Abstract
The Mre11/Rad50/Nbs1 (MRN) complex is required for eukaryotic DNA double-strand break (DSB) repair and meiotic recombination. We cloned the Coprinus cinereus rad50 gene and showed that it corresponds to the complementation group previously named rad12, identified mutations in 15 rad50 alleles, and mapped two of the mutations onto molecular models of Rad50 structure. We found that C. cinereus rad50 and mre11 mutants arrest in meiosis and that this arrest is Spo11 dependent. In addition, some rad50 alleles form inducible, Spo11-dependent Rad51 foci and therefore must be forming meiotic DSBs. Thus, we think it likely that arrest in both mre11-1 and the collection of rad50 mutants is the result of unrepaired or improperly processed DSBs in the genome and that Rad50 and Mre11 are dispensable in C. cinereus for DSB formation, but required for appropriate DSB processing. We found that the ability of rad50 mutant strains to form Rad51 foci correlates with their ability to promote synaptonemal complex formation and with levels of stable meiotic pairing and that partial pairing, recombination initiation, and synapsis occur in the absence of wild-type Rad50 catalytic domains. Examination of single- and double-mutant strains showed that a spo11 mutation that prevents DSB formation enhances axial element (AE) formation for rad50-4, an allele predicted to encode a protein with intact hook region and hook-proximal coiled coils, but not for rad50-1, an allele predicted to encode a severely truncated protein, or for rad50-5, which encodes a protein whose hook-proximal coiled-coil region is disrupted. Therefore, Rad50 has an essential structural role in the formation of AEs, separate from the DSB-processing activity of the MRN complex.
MEIOSIS is the unique type of cell division that results in the production of haploid gametes. During prophase I, homologs condense, pair, and recombine. At least one recombination event per homologous pair becomes a crossover, which, in combination with sister-chromatid cohesion, creates a chiasma that holds homologs together as they attach to the metaphase I spindle. Release of sister-chromatid cohesion along chromosome arms allows anaphase I separation of homologs, and release of centromere cohesion allows anaphase II separation of sister chromatids.
Meiotic recombination events initiate with a Spo11-catalized DNA double-strand break (DSB), and some of the proteins required for meiotic recombination have been recruited from mitotic DNA repair pathways. A primary example is the complex of proteins referred to as Mre11, Rad50, and Nbs1 (MRN). Evidence from Saccharomyces cerevisiae, in which the Nbs1 ortholog is called Xrs2, showed that the MRN complex is necessary for meiotic DSB formation by Spo11 (Alani et al. 1990; reviewed in Borde 2007). However, in other species different results have been obtained. In Caenorhabditis elegans, Rad50 facilitates DSB formation by Spo11 but is apparently not strictly required for Spo11 activity (Hayashi et al. 2007). In Schizosaccharomyces pombe (Young et al. 2004), Arabidopsis thaliana (Puizina et al. 2004), and Drosophila melanogaster (Mehrotra and McKim 2006) the MRN complex is required for break processing but not break formation.
