungan, J. Savard, K. Snow, L. R. Crumly et al., 2005. Genetic linkage maps of the red flour beetle, Tribolium castaneum, based on bacterial artificial chromosomes and expressed sequence tags. Genetics 170 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen, M. D., T. Kimzey, T. D. Shippy, S. J. Brown, R. E. Denell et al., 2007. piggyBac-based insertional mutagenesis in Tribolium castaneum using donor/helper hybrids. Insect Mol. Biol. 16 265–275. [DOI] [PubMed] [Google Scholar]
- Maderspacher, F., G. Bucher and M. Klingler, 1998. Pair-rule and gap gene mutants in the flour beetle Tribolium castaneum. Dev. Genes Evol. 208 558–568. [DOI] [PubMed] [Google Scholar]
- Miller, S. C., S. J. Brown and Y. Tomoyasu, 2008. Larval RNAi in Drosophila? Dev. Genes Evol. 218 505–510. [DOI] [PubMed] [Google Scholar]
- Misquitta, L., and B. M. Paterson, 1999. Targeted disruption of gene function in Drosophila by RNA interference (RNA-i): a role for nautilus in embryonic somatic muscle formation. Proc. Natl. Acad. Sci. USA 96 1451–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, L. M., R. Booker and L. M. Riddiford, 1991. Isolation and embryonic expression of an abdominal-A-like gene from the lepidopteran, Manduca sexta. Development 112 119–129. [DOI] [PubMed] [Google Scholar]
- Nie, W., B. Stronach, G. Panganiban, T. Shippy, S. Brown et al., 2001. Molecular characterization of Tclabial and the 3′ end of the Tribolium homeotic complex. Dev. Genes Evol. 211 244–251. [DOI] [PubMed] [Google Scholar]
- Park, T., 1934. Observations on the general biology of the flour beetle Tribolium confusum. Q. Rev. Biol. 9 36–54. [Google Scholar]
- Phelps, C. B., and A. H. Brand, 1998. Ectopic gene expression in Drosophila using GAL4 system. Methods 14 367–379. [DOI] [PubMed] [Google Scholar]
- Richards, S., R. A. Gibbs, G. M. Weinstock, S. J. Brown, R. Denell et al., 2008. The genome of the model beetle and pest Tribolium castaneum. Nature 452 949–955. [DOI] [PubMed] [Google Scholar]
- Roberts, P. A., 1970. Screening for X-ray-induced crossover suppressors in Drosophila melanogaster: prevalence and effectiveness of translocations. Genetics 65 429–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, G. M., and A. C. Spradling, 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218 348–353. [DOI] [PubMed] [Google Scholar]
- Sander, K., 1976. Specification of the basic body pattern in insect embryogenesis. Adv. Insect Physiol. 12 125–128. [Google Scholar]
- Schroder, R., A. Beermann, N. Wittkopp and R. Lutz, 2008. From development to biodiversity: Tribolium castaneum, an insect model organism for short germband development. Dev. Genes Evol. 218 119–126. [DOI] [PubMed] [Google Scholar]
- Shippy, T. D., S. J. Brown and R. E. Denell, 2000. a Maxillopedia is the Tribolium ortholog of proboscipedia. Evol. Dev. 2 145–151. [DOI] [PubMed] [Google Scholar]
- Shippy, T. D., J. Guo, S. J. Brown, R. W. Beeman and R. E. Denell, 2000. b Analysis of maxillopedia expression pattern and larval cuticular phenotype in wild-type and mutant Tribolium. Genetics 155 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippy, T. D., M. Ronshaugen, J. Cande, J. He, R. W. Beeman et al., 2008. Analysis of the Tribolium homeotic complex: insights into mechanisms constraining insect Hox clusters. Dev. Genes Evol. 218 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloff, A., 1966. The Genetics of Tribolium and Related Species. Academic Press, New York.
- Sokoloff, A., 1972. The Biology of Tribolium, Vol. 1. Clarendon Press, Oxford.
- Sokoloff, A., 1974. The Biology of Tribolium, Vol. 2. Clarendon Press, Oxford.
- Sokoloff, A., 1977. The Biology of Tribolium, Vol. 3. Clarendon Press, Oxford.
- Stauber, M., H. Jackle and U. Schmidt-Ott, 1999. The anterior determinant bicoid of Drosophila is a derived Hox class 3 gene. Proc. Natl. Acad. Sci. USA 96 3786–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, J. J., S. J. Brown, R. W. Beeman and R. E. Denell, 1991. A deficiency of the homeotic complex of the beetle Tribolium. Nature 350 72–74. [DOI] [PubMed] [Google Scholar]
- Stuart, J. J., S. J. Brown, R. W. Beeman and R. E. Denell, 1993. The Tribolium homeotic gene Abdominal is homologous to abdominal-A of the Drosophila bithorax complex. Development 117 233–243. [DOI] [PubMed] [Google Scholar]
- Sulston, I. A., and K. V. Anderson, 1996. Embryonic patterning mutants of Tribolium castaneum. Development 122 805–814. [DOI] [PubMed] [Google Scholar]
- Tomoyasu, Y., and R. E. Denell, 2004. Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev. Genes Evol. 214 575–578. [DOI] [PubMed] [Google Scholar]