Abstract
The mating-type bias (mat-bias) of gene conversion was previously described as a phenomenon in which the number of prototrophic recombinants in an ura4A heteroallelic two-factor cross relates to the mating types of the parents. We show now that the mat-bias is restricted neither to ura4A nor to recombination hotspots, but occurs at other genomic loci, too. It is specific for gene conversion and absent in azygotic meiosis. Thus, the mat-bias must originate from mating-type-specific “imprinting” events before karyogamy takes place. Structural variations of the mating-type locus, such as h+N, h+S, h−S, h+smtΔ, or h−smtΔ, showed mat-bias manifestation. Mutations in genes coding for histone acetylase (gcn5, ada2) and histone deacetylase (hos2, clr6) activities smooth or abolish the mat-bias. In addition, the mat-bias depends on the presence of Swi5. We propose a new role for Swi5 and the histone acetylation status in mat-bias establishment through directionality of repair from the intact chromatid to the broken chromatid.
THE main function of homologous recombination in mitotic cells is the repair of DNA damages such as double-strand breaks (DSBs) or single-strand nicks. Such DNA liaisons are generated by DNA-damaging agents or are the result of replication errors, e.g., when replication forks collapse. During meiosis, repair of programmed DSBs initiate crossover formation and chiasma, which are necessary for proper homologous chromosome alignment and segregation at meiosis I, as well as providing the basis for evolution. Meiotic recombination intermediates involve either a homologous chromatid or the sister chromatid as repair partner. It was shown at the meiotic recombination hotspot mbs1 in Schizosaccharomyces pombe that intermediates involving the sister chromatid outnumber the homologous chromatid as repair partner (Cromie et al. 2006).
Meiotic DSBs are formed by the conserved, topoisomerase-IV-like protein Spo11 (Keeney and Kleckner 1995). A Spo11 homo-dimer forms covalent bonds to the DSB, which are soon after removed by endo-nucleolytic cuts in the DNA, leaving ssDNA ends behind (Neale and Keeney 2006). In S. pombe (and in other eukaryotes), such ssDNA ends are substrates for two recombinases, Rad51 and the meiosis-specific Dmc1 (Grishchuk and Kohli 2003). In contrast to their bacterial homolog recA, Rad51 and Dmc1 are considerably less efficient in nucleoprotein filament formation and strand exchange formation (Sung 1997). They need accessory proteins, like Rad55/57 as well as Swi5/Sfr1 to facilitate their binding to ssDNA ends, which are coated by replication protein A (Haruta et al. 2006). In addition to its role in homologous recombination repair, Swi5 is also involved in mating-type switching. It forms an alternative complex with Swi2 instead of Sfr1, which directs the repair of the processed imprint, a specific DNA liaison at the smt locus next to the expressed mat1 cassette, toward the nonexpressed mating-type cassette of the opposite cell type (Jia et al. 2004). This directionality of recombination depends on the association of Swi2 with the heterochromatin of the mating-type region, which specifically occurs in the h− cell type.
Local chromatin structure plays an important role in the occurrence of DSB and can be altered by post-translational modification of histones and other chromatin-related proteins or by nucleosome-remodeling complexes. Histone modifications change the interactions among histones or with DNA (Strahl and Allis 2000) and thus affect precise localization of DSBs. The hotspot activity of ade6-M26 in fission yeast, e.g., is accompanied with remodeling of the chromatin (Mizuno et al. 1997) by cooperative action of the histone acetyltransferase (HAT) Gcn5 and the ATP-dependent chromatin-remodeling factor Snf22 (Yamada et al. 2004). Histone deacetylases (HDACs) and histone methylases, on the other hand, have been found in repression of meiotic DSB formation: the HDAC mutant rdp3 as well as the histone methylase mutant set2 in Saccharomyces cerevisiae increase DSB formation at the HIS4 recombination hotspot considerably (Merker et al. 2008).
The fission yeast ura4A hotspot was discovered when the ura4+ gene was integrated 15 kb centromere-proximal to the ade6 gene as a genetic marker for characterizing another meiotic recombination hotspot, ade6-M26 (Grimm et al. 1994). In tetrad analysis of a heterogenous cross (wild type × ura4A), 3.7% non-Mendelian segregants (NMS) were found (Zahn-Zabal et al. 1995). The hotspot activity of ura4 is context specific since, at its original locus, ura4 shows low recombination frequencies (Baur et al. 2005). The study of 12 spontaneous ura4A mutations revealed a gradient of gene conversion ranging from 18% NMS at the 5′-end to 6% at the 3′-end (Baur et al. 2005). In yeast, such a gene conversion polarity is explained by DSBs in the promotor region (Lichten and Goldman 1995; Liu et al. 1995; Paques and Haber 1999; Petes 2001). A site of meiotic DSBs has been detected ∼500 bp upstream of the ura4A start codon (Gregan et al. 2005; Sakem 2005), which is in accordance with the observed 5′–3′ gene conversion polarity and the location of DNA sequences precipitated by Rec12 (Ludin et al. 2008). Additionally, tetrad analysis of the two-factor cross, h+ ura4A-13 × h− ura4A-10, demonstrated a predominance of co-conversions in >70% of NMS (Baur et al. 2005), suggesting that heteroduplex DNA is mostly formed over both alleles.
Another phenomenon at the ura4A hotspot has been reported: the mating-type-related bias (mat-bias) of gene conversion (Baur et al. 2005). The ura4A allele nearer to the DSB formation site (ura4A-13) in the h+ parent acts twice as often as acceptor of information compared with ura4A-13 in the h− parent. Such a bias must result from a “memory effect” on the chromosomes before cells mate and recombination occurs. In this work, we report a follow-up study on this subtle and intriguing phenomenon.
MATERIALS AND METHODS
Strains, media, and general genetic methods:
The ura4A-10 and ura4A-13 strains were described in Baur et al. (2005) and, together with newly constructed strains, are listed in Table 1. Strains with the genotypes gcn5Δ, clr6-1, and clr3Δ were provided by Anthony Wright, ada2Δ by Kunihiro Ohta, hos2Δ by Karl Ekwall, h+S by Olaf Nielsen, swi5Δ and mat1-M smtΔ by Gerald Smith, swi2Δ by Shiv Grewal, and mat1-P smtΔ by Amar Klar. When necessary, the presence of the ura4A cassette was detected by polymerase chain reaction (PCR) from genomic DNA using primers KL1 and KL4 as described by Baur et al. (2005).
TABLE 1.