In all eukaryotic organisms studied, mre11, rad50, and xrs2/nbs1 mutants exhibit meiotic recombination defects and radiation sensitivity (Cox and Parry 1968; Game et al. 1980; Malone and Esposito 1981; Malone 1983; Alani et al. 1990; Ivanov et al. 1992; Ajimura et al. 1993; Ramesh and Zolan 1995; Tavassoli et al. 1995; Carney et al. 1998; Chamankhah et al. 1998; Matsuura et al. 1998; Varon et al. 1998; Stewart et al. 1999; Gerecke and Zolan 2000; Gallego et al. 2001; Hartsuiker et al. 2001; Pitts et al. 2001; Hayashi et al. 2007). The members of the Mre11 complex have been grouped by epistasis analysis (Game et al. 1980; Malone and Esposito 1981; Ivanov et al. 1992; Ajimura et al. 1993; Valentine et al. 1995) and have been shown to directly interact (Johzuka and Ogawa 1995; Petrini et al. 1995; Dolganov et al. 1996; Trujillo et al. 1998; Varon et al. 1998). Mammalian deletion mutants of the Mre11 complex exhibit embryonic lethality (Xiao and Weaver 1997; Luo et al. 1999; Yamaguchi-Iwai et al. 1999; Zhu et al. 2001). Viable mutations in human homologs of the MRN complex invariably result in syndromes characterized by a loss of fertility, and cancer early in life (Carney et al. 1998; Matsuura et al. 1998; Varon et al. 1998; Stewart et al. 1999; Pitts et al. 2001; Bender et al. 2002). The biochemical properties of the individual proteins composing this complex have been characterized in mammals and yeast (reviewed in Williams et al. 2007). Mre11 binds DNA and exhibits in vitro ssDNA endonuclease and Mn2+-dependent 3′–5′ double-stranded DNA exonuclease activities (Paull and Gellert 1998). Both Mre11 nuclease activity and homodimerization are important for DSB repair (Williams et al. 2008). Rad50 is an ATP-dependent DNA binding protein that forms a multimer (Raymond and Kleckner 1993; Hopfner et al. 2000a) and has both ATPase and adenylate kinase activities that are important for protein function (Bhaskara et al. 2007). Rad50 is furthermore a prototype for a superfamily of SMC proteins and ABC transporters (Hopfner and Tainer 2003). The Nbs1 protein serves to localize the complex in the nucleus (Tauchi et al. 2001), modifies MRN complex activity (Paull and Gellert 1999), and plays a primary role in signal transduction after DNA damage (Lee and Paull 2004, 2005).
Partial Pyrococcus furiosus Rad50/Mre11 complexes have been crystallized by Hopfner et al. (2000b, 2001). Rad50 has two α-helical coiled-coil domains separated by a flexible hinge (referred to as a hook) harboring a semi-zinc finger at the center. The N and C termini of the protein are globular in nature and together compose a functional ATP-binding site. They are brought into contact with each other by an intertwining of the N- and C-terminal coils. The two interwoven coils form a flexible rod, creating a binding site for Mre11 made up of acidic residues just adjacent to the globular domain (Hopfner et al. 2001). Crystallographic and biochemical data (Hopfner et al. 2002a) have confirmed a role in protein dimerization for the conserved semi-zinc finger in the hook region (Sharples and Leach 1995). Current models (Hopfner et al. 2002a,b; Williams and Tainer 2005) suggest that hook interactions of Rad50 molecules allow bridging of either sister chromatids or DSB ends. These Rad50 molecules are also held together with the aid of two dimerized Mre11 molecules at each opposing head of the complex (Williams et al. 2008). Recent atomic force microscopy studies (Moreno-Herrero et al. 2005) showed that MRN complexes change conformation upon DNA binding. The unbound complexes exhibit greater flexibility of the coiled-coil regions, which may favor intracomplex interactions, whereas the coiled coils of DNA-bound complexes are more rigid and parallel, thus favoring intercomplex interactions that would facilitate complex ability to bridge and stabilize DNA ends (Williams and Tainer 2005).
Meiotic defects in rad50 mutants (reviewed in Borde 2007; Cherry et al. 2007) include failure of Spo11-dependent DSB formation or processing (Alani et al. 1990; Young et al. 2004; Mehrotra and McKim 2006; Hayashi et al. 2007), incomplete formation of axial elements (AE) and the synaptonemal complex (SC) (Alani et al. 1990), and the formation of nonviable products (Alani et al. 1990; Young et al. 2004). Thus, it has a central role in meiotic recombination and chromosome metabolism.