S. pombe strains
| Designationa | Genotype |
|---|---|
| EPY1 | h−ura4-D18 ura4A-10 leu2-120 |
| EPY2 | h+ ura4-D18 ura4A-10 leu2-120 |
| EPY3 | h−ura4-D18 ura4A-13 lys7-2 |
| EPY4 | h+ ura4-D18 ura4A-13 lys7-2 |
| EPY5 | h−ura4-D18 ura4A-13 |
| EPY6 | h+ ura4-D18 ura4A-13 |
| EPY7 | h−ura4-D18 ura4A-13 gcn5∷KanMX4 |
| EPY8 | h+ ura4-D18 ura4A-13 gcn5∷KanMX4 |
| EPY9 | h−ura4-D18 ura4A-13 hos2∷LEU2+ leu1-32 |
| EPY10 | h+ ura4-D18 ura4A-13 hos2∷LEU2+ leu1-32 |
| EPY11 | h−ura4-D18 ura4A-10 |
| EPY12 | h+ ura4-D18 ura4A-10 |
| EPY13 | h−ura4-D18 ura4A-10 gcn5∷KanMX4 |
| EPY14 | h+ ura4-D18 ura4A-10 gcn5∷KanMX4 |
| EPY15 | h−ura4-D18 ura4A-10 hos2∷LEU2+ leu1-32 |
| EPY16 | h+ ura4-D18 ura4A-10 hos2∷LEU2+ leu1-32 |
| EPY17 | h+S ura4-D18 ura4A-13 |
| EPY18 | h+S ura4-D18 ura4A-10 |
| EPY25 | h+ ura4-D18 ura4A-10 lys1-131 |
| EPY27 | h+ ura4-D18 ura4A-13 lys1+∷matPc |
| EPY28 | h+ ura4-D18 ura4A-10 lys1+∷matPc |
| EPY29 | h+ ura4-D18 ura4A-13 smtΔ |
| EPY30 | h+ ura4-D18 ura4A-10 smtΔ |
| EPY31 | h+ ura4-D18 ura4A-13 ade6-M216 |
| EPY32 | h−ura4-D18 ura4A-13 ade6-M216 |
| EPY33 | h+ ura4-D18 ura4A-10 ade6-M210 |
| EPY34 | h−ura4-D18 ura4A-10 ade6-M210 |
| EPY35 | h−ura4-D18 ura4A-13 lys1-131 |
| EPY36 | h−ura4-D18 ura4A-10 lys1-131 |
| EPY37 | h+ ura4-D18 ura4A-13 lys1-131 |
| EPY43 | h+ ura4-D18 ura4A-13 swi5-201∷KanMX6 |
| EPY44 | h−ura4-D18 ura4A-13 swi5-201∷KanMX6 |
| EPY45 | h+ ura4-D18 ura4A-10 swi5-201∷KanMX6 |
| EPY46 | h−ura4-D18 ura4A-10 swi5-201∷KanMX6 |
| EPY47 | h+ ura4-D18 ura4A-13 ada2∷KanMX |
| EPY48 | h−ura4-D18 ura4A-13 ada2∷KanMX |
| EPY49 | h+ ura4-D18 ura4A-10 ada2∷KanMX |
| EPY50 | h−ura4-D18 ura4A-10 ada2∷KanMX |
| EPY51 | h−ura4-D18 ura4-A13 smtΔ |
| EPY52 | h−ura4-D18 ura4-A10 smtΔ |
| EPY53 | h−ura4-D18 ura4A-13 swi2∷KanMX6 |
| EPY54 | h+ ura4-D18 ura4A-13 swi2∷KanMX6 |
| EPY55 | h−ura4-D18 ura4A-10 swi2∷KanMX6 |
| EPY56 | h+ ura4-D18 ura4A-10 swi2∷KanMX6 |
| EPY57 | h−ura4-D18 ura4A-13 lys1+∷matMc |
| EPY58 | h−ura4-D18 ura4A-10 lys1+∷matMc |
| EPY63 | h−ura4-D18 ura4A-10 gcn5∷KanMX4 ada2∷KanMX |
| EPY64 | h+ ura4-D18 ura4A-10 gcn5∷KanMX4 ada2∷KanMX |
| EPY65 | h−ura4-D18 ura4A-13 gcn5∷KanMX4 ada2∷KanMX |
| EPY66 | h+ ura4-D18 ura4A-13 gcn5∷KanMX4 ada2∷KanMX |
| EPY75 | h+ ura4-D18 ura4A-13 clr6-1 |
| EPY76 | h−ura4-D18 ura4A-13 clr6-1 |
| EPY77 | h+ ura4-D18 ura4A-10 clr6-1 |
| EPY78 | h−ura4-D18 ura4A-10 clr6-1 |
| EPY85 | h+ ura4-D18 ura4A-13 clr3Δ |
| EPY86 | h−ura4-D18 ura4A-13 clr3Δ |
| EPY87 | h+ ura4-D18 ura4A-10 clr3Δ |
| EPY88 | h−ura4-D18 ura4A-10 clr3Δ |
| EPY89 | h+ ura4-D18 ura4A-10 lys1+∷matPi |
| EPY90 | h−ura4-D18 ura4A-10 lys1+∷matMi |
| EPY91 | h−ura4-D18 ura4A-13 lys1+∷matMi |
| EPY92 | h+ ura4-D18 ura4A-13 lys1+∷matPi |
| KLY386 | h+ ura4-D18 ura4A-10 lys1-131 swi5-201∷KanMX6 |
| KLY387 | h+ ura4-D18 ura4A-13 lys1-131 |
| KLY388 | h+ ura4-D18 ura4A-10 lys1-131 |
| KLY389 | h+ ura4-D18 ura4A-13 lys1-131 lys1+∷swi5 |
| KLY390 | h+ ura4-D18 ura4A-10 lys1-131 lys1+∷swi5 |
| KLY391 | h+ ura4-D18 ura4A-10 lys1-131 swi5-201∷KanMX6 lys1+∷swi5 |
| MR437 | h− ade6-D20 ura4-D18 his3-D1 dmc1∷ura4 rad51∷his3 |
| 40178 | h− ade3-58 |
| 40543 | h+ ade3-58 |
| 139-5538 | h− ade4-51 |
| 142-5665 | h+ ade4-51 |
| 139-5527 | h− ade4-M17 |
| 50-1983 | h+ ade4-M17 |
| 6-215 | h− ade6-469 |
| 6-216 | h+ ade6-469 |
| 5-165 | h− ade6-M26 |
| 5-166 | h+ ade6-M26 |
| 5-167 | h− ade6-M375 |
| 5-168 | h+ ade6-M375 |
| 51-2010 | h− ade7-152 |
| AG121 | h+ ade7-152 |
| AG161 | h− ade7-50 |
| AG242 | h+ ade7-50 |
| 39-1147 | h− cdc1-7 |
| 152-6045 | h+ cdc1-7 |
| 23407 | h− his1-102 |
| 23773 | h+ his1-102 |
| 4-120 | h− pro1-1 |
| 4-121 | h+ pro1-1 |
| 4-126 | h− ura1-61 |
| 4-127 | h+ ura1-61 |
| 145-5768 | h− ura4-D18 sod2∷ura4 |
| 145-5769 | h+ ura4-D18 sod2∷ura4 |
| 144-5765 | h− ura4-D18 lys3-37 |
| 145-5756 | h+ ura4-D18 lys3-37 |
All strains marked with EPY and KLY were constructed for this study. All other strains were from the strain collection in Bern.