We cloned the rad50 gene and examined members of the MRN complex in the basidiomycete Coprinus cinereus. C. cinereus has naturally synchronous meiosis (Raju and Lu 1970), well-developed cytogenetics (Pukkila et al. 1984; Pukkila and Lu 1985; Seitz et al. 1996; Li et al. 1999), and an annotated genomic sequence (Stajich et al. 2006). In screens for mutants defective in both the survival of gamma radiation and meiosis (Zolan et al. 1988; Valentine et al. 1995), we identified four complementation groups, termed rad3, rad9, rad11, and rad12. These four genes are all part of the same gamma radiation survival pathway (Valentine et al. 1995; Cummings et al. 2002). The gene rad11 was cloned and found to encode the C. cinereus ortholog of Mre11 (Gerecke and Zolan 2000). Therefore, we reasoned that one of the other genes of the group would encode the C. cinereus ortholog of Rad50. Here we present our finding that the C. cinereus gene initially named rad12 (Ramesh and Zolan 1995) encodes Rad50 and examine critical features of the role of Rad50 in meiosis.
MATERIALS AND METHODS
Strains and culture conditions:
The wild-type dikaryon used for the Rad51 time-course experiment shown in Figure 4 has been previously described (Valentine et al. 1995). rad50 mutant strains were generated in five separate mutant screens (Zolan et al. 1993, 1995; Ramesh and Zolan 1995; Valentine et al. 1995), all starting with the strain Java-6 (Binninger et al. 1987). Strain Okayama-7 (Wu et al. 1983) was used for all outcrosses and backcrosses. Of the rad50 strains examined in this study, rad50-1, rad50-2, rad50-3, rad50-4, rad50-5, rad50-6, rad50-8, rad50-14, and rad50-15 were outcrossed and then backcrossed four times; rad50-10, rad50-11, and rad50-16 were outcrossed and then backcrossed once; and rad50-7, rad50-9, rad50-12, and rad50-13 were outcrossed only.
Figure 4.—
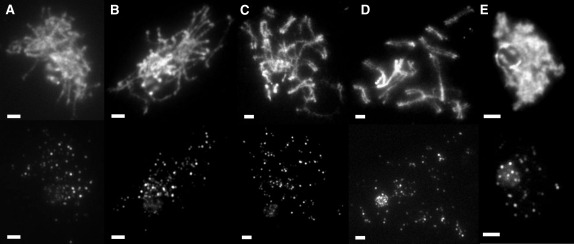
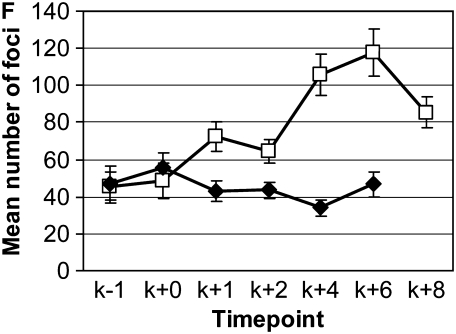
Anti-Rad51 staining in wild type and spo11-1. (A–E) Representative images of wild-type meiotic time course. (A) Preleptotene image taken 1 hr after karyogamy (K + 1). (B) Leptotene image taken at K + 2. (C) Zygotene image taken at K + 4. (D) Pachytene image taken at K + 6. (E) Diplotene image taken at K + 8. Bars, 2 μm for all images. (F) Mean number of Rad51 foci in spo11-1 (♦) and in wild type (□). Error bars show 95% confidence intervals.
The rad50-1;spo11-1, rad50-4;spo11-1, rad50-5;spo11-1, rad50-8;spo11-1, and mre11-1;spo11-1 strains were generated by crossing a rad50 or a mre11 single mutant to R126-49 [spo11-1 (AmBm); Cummings et al. 1999; Celerin et al. 2000]. Double mutants were identified by subjecting progeny to a radiation-sensitivity chunk test (Zolan et al. 1988) to test for the rad50 or the mre11 mutation. All strains except those used to construct the mre11-1;spo11-1 double mutant were then mated to spo11-1 strains with A42B42 or A43B43 mating types. Strains that mated to spo11-1;A43B43 only, fruited white (sporeless), and were radiation sensitive were scored as A42B42 double mutants. These in turn were mated to strain Okayama-7 (A43B43) to generate double mutants, sibling single mutants, and wild-type controls with both A42B42 and A43B43 mating types. Compatible monokaryons were mated and used for further analysis. For construction of the mre11-1;spo11-1 double mutant, progeny were screened for hygromycin resistance as a marker for the insertional mutation in spo11-1 (Celerin et al. 2000). Strain genotype was confirmed by sequencing and/or diagnostic restriction digests of PCR-amplified fragments containing the mutated region. Mating pairs were identified as previously described (Zolan et al. 1988).