The synthetic growth medium (GMA) consisted of 0.17% Difco nitrogen base without amino acids, 0.375% sodium glutamate, 1% glucose, and 2% agar. Standard minimal medium (MMA), yeast extract agar (YEA), malt extract agar (MEA), and the general genetic methods were described by Gutz et al. (1974) and Baur et al. (2005). Supplements were added to the media at 0.01% (w/v). The “+ all” abbreviation stands for a mix of adenine, uracil, histidine, lysine, and leucine, each to a final concentration of 0.01% (w/v) in the medium.
Sequencing of ade7 mutations:
Total genomic DNA was isolated from haploid strains ade7-50 (AG161) and ade7-152 (51-2010) (Table 1) by the small-scale method (Wright et al. 1986). These genomic DNAs were used as targets to generate 1316-bp-long PCR fragments containing the entire ade7 ORF with the two oligonucleotide primers EP5 (5′-AATTGTAATTGCTTTTGTTTCTCTG-3′) and EP6 (5′-ATCGCCAAAACACCGTTAAC-3′). DNA sequencing was performed on these PCR fragments with the Big Dye Terminator v.3.1 Cycle sequencing kit from Applied Biosystems and the oligonucleotide primers EP7 (5′-GCAATAGCATTTAAAGTCGATTTAC-3′), EP8 (5′-ATACCTCATCAACAAGCACAATC-3′), EP9 (5′-ATAGCTTCGATAGGAAGAATCTTG-3′), EP10 (5′-TTAAACCTTCATTTCTACATCAAG-3′), and EP11 (5′-GAATTAGCCAAACAAGTTGC-3′).
Random spore analysis:
The frequency of prototrophic recombinants was determined in heteroallelic crosses between mutant strains listed in Table 1. Cell material of both parental strains was mixed in water and plated as a dense lawn on MEA. Incubation was for 3 days at 25°. To kill vegetative cells, the sporulated material was treated overnight at 30° with an aqueous snail enzyme solution [1:1000 (v/v) Helix pomatia juice, Biosepra]. For titer estimation of prototrophic recombinants and total spores, samples of appropriately diluted spore suspensions were spread on GMA and on GMA with the required supplements for the respective mutations. The incubation was 5 days at 30°.
Construction of diploid strains for studying intragenic recombination at ura4A after azygotic meiosis:
Parental strains carrying lys7-2, ura4A-13 and leu2-120, ura4A-10, respectively, were crossed on MEA medium (EPY1 × EPY4 and EPY2 × EPY3) (Table 1). After 25 hr of incubation at 25°, cross material was streaked on MMA + ura and incubated at 30°. After 4–5 days diploid colonies prototrophic for leucine and lysine and growing with darkly pink pigment on YEA with phloxin B were picked and put onto MEA medium and incubated for 3 days at 25°. The sporulated material was used for random spore analysis as described above.
To check intragenic recombination at ura4A after a small number of diploid generation cycles, parental haploid strains (EPY31 and EPY34; see Table 1) were precultured in 0.5% yeast extract, 3% glucose medium and reinoculated to liquid SPO (7.3 mm KH2PO4, 1% glucose, 4.2 mm pantothenic acid, 81.2 mm nicotinic acid, 55.5 mm inositol, 40.8 μm biotin) medium overnight. The two parental strains were then crossed on MEA medium for 6 hr at 25°. This short mating time was needed to circumvent the problem of asynchronous mating. Cross material was collected and centrifuged at 1000 rpm in a 7–30% lactose gradient. Fractions (1.5 ml) were taken from the supernatant (control) and from the zygote-enriched pellet. They were plated separately to fully supplemented medium (YEA + all) for 20 hr at 30° to allow for about five mitotic divisions of the diploids. Longer incubation times bore the risk of having secondary mating and zygote formation of the remaining haploid cells. The cell material was then transferred to sporulation medium (MEA) for 10 hr at 25°. The mix of sporulated diploids and mated and unmated haploid cells was treated with snail enzyme, and recombination analysis was performed. This diploid propagation yielded low spore counts and was thus performed only with type IV crosses.
Construction of strains with mating-type gene insertions:
Integration plasmids pAY164, pAY165, pAY161, and pAY166 containing the mat-Pc, mat-Mc, mat-Pi, and mat-Mi genes, respectively, as well as an N-terminal truncated lys1 gene, were obtained from Ayumi Yamamoto (Yamamoto and Hiraoka 2003). The plasmids were transformed into lys1-131 mutants and integrative transformants were selected for lysine prototrophy (EPY25, EPY 35, EPY36, and EPY37; see Table 1).
Southern blot analysis of mating-type regions in different strains:
As +N mating type, we used the wild-type heterothallic strain h+ 975; as h−S wild-type, heterothallic strain h− 972; as h+S, strain EPY24; and for mat1-M smtΔ and mat1-P smtΔ, strains EPY51 and EPY29, respectively (Table 1). The PCR fragments amplified with the primers EP12 (5′-TAGCCATACAACCTAATAGCCAG-3′) and EP13 (5′-ACAGATATAAATCGCAATACATCG-3′) gave the probe for the mat1-M region of the h−S mating type. Primers EP14 (5′-ACCTAATAGCCAGAAACGCC-3′) and EP15 (5′-GGTCGTAAAACTCAGAGTTAGAGG-3′) were used for generating the PCR fragment for the mat1-P region probe specific for h+S mating type.
Genomic DNA of different mating-type strains was digested with the restriction enzyme HindIII or double digested with HindIII and BamHI (both New England Biolabs). The digested DNA was migrated and blotted to Hybond-N+ nylon membrane (Genescreen Plus). The two different probes were labeled with [α-32P]dCTP by using Ready-to-Go DNA labeling beads (Amersham). The hybridized fragments were visualized with a PhosphoImager.
Cloning of swi5:
A 1-kb DNA fragment of the swi5 gene, including its 5′- and 3′-UTRs, was amplified by PCR fromgenomic wild-type DNA with the primers KL243 (5′-GAAGGCAATTTGTACATGG-3′) and KL244 (5′-CGAGCCTTAATACTAATACACTAGC-3′) and cloned into the pJET vector (FERMENTAS). A XhoI/XbaI fragment of this subclone was then ligated into the XhoI/XbaI-digested pAY164 to make pKL13. The integrity of the swi5 gene was confirmed by complementing the UV sensitivity of swi5Δ in a swi5 deletion mutant (KLY386) transformed with integrative pKL13 (KLY391, supplemental Figure S4).
RESULTS AND DISCUSSION
The mat-bias is a general phenomenon with region-specific variation:
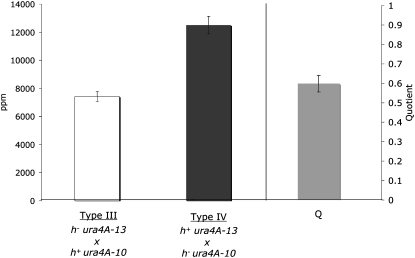
The mat-bias of gene conversion was previously described as a phenomenon at the ura4A recombination hotspot, in which a heteroallelic cross of the most 5′ allele, ura4A-13 (C to A at position 562), with the most 3′ allele, ura4A-10 (C to T at position 1167), results in about twice as many prototrophs when the ura4A-13 allele enters the cross with the h+ parent (type IV cross) than with the h− parent (type III cross) (Baur et al. 2005). The mat-bias was defined as the quotient (Q) of the prototrophs per million (ppm) viable spores of the type III and type IV crosses.
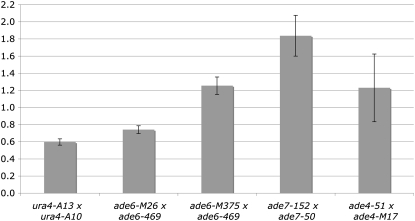
In this study, we first confirmed the mat-bias at ura4A by random spore analysis of two-factor crosses with ura4-A13 and ura4-A10 (Figure 1, supplemental Table S1). Almost twice the amount of prototrophs were found in the type IV cross as in the type III cross (Q = 0.59, Figure 1, supplemental Table S1). To address the question whether the mat-bias is restricted to the ura4A hotspot or is a general phenomenon, heterozygous two-factor crosses with ade6-M26 (G to T at position 135) and ade6-469 (C to T at position 1467) were carried out and revealed a slightly lower, but significant, mat-bias of 0.74 (Figure 2, supplemental Table S1). Again the 5′ allele ade6-M26 in the h+ parent was more often converted to wild type than ade6-M26 in the h− parent. Tetrads from heterozygous ade6-M26 crosses (Gutz 1971), where ade6-M26 was in the cross with either h+ or h−, confirmed this result (see Table 2a in Gutz 1971: 6.06% vs. 3.13% aberrant tetrads, respectively).
Figure 1.—
The mat-bias at ura4A. The open bar indicates the average of prototrophs per million viable spores of at least four independent type III crosses: h− ura4A-13 (EPY5) × h+ ura4A-10 (EPY12) or type IV crosses: h+ ura4A-13 (EPY6) × h− ura4A-10 (EPY11). The shaded bar represents the mean quotient of type III and type IV (in ppm), which was determined as the average of the quotients of all type III/type IV values. Error bars stand for standard error on the mean.
Figure 2.—
The mat-bias at different genomic loci. Shaded bars indicate the ppm quotient average of type III and type IV crosses, which were independently repeated at least four times (supplemental Table S1). The y-axis represents the quotient of type III and type IV (in ppm) crosses. Type III crosses were EPY5 × EPY12, 5-165 × 6-216, 5-167 × 6-216, 51-2010 × AG242, and 139-5538 × 50-1983, respectively. Type IV crosses were EPY6 × EPY11, 5-166 × 6-215, 5-168 × 6-216, AG121 × AG161, and 142-5665 × 139-5527, respectively. Error bars stand for standard error on the mean.
The observation at ura4A and ade6-M26 might indicate that the mat-bias is a characteristic feature of hotspots. To check that conjecture, crosses were also performed with ade6-M375 (G to T at position 132). A slight, but statistically significant, inverted mat-bias was detected (Q = 1.26, Figure 2, supplemental Table S1). Tetrads from heterozygous ade6-M375 crosses (Gutz 1971), however, where the ade6-M375 parent was either h+ or h−, suggested no bias (see Table 2a in Gutz 1971: 0.88% vs. 0.87% aberrant tetrads, respectively). Although in this previous tetrad analysis the mat-bias was readily found in crosses with ade6-M26 and was not detected with ade6-M375, the amount of dissected tetrads did not support statistical significance (Gutz 1971).
An inverted, but strong, mat-bias was also observed at the ade7 gene on chromosome II (Q = 1.82, Figure 2, supplemental Table S1) with the alleles ade7-152 (G to A at position 3) and ade7-50 (G to C at position 780). This could be explained by the presence of an initiation event near the 3′-end of ade7. Interestingly, a peak of Rec12-DNA linkage was found in a 3′ probe of the ade7 gene in Rec12 chromatin immunoprecipitation experiments (Cromie et al. 2007; supplemental data set S1), indicating the presence of a meiotic DSB formation site. Comparison of recombination density (supplemental Table S1), which could be calculated for sequenced alleles at ura4A, ade6, and ade7, shows a similar density for ade7 and ade6-M375 crosses. This suggests that the mat-bias is not restricted to hotspots. It might be more pronounced in loci with steeper gene conversion polarity, probably due to located meiotic DSBs. In loci with less distinct gene conversion polarities, which might originate from a more basal distribution of meiotic DSBs, a mat-bias will not be observed, as in the ade4 gene on chromosome I (Q = 1.18, Figure 2, supplemental Table S2) with no pronounced Rec12-DNA linkage sites nearby (Cromie et al. 2007; supplemental data set S1).
If the mat-bias extent correlates with the slope of gene conversion polarity, it is only reasonable that it will not be detectable in intergenic recombination assays. Indeed, no significant differences of genetic distance (in centimorgans) in the tested intervals leu2-lys7, lys3-ura1, pro1-ura1, and pro1-ade3 on chromosome I were found between the two cross types (Table 2). This led us to conclude that the mat-bias is a phenomenon of gene conversion, not of crossing over.
TABLE 2.
Intergenic recombination in intervals on chromosome I
| Interval | Cross typea | Colonies tested | P1 (− +) | P2 (+ −) | R1 (− −) | R2 (+ +) | Recombination frequencyb (%) | Distancec (cM) | χ2 testd |
|---|---|---|---|---|---|---|---|---|---|
| Zygotic meiosis | |||||||||
| lys3-37e | III | 447 | 171 | 195 | 42 | 39 | 18.1 | 22.5 | 2.3 |
| × ura1-61 | IV | 448 | 166 | 185 | 45 | 52 | 21.7 | 28.4 | |
| ura1-61 | III | 448 | 173 | 155 | 49 | 71 | 26.8 | 38.4 | 5.6 |
| × pro1-1 | IV | 448 | 176 | 174 | 50 | 48 | 21.9 | 28.8 | |
| pro1-1 | III | 448 | 136 | 161 | 81 | 70 | 33.7 | 56.0 | 0.9 |
| × ade3-58 | IV | 448 | 146 | 150 | 78 | 74 | 33.9 | 56.7 | |
| leu2-120 | III | 224 | 101.5 | 99 | 8.5 | 15 | 10.5 | 11.8 | 2.2 |
| × lys7-2 | IV | 224 | 91 | 111 | 11 | 11 | 9.8 | 10.9 | |
| Azygotic meiosis | |||||||||
| leu2-120 | III | 224 | 101.5 | 96.5 | 8.5 | 17.5 | 11.6 | 13.2 | 4.1 |
| × lys7-2 | IV | 224 | 89 | 112 | 12 | 11 | 10.3 | 11.5 | |
In type III crosses, the mutation stated first is in the h− strain; in type IV crosses, it is in the h+ strain.