Spore viability:
Spore viabilities were determined using the spotted drop method described in Celerin et al. (2000). Spores were serially diluted and 10-μl drops were spotted onto a gridded plate. Five to seven mushroom caps were counted per strain and analyzed using the ANOVA test (SPSS 10.0 for the Macintosh).
Degenerate PCR amplification of Ccrad50:
Rad50 homologs from other organisms were aligned using ClustalX (Thompson et al. 1997). Regions of high conservation were used as the basis for the design of degenerate primers. rad50 was amplified from C. cinereus genomic DNA using a degenerate primer pair: F1, 5′ AAR ACN ACN ATH ATH GAR TGY YTN 3′, and R1, 5′ GGG NWS RTC YTC YTG RTG RCA RAA D 3′, corresponding to amino acids 38–46 and 158–165 of CcRad50. A product corresponding to rad50 was used to probe a chromosome 8- and 9-enriched cosmid library of strain Okayama-7 (Zolan et al. 1992). Three cosmids were isolated: 3B6, 5F8, and 23G1. The cosmid 23G1 was found to contain a complete copy of rad50 through subsequent sequencing analysis.
RFLP analysis:
Strain Okayama-7 and a fourth-generation backcrossed rad12-4 strain (rad12-4;4-1) were mated. Progeny were analyzed as described in Gerecke and Zolan (2000) except that all genomic DNA was digested with EcoRI and probed with a 3B6 cosmid containing Ccrad50.
Transformation rescue assay:
Transformation rescue of oidial protoplasts of the C. cinereus rad50-4 mutant was performed as described (Binninger et al. 1987; Zolan et al. 1992). A rad50-4;trp1-1,1-6;ade8 strain was transformed with 5 μg of cosmid 23G1. This cosmid also contains the trp1 gene, and tryptophan prototrophs were selected and assayed as described in Gerecke and Zolan (2000) except that the rescue of meiotic defects was assayed through a mating with a compatible rad50-4 strain (rad50-4;4-2).
Sequencing of wild-type rad50:
Plasmids containing subclones of genomic DNA of rad50 were isolated from Escherichia coli and initially sequenced with M13F/R or T3/T7 vector primers. Primers for a sequencing walk were based upon the previous sequence data and designed using the OLIGO 5.0 program. Primers for internal sequencing and amplification were made by Genosys or by MWG-Biotech. PCR products were sequenced using standard techniques. Sequences were edited and made contiguous by Sequencher v.3.1.1 (Gene Codes), and translated by DNASIS v.2.0 (Hitachi Software Engineering). Predicted proteins were identified using the blastx program in BLAST (Altschul et al. 1990) and alignments were performed using Clustal X (Thompson et al. 1997). Coiled structure of Rad50 was predicted using the MultiCoils program (Wolf et al. 1997).
Sequencing of rad50 cDNA:
First-strand cDNA synthesis using reverse transcriptase (SuperScript II; GibCo-BRL Life Technologies, Gaithersburg, MD) was performed on poly(A)+ mRNA isolated from mushroom caps at 1 hr after karyogamy (K + 1). A gene-specific amplification was then performed using a rad50 primer pair. The product was run on an agarose gel and Southern blotted. It was then probed with an amplified piece of rad50. A band that hybridized to rad50 DNA was gel extracted and cloned into a TOPO-TA vector. The resulting clones were sequenced using vector primers and shown to correspond to a 1.1-kb fragment of 3′ rad50 cDNA. A reverse primer was made, extending across 10 bp of exon sequence immediately following the 3′ and 5′ flank of intron 17. In this way we eliminated amplification of genomic sequence. This primer was used to extend the oligo (dT) first-strand cDNA synthesis reaction farther. This first-strand synthesis reaction was used as a template for an amplification with a forward primer across intron 15. This fragment was sequenced. A new first-strand cDNA synthesis reaction was then performed, using a reverse primer across intron 15 and poly(A)+ mRNA isolated from mushroom caps at K + 6. This first-strand reaction was amplified (Advantage 2.0 kit, BD Biosciences–Clontech) with a forward primer from the beginning of the rad50 genomic sequence. This product was also sequenced. Several primers in the 5′-UTR were used to amplify the 5′ end of rad50 with a reverse primer that spanned the fourth intron. These products were sequenced. Sequences were made contiguous using the Sequencher program v.3.1.1 (Gene Codes).