Recombination frequency, R = R1 + R2/(R1 + R2 + P1 + P2), where R signifies recombinants and P parentals.
Genetic distance = −50 ln(1 − 2[R]).
For significance at the 0.05 level, χ2 should be ≥7.82 (3 d.f.).
The data (except the interval leu2-lys7) were obtained from E. Lehmann and were published elsewhere (Ludin et al. 2008); the values for leu2-120-lys7-2 are the average of two independent crosses.
The mat-bias is absent in azygotic meiosis:
S. pombe naturally exists as haploid cellular organism but can be propagated to live in a diploid cell cycle state. For that, zygotes are transferred to nitrogen-rich medium shortly after mating has taken place. Such diploids can be kept in this state by the use of heterozygous markers (see materials and methods). The transfer of the diploid cells to nitrogen-poor medium then induces azygotic meiosis. We wondered whether the mat-bias is still present in azygotic meiosis of diploids after the chromosomes of the original h+ and h− parent are well mixed and replicated through several diploid cell cycles. After an estimated 52 and 70 mitotic divisions of four diploid isolates, each originating from either type III or type IV crosses, the mat-bias, measured at the ura4A locus, could no longer be detected (Q = 1.04 and 1.05, respectively; Table 3). The amount of prototrophs in these azygotes decreased to the type III level in zygotic meiosis.
TABLE 3.
No mat-bias in azygotic meiosis
| Type IIIa | Type IVa | ||
|---|---|---|---|
| Generations | (ppmb ± SEM) | (ppmb ± SEM) | Qc |
| NAa | 6170 | 11,480 | 0.54 |
| 5d | ND | 6340 ± 760 | NA |
| 52e | 8070 ± 540 | 7770 ± 400 | 1.04 |
| 70e | 7050 ± 400 | 6690 ± 280 | 1.05 |
Control: zygotic cross EPY3 × EPY2.
Prototrophs per million viable spores was determined in four independent diploids, SEM, standard error of the mean.
Q, ppm of type III cross divided by ppm of type IV cross.
Diploid derived from cross EPY31 × EPY34.
Diploids derived from crosses EPY2 × EPY3 (type III) or EPY1 × EPY4 (type IV).
Since the mat-bias was gone after 50 and 72 diploid generations, we wondered how long such a difference in the “epigenetic state” of chromatin is retained after mating. After a short mass mating (6 hr on MEA medium) and diploid enrichment through a lactose gradient, diploids were grown for five generations on rich medium before transfer to nitrogen-poor medium (see materials and methods). Due to technical difficulties, only diploids derived from the type IV cross were analyzed. The frequency of intragenic recombination at ura4A after five diploid generations was comparable to the values obtained after 52 and 70 generations (Table 3). Thus, the mat-bias at ura4A had vanished after few diploid generations.
We also tested the level of intergenic recombination in the leu2-lys7 interval after 52 diploid generations. It was comparable to the recombination frequency in zygotic meiosis, and no differences between type III and type IV crosses were observed (Table 2).
Absence of chromatin-remodeling proteins affects the level of recombination at ura4A and the mat-bias:
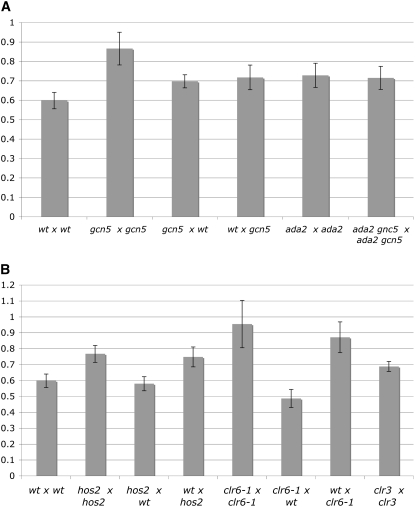
The loss of the mat-bias during azygotic meiosis could indicate that the chromatin of an h+ strain is different from the chromatin of an h− strain. To check the role of chromatin structure in the mat-bias more directly, we tested different histone acetyltransferase and histone deacetylase mutants, namely members of histone acetyltransferase complexes gcn5 and ada2 and members of histone deacetylases hos2, clr6, and clr3, for the presence or absence of the mat-bias at ura4A.
Gcn5 and Ada2 belong to the SAGA group of acetyltransferases and are required for full activation of chromatin changes at ade6-M26. gcn5 and ada2 mutants reduce prototroph frequency in crosses with ade6-M26 × ade6-469 by two- to fourfold (Hirota et al. 2008). In crosses homozygous for gcn5Δ, the mat-bias was greatly diminished (Figure 3A, supplemental Table S2). The level of prototrophs in the type IV cross dropped slightly below the values of the type III crosses in the wild type. We also checked the mat-bias in crosses heterozygous for gcn5Δ. In both configurations, a mat-bias was observed, although less pronounced than in the wild type. Similar values were found in ada2Δ homozygous crosses and in ada2Δ gcn5Δ homozygous crosses (Figure 3A, supplemental Table S2). This genetic relationship led us to conclude that, with respect to mat-bias establishment, ada2 might act upstream of gcn5, but both act in the same pathway.
Figure 3.—
The mat-bias in HAT and HDAC mutants. Shaded bars indicate the ppm quotient average of type III and type IV crosses in HAT mutants (A) or in HDAC mutants (B), which were independently repeated at least three times (supplemental Table S2). The y-axis represents the quotient of type III and type IV (in ppm) crosses. Type III crosses were (A) EPY5 × EPY12, EPY7 × EPY14, EPY5 × EPY14, EPY7 × EPY12, EPY48 × EPY49, and EPY65 × EPY64, respectively; (B) EPY5 × EPY12, EPY9 × EPY16, EPY9 × EPY12, EPY5 × EPY16, EPY76 × EPY77, EPY76 × EPY12, EPY5 × EPY77, and EPY86 × EPY87, respectively. Type IV crosses were (A) EPY6 × EPY11, EPY8 × EPY13, EPY8 × EPY11, EPY6 × EPY13, EPY47 × EPY50, and EPY66 × EPY63, respectively; (B) EPY6 × EPY11, EPY10 × EPY15, EPY10 × EPY11, EPY6 × EPY15, EPY75 × EPY78, EPY75 × EPY11, EPY6 × EPY78, and EPY85 × EPY88, respectively. Error bars stand for standard error on the mean.
S. pombe has several HDACs, among them hos2, clr6, and clr3, which basically counteract the activity of HAT complexes. They often are transcriptional corepressors, but some HDACs promote transcription (Pijnappel et al. 2001; Wiren et al. 2005). Hos2 affects mostly the 5′ coding regions of growth-related genes, promoting their expression by deacetylation, primarily of H4K16Ac in their ORF (Wiren et al. 2005). In hos2Δ homozygous crosses the mat-bias was largely diminished (Q = 0.77, Figure 3B) without changing the level of overall recombination frequencies greatly (supplemental Table S2). In clr6-1 homozygous crosses, the overall prototroph frequency was reduced in the type III cross 2.4-fold and the type IV crosses 3.8-fold, thus eliminating the mat-bias (Q = 0.95). Interestingly, heterozygous hos2Δ crosses (h+ hos2+ × h− hos2Δ) as well as heterozygous clr6-1 crosses (h+ clr6+ × h− clr6-1) showed wild-type mat-biases of 0.58 and 0.48, whereas heterozygous crosses involving hos2Δ or clr6-1 parents with h− cell type (h+ hos2Δ × h− hos2+ and h+ clr6-1 × h− clr6+) displayed mat-biases similar to the homozygous crosses (Q = 0.74, and Q = 0.87, Figure 3B, supplemental Table S2). Thus, the role of these two HDACs in mat-bias establishment is more pronounced in h+ strains. Homozygous crosses with clr3Δ did not change the mat-bias as hos2Δ and clr6-1 did (Q = 0.69, Figure 3B, supplemental Table S2).
The mat-bias does not depend on the structure of the mating-type region:
What is the difference in cells expressing h+ vs. h− cell-type information, except the expression of the cell-type-specific genes? The strains initially used were stable heterothallic strains h+N and h−S. The main difference between these strains is the genomic structure of the mating-type region (supplemental Figure S1). h+N contains a duplication of the mat1 cassette separated by the K region that normally lies between mat2-P and mat3-M (Beach and Klar 1984). The K-region is essential for transcriptional silencing and suppression of recombination therein (Grewal and Klar 1997), but is dispensable for growth. The h−S strain contains a fusion of the mat2-P and mat3-M cassettes along with a deletion of the K-region (Beach and Klar 1984).
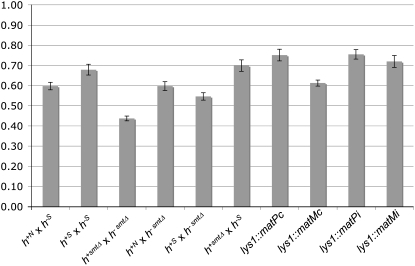
To check whether the loss of the K-region or other mating-type structures affects the mat-bias at ura4A, we measured recombination in strains with different mating-type structures, namely h+S, mat1-P smtΔ, and mat1-M smtΔ, respectively (supplemental Figure S1), and combined them with the previously used strains h+N and h−S. The mating-type structures of all strains were confirmed by Southern blot (supplemental Figure S2). The mat-biases from any combinations showed variations but were detected with statistical significance in all cases (Figure 4, supplemental Table S3).
Figure 4.—
The mat-bias does not depend on mating-type structure. Shaded bars indicate the ppm quotient average of type III and type IV crosses (y-axis) with strains of diverse mating-type structures and additional mating-type genes. Crosses were repeated at least three times (supplemental Table S3). Type III crosses were EPY5 × EPY12, EPY5 × EPY18, EPY51 × EPY30, EPY51 × EPY12, EPY51 × EPY18, EPY5 × EPY30, EPY5 × EPY28, EPY57 × EPY12, EPY5 × EPY89, and EPY91 × EPY12, respectively. Type IV crosses were EPY6 × EPY11, EPY17 × EPY11, EPY29 × EPY52, EPY6 × EPY52, EPY17 × EPY52, EPY29 × EPY11, EPY27 × EPY11, EPY6 × EPY58, EPY92 × EPY11, and EPY6 × EPY90, respectively. Error bars stand for standard error on the mean.
Since all tested mating-type locus structures show mat-bias manifestation, we decided to check whether additional expression of genes from the mat1 cassette could influence the mat-bias. The h− cell type specifically expresses two genes, matMc and matMi, whereas the h+ strain expresses matPc and matPi. The matPc and matMc genes are responsible for pheromone and receptor synthesis and the following conjugation. The entry into meiosis requires the expression of matPi and matMi genes, which code for transcriptional co-activators of mei3 (Willer et al. 1995). In the diploid P/M cells, matPi and matMi are not expressed during vegetative growth (Kelly et al. 1988). The ectopic insertions of matPc, matMc, matPi, and matMi genes at the lys1 locus have been shown to lead to expression of the corresponding proteins but the level of protein expression was not determined (Yamamoto and Hiraoka 2003). Heterozygous crosses with corresponding insertions (h+N strains with integrated matPc or matPi and h−S strains with integrated matMc or matMi) did not affect the mat-bias (Figure 4, supplemental Table S3). We did not test co-integration of matPc and matPi or matMc and matMi, respectively, to check for cooperative action of both activators.
Role of Swi5 and Swi2 in mat-bias establishment:
If the mat-bias were an indirect consequence of expression of the regulatory proteins encoded at the mat1 locus, the identification of differentially expressed genes in the two heterothallic strains would be the basis for finding regulators of the mat-bias. Among cell-type-specific genes coding for pheromone synthesis or pheromone receptors or generally involved in the mating process, few genes were detected that have other obvious functions (Mata and Bahler 2006). Among them is swi5, which showed a slightly higher expression in M-cells (1.8-fold) than in P-cells in the Ste11 overexpression background. Swi5 has a role in meiotic DNA repair in complex with other recombination proteins (reviewed in Akamatsu et al. 2003; Ellermeier et al. 2004; Haruta et al. 2008) and participates with Swi2 in a complex involved in DNA break repair for mating-type switching (Akamatsu et al. 2003). Swi2 exhibits a cell-type-specific chromatin localization pattern in the mating-type region and recruits Swi5 to this region as well (Jia et al. 2004). This Swi2/Swi5 loading determines long-range intrachromosomal interactions leading to correct donor choice.
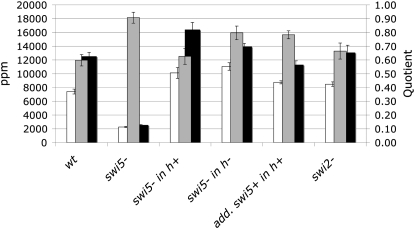
We checked the mat-bias at ura4A in swi5 and swi2 deletion mutants. Neither the mat-bias nor the prototroph frequencies changed in crosses with homozygous swi2Δ (Figure 5, supplemental Table S4). Homozygous swi5Δ crosses, however, showed a threefold reduction of prototroph frequencies at ura4A for type III crosses and a sixfold reduction for type IV crosses (Figure 5, supplemental Table S4). Thus the mat-bias was erased in swi5Δ (Q = 0.91). In heterozygous swi5Δ crosses, where the swi5Δ parents were h+, the mat-bias was not much different from the wild-type level (Q = 0.63), whereas in the opposite configuration (swi5Δ parents were h−), the mat-bias was reduced (Q = 0.80). It was also reduced in crosses with additional copies of swi5 integrated into h+ swi5+ strains (Q = 0.78).