Sequencing of mutant rad50 alleles:
Primers for internal sequencing and amplification were based upon the wild-type sequence data and designed using the OLIGO 5.0 program. Overlapping regions of DNA for sequencing original rad50 mutant strains were amplified by PCR and directly sequenced. Template DNA for amplification reactions consisted of rad50 mutant genomic DNA extracted from vegetative tissue (Zolan and Pukkila 1986). PCR products were desalted using the QIAquick PCR purification kit or extracted from agarose gels using the QIAEX II kit (QIAGEN, Valencia, CA). Sequencing and analysis were performed using standard procedures.
RT–PCR on Ccrad50 mutants:
RT–PCR was performed on RNA extracted from heterozygotes of rad50 mutant and wild-type mushrooms. The two strains, rad50-1 and rad50-4, were mated to strain PJP 307, a meiotically wild-type strain. The resulting dikaryons were subcultured twice and poly(A)+ RNA was extracted from meiotic tissue at K + 6 (RNeasy plant mini kit, QIAGEN; PolyATtract System 1000, Promega, Madison, WI). First-strand cDNA was made using the Superscript kit (GibCo-BRL Life Technologies). For rad50-1, primers were designed to be cDNA specific and to span introns 3 and 5. For rad50-4, a cDNA-specific forward primer spanning intron 19 was used in combination with a 3′-UTR reverse primer or an intron 22-spanning cDNA-specific primer. PCR amplification and sequencing were performed by standard techniques.
Protein modeling of CcRad50 mutants:
Protein models were based on the structures of the P. furiosus Rad50 protein (RCSB codes 1F2U and 1L8D) (Hopfner et al. 2000b, 2002a). CcRad50, CcRad50-4, and CcRad50-5 models were created with MOLSCRIPT (Kraulis 1991) and PYMOL (http://www.pymol.org).
Electron microscopy:
Chromosome spreads were performed as described by Pukkila et al. (Pukkila and Lu 1985; Pukkila et al. 1992). Silver nitrate staining of meiotic nuclei was performed as per Seitz et al. (1996). Grids were prepared as per Pukkila et al. (1992) and viewed with a JEOL-1010 electron microscope. Images of stained nuclei were scanned into a computer and analyzed using Adobe Photoshop 7.0 (Adobe Systems), National Institutes of Health (NIH) Image 1.63 (Wayne Rasband, NIH), and Microsoft Excel '98.
Epistasis analysis of late meiotic phenotypes:
Slices of cap tissue were taken at various time points after karyogamy (as defined in Seitz et al. 1996), and veil cells were peeled and discarded. The tissue was stored in 70% ethanol, 30% NS buffer (20 mm Tris–HCl, pH 7.5, 0.25 m mannitol, 1 mm EDTA, 1 mm MgCl2, 0.1 mm CaCl2) at 4°. Single gill layers of meiotic cells were dissected and mounted onto ProbeOn Plus (Fisher Scientific, Pittsburgh) microscope slides with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Vectashield, Vector Laboratories, Burlingame, CA) and examined. For the rad50 experiments (Figure 3, A–C), slides were examined using a Nikon Microphot-FXA microscope with a Ludl filterwheel. Images of stained nuclei were captured using a Photometrics cooled digital CCD camera system and Quips XL SmartCapture/IP Lab Spectrum imaging software (Vysis). For the mre11-1 experiments (Figure 3D), images were taken using a Nikon E800 fluorescence microscope, driven by the Metamorph imaging system (Molecular Devices). Images were taken as z-series stacks using a step size of 0.09 μm, capturing all planes in which the tissue sample was in focus. Each stack was then flattened into a single image, using the maximum pixel brightness.