Figure 5.—
The mat-bias in swi5 mutants, swi5 duplication strains, and swi2 mutants. Open bars indicate the ppm average of at least four independent type III crosses (primary y-axis): EPY5 × EPY12, EPY44 × EPY45, EPY5 × EPY45, EPY44 × EPY12, EPY5 × KLY390, and EPY53 × EPY56, respectively. Solid bars indicate the ppm average of at least three independent type IV crosses: EPY6 × EPY11, EPY43 × EPY46, EPY43 × EPY11, EPY6 × EPY46, KLY389 × EPY11, and EPY54 × EPY55, respectively. Shaded bars represent the mean quotient of ppm of type III and ppm of type IV crosses (secondary y-axis). Error bars stand for standard error on the mean.
From these results, we can conclude that mat-bias establishment depends on swi5 and that this dependency is more eminent in strains of the h− cell type. The fact that a wild-type mat-bias was found in swi2Δ crosses led us to believe that the Swi2/Swi5 subcomplex is not involved in this phenomenon. Swi5 also interacts with Sfr1, which shows homology to Swi2 (Haruta et al. 2008). In vitro DNA strand-exchange assays with purified Swi5/Sfr1 complexes have shown that Swi5/Sfr1 stimulates the ssDNA-dependent ATPase activities of Rhp51 and Dmc1 during DNA strand exchange. Indeed, the reduced prototroph frequencies in the swi5Δ homozygous crosses (Figure 5, supplemental Table S4) support its importance in recombination, as was previously shown (Ellermeier et al. 2004). It was further suggested that Swi5/Sfr1 might have both ssDNA- and dsDNA-binding activities (Haruta et al. 2006). From which strand—the broken chromatid (ssDNA) or the repair partner (dsDNA)—this activity would support Rhp51 or Dmc1 remains to be established. However, the alternative role of Swi5 in mating-type switching—where Swi5/Swi2 is bound to mating-type heterochromatin probably through the DNA-binding motif of Swi2 and thus directs the repair of the cleaved imprint (gene conversion) toward the repair partner—presents the possibility that Swi5/Sfr1 bound to the unbroken chromatid could pull the Rhp51 or Dmc1 recombination machinery toward the repair partner.
How is the mat-bias established?
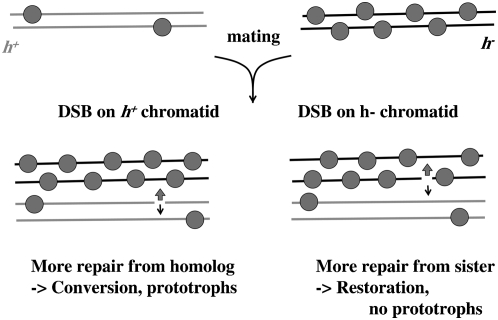
The mat-bias reflects a subtle phenomenon, which may have its roots in tiny differences of chromatin states of the two cell types h+ and h−. How can these tiny differences affect the outcome of recombination? Because of gene conversion polarity at ura4A, uracil prototrophs arise in type IV crosses mainly by DSBs in the h+ chromatid, whereas in type III crosses, DSBs must occur in the h− chromatid. One obvious explanation for the mat-bias would be that the h+ chromatid gets more DSBs than the h− chromatid. Such a bias could depend on the chromatin state (histone acetylation/deacetylation). The weaker or erased mat-biases in homozygous crosses of gcn5, ada2, clr6, and hos2 mutants confirm such a supposition (Figure 3). Heterozygous crosses with hos2Δ or clr6-1 in the h+ parent gave higher ppm levels in the type III cross than the opposite heterozygous cross (supplemental Table S2), and thus an alleviated mat-bias results. This higher amount of prototrophs must be explained by more DSBs in the h− chromatid compared to wild type. However, the fact that reduced deacetylation of histones in the h+ chromatid would rather render the chromatin open and thus accessible for DSBs, led us to believe that hos2 and clr6 mutants would rather increase the amount of DSBs in the h+ chromatid than stimulate DSB formation in the h− chromatid. In addition, the loss of the mat-bias in a swi5 mutant would hardly support the model of DSB formation bias: How would a protein involved in assisting recombination complexes stimulate the formation of DBSs in the h+ chromatid, especially, when its expression is stronger in the h− strain?
We propose an alternative model, which explains the mat-bias by a repair partner choice, balanced by subtle differences of Swi5 associated with chromatin in h+ vs. h− cells (Figure 6). In h− cells, more Swi5 is associated with chromatin than in h+ cells (higher swi5 mRNA levels in h− cells). DSBs occurring in the h+ chromatid would more quickly be repaired through the homolog than the sister because the h− chromatid is decorated with higher amounts of Swi5 and would thus more likely attract the recombination machinery. A gene conversion event would result from this repair partner. Such directionality in intrachromatid recombination by Swi5/Swi2 was shown in mating-type switching (Jia et al. 2004). The association of Swi5 to chromatin in this case would depend on Sfr1 (proposed to have a dsDNA-binding activity; Haruta et al. 2008). DSBs in the h− chromatid, on the other hand, would more often be repaired through the sister chromatid, attracted by the Swi5 presence. This would result in restoration of the mutant ura4-A13 allele, so there would be no yield of prototrophs. In a homozygous swi5Δ cross, the choice of repair partner is no longer biased toward the h− chromatid since no Swi5 is present on either chromatid (supplemental Figure S3A). In heterozygous swi5Δ crosses with swi5Δ in the h+ strain, the mat-bias would be detected since the h− chromatid is decorated with Swi5 and thus directs repair to the homolog when DSBs occur in the h+ chromatid or to the sister when DSBs occur in the h− chromatid (supplemental Figure S3B). Is swi5Δ combined with h−, the mat-bias will be attenuated because now the DSBs in the h− chromatid would no longer prefer the sister (no Swi5 decoration) but rather the homolog (wild-type h+ Swi5 decoration; supplemental Figure S3C).
Figure 6.—
Mat-bias model. See text.
The specific expression of mating-type-specific factors must be the basis for the mat-bias. Why Swi5 is more strongly expressed in h− cells might be explained by its function in mating-type switching. The distribution of Swi2/Swi5 along the 15-kb-long heterochromatin of the mating-type region in homothallic h− cells might call for a stronger expression of Swi5. The mat-bias thus might be only a side product of this expression bias. Indeed, an additional swi5 gene in h+ crossing partners, minimizing the swi5 expression bias between the two mating types, reduces the mat bias, although it is not completely gone (Figure 5). How well Swi5 is associated with chromatin might depend on the chromatin state. We cannot distinguish whether the influence of the HATs or HDACs on the mat-bias depends on Swi5 presence or whether Swi5 and the histone acetylation state contributes synergistically to mat-bias establishment. Whatever influences the delicate balance of repair partner choice influences the mat-bias.
Acknowledgments
We thank A. Yamamoto and Y. Hiraoka for providing plasmids and E. Lehmann for intergenic recombination data. This work was supported by the Swiss National Science Foundation (SNF); K.L. was supported by a SNF Marie-Heim-Vögtlin fellowship and the Hochschulstiftung/Burgergemeinde Bern.
References
- Akamatsu, Y., D. Dziadkowiec, M. Ikeguchi, H. Shinagawa and H. Iwasaki, 2003. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc. Natl. Acad. Sci. USA 100 15770–15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur, M., E. Hartsuiker, E. Lehmann, K. Ludin, P. Munz et al., 2005. The meiotic recombination hot spot ura4A in Schizosaccharomyces pombe. Genetics 169 551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach, D. H., and A. J. Klar, 1984. Rearrangements of the transposable mating-type cassettes of fission yeast. EMBO J. 3 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie, G. A., R. W. Hyppa, A. F. Taylor, K. Zakharyevich, N. Hunter et al., 2006. Single Holliday junctions are intermediates of meiotic recombination. Cell 127 1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie, G. A., R. W. Hyppa, H. P. Cam, J. A. Farah, S. I. Grewal et al., 2007. A discrete class of intergenic DNA dictates meiotic DNA break hotspots in fission yeast. PLoS Genet. 3 e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier, C., H. Schmidt and G. R. Smith, 2004. Swi5 acts in meiotic DNA joint molecule formation in Schizosaccharomyces pombe. Genetics 168 1891–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan, J., P. K. Rabitsch, B. Sakem, O. Csutak, V. Latypov et al., 2005. Novel genes required for meiotic chromosome segregation are identified by a high-throughput knockout screen in fission yeast. Curr. Biol. 15 1663–1669. [DOI] [PubMed] [Google Scholar]
- Grewal, S. I., and A. J. Klar, 1997. A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics 146 1221–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm, C., J. Bahler and J. Kohli, 1994. M26 recombinational hotspot and physical conversion tract analysis in the ade6 gene of Schizosaccharomyces pombe. Genetics 136 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk, A. L., and J. Kohli, 2003. Five RecA-like proteins of Schizosaccharomyces pombe are involved in meiotic recombination. Genetics 165 1031–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz, H., 1971. Site specific induction of gene conversion in Schizosaccharomyces pombe. Genetics 69 317–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz, H., U. Leupold and N. Loprieno, 1974. Schizosaccharomyces pombe Handbook of Genetics, pp. 395–446, edited by R. C. King. Plenum, New York.
- Haruta, N., Y. Kurokawa, Y. Murayama, Y. Akamatsu, S. Unzai et al., 2006. The Swi5-Sfr1 complex stimulates Rhp51/Rad51- and Dmc1-mediated DNA strand exchange in vitro. Nat. Struct. Mol. Biol. 13 823–830. [DOI] [PubMed] [Google Scholar]
- Haruta, N., Y. Akamatsu, Y. Tsutsui, Y. Kurokawa, Y. Murayama et al., 2008. Fission yeast Swi5 protein, a novel DNA recombination mediator. DNA Repair 7 1–9. [DOI] [PubMed] [Google Scholar]
- Hirota, K., K. I. Mizuno, T. Shibata and K. Ohta, 2008. Distinct chromatin modulators regulate the formation of accessible and repressive chromatin at the fission yeast recombination hotspot ade6–M26. Mol. Biol. Cell 19 1162–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, S., T. Yamada and S. I. Grewal, 2004. Heterochromatin regulates cell type-specific long-range chromatin interactions essential for directed recombination. Cell 119 469–480. [DOI] [PubMed] [Google Scholar]
- Keeney, S., and N. Kleckner, 1995. Covalent protein-DNA complexes at the 5′ strand termini of meiosis-specific double-strand breaks in yeast. Proc. Natl. Acad. Sci. USA 92 11274–11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, M., J. Burke, M. Smith, A. Klar and D. Beach, 1988. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J. 7 1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten, M., and A. S. Goldman, 1995. Meiotic recombination hotspots. Annu. Rev. Genet. 29 423–444. [DOI] [PubMed] [Google Scholar]
- Liu, J., T. C. Wu and M. Lichten, 1995. The location and structure of double-strand DNA breaks induced during yeast meiosis: evidence for a covalently linked DNA-protein intermediate. EMBO J. 14 4599–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludin, K., J. Mata, S. Watt, E. Lehmann, J. Bahler et al., 2008. Sites of strong Rec12/Spo11 binding in the fission yeast genome are associated with meiotic recombination and with centromeres. Chromosoma 117 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata, J., and J. Bahler, 2006. Global roles of Ste11p, cell type, and pheromone in the control of gene expression during early sexual differentiation in fission yeast. Proc. Natl. Acad. Sci. USA 103 15517–15522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker, J. D., M. Dominska, P. W. Greenwell, E. Rinella, D. C. Bouck et al., 2008. The histone methylase Set2p and the histone deacetylase Rpd3p repress meiotic recombination at the HIS4 meiotic recombination hotspot in Saccharomyces cerevisiae. DNA Repair 7 1298–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, K., Y. Emura, M. Baur, J. Kohli, K. Ohta et al., 1997. The meiotic recombination hot spot created by the single-base substitution ade6–M26 results in remodeling of chromatin structure in fission yeast. Genes Dev. 11 876–886. [DOI] [PubMed] [Google Scholar]
- Neale, M. J., and S. Keeney, 2006. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques, F., and J. E. Haber, 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes, T. D., 2001. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2 360–369. [DOI] [PubMed] [Google Scholar]
- Pijnappel, W. W., D. Schaft, A. Roguev, A. Shevchenko, H. Tekotte et al., 2001. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 15 2991–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakem, B., 2005. Competition between the meiotic recombination hotspots ade6-M26 and ura4A of Schizosaccharomyces pombe and analysis of DNA double strand breaks at ura4A. Ph.D. Thesis, University of Bern, Bern, Switzerland.
- Strahl, B. D., and C. D. Allis, 2000. The language of covalent histone modifications. Nature 403 41–45. [DOI] [PubMed] [Google Scholar]
- Sung, P., 1997. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 11 1111–1121. [DOI] [PubMed] [Google Scholar]
- Willer, M., L. Hoffmann, U. Styrkarsdottir, R. Egel, J. Davey et al., 1995. Two-step activation of meiosis by the mat1 locus in Schizosaccharomyces pombe. Mol. Cell. Biol. 15 4964–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiren, M., R. A. Silverstein, I. Sinha, J. Walfridsson, H. M. Lee et al., 2005. Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. EMBO J. 24 2906–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, A. P. H., K. Maundrell and S. Shall, 1986. Transformation of Schizosaccharomyces pombe by non-homologous unstable integration of plasmids in the genome. Curr. Genet. 10 503–508. [DOI] [PubMed] [Google Scholar]
- Yamada, T., K. Mizuno, K. Hirota, N. Kon, W. P. Wahls et al., 2004. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 23 1792–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, A., and Y. Hiraoka, 2003. Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. EMBO J. 22 2284–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn-Zabal, M., E. Lehmann and J. Kohli, 1995. Hot spots of recombination in fission yeast: inactivation of the M26 hot spot by deletion of the ade6 promoter and the novel hotspot ura4-aim. Genetics 140 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]