Figure 3.—
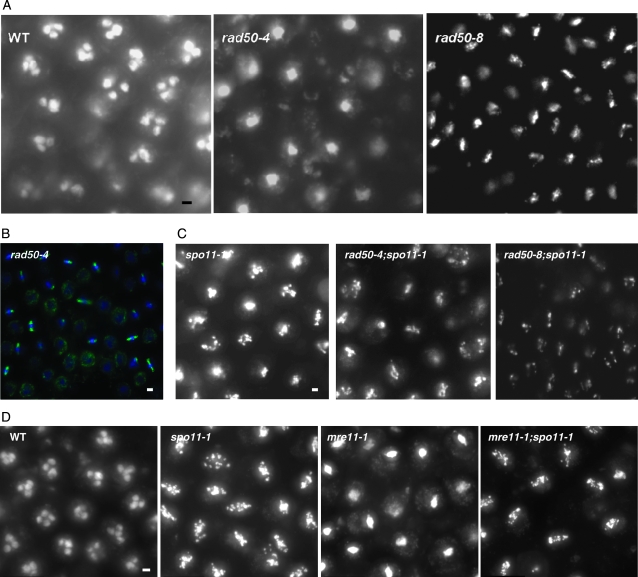
rad50 mutant and mre11-1 arrests are Spo11 dependent. (A–C) Samples were taken from mushrooms 12 hr after karyogamy, fixed, stained with DAPI, and photographed. (A) Wild-type and rad50-4 and rad50-8 single mutants. (B) rad50-4 stained with DAPI and with spindles detected using mouse anti-α-tubulin antibody, as described in Celerin et al. (2000). (C) A spo11-1 single mutant compared with rad50-4;spo11-1 and rad50-8;spo11-1 double mutants. (D) Wild-type, spo11-1, and mre11-1 single mutants compared to mre11-1;spo11-1. Tissue samples, taken at K + 10, were stained with DAPI and photographed as a z-stack series of images. Image stacks were flattened into a single image as described in the text. Bars, 2 μm.
Rad51 protein expression and purification:
We constructed a Rad51 protein expression construct by cloning the full-length rad51 cDNA (Stassen et al. 1997) into the pET-30 expression vector (Novagen), which was then transformed into BL21(DE3) cells for expression. Protein expression conditions are similar to Novagen company literature and alterations are described in full in supplemental methods.
Anti-Rad51 antibody production:
Purified Rad51 in SDS–PAGE gel slices was sent to Cocalico Biologicals (Reamstown, PA) and used for immunization of rabbits according to their injection schedule, using the acrylamide as the adjuvant.
Affinity purification of anti-Rad51 antibody:
To remove vector-encoded tags, serum was adsorbed using an affinity column made with a portion of Mre11 expressed in the same vector. Anti-Rad51 antibody was then purified using a second affinity column. A detailed description is in supplemental methods.
Commercial antibodies:
Anti-human Rad51 was obtained from Calbiochem (San Diego; La Jolla, CA) (PC130). Purified nonspecific IgG from rabbit serum was obtained from Sigma (St. Louis) (18140).
Immunostaining of chromosome spreads:
Staining of chromosome spreads was performed using the following protocol, which was adapted from Sym et al. (1993). Chromosome spreads were obtained as described (Pukkila et al. 1992; Seitz et al. 1996) except that we used uncharged slides (VWR), cleaned with Sparkle glass cleaner (A. J. Funk and Co.). Slides with chromosome spreads were wetted in PBS, pH 7.4, for 10 min. The slides were then blocked with 100 μl of PBS containing 5% BSA and 1% normal goat serum (Vector Laboratories catalog no. S-1000) twice for 15 min each at room temperature, under a parafilm coverslip. The slides were incubated with 100 μl anti-Rad51 antibody diluted to a final concentration of 1 μg/ml in PBS containing 5% BSA, covered with a parafilm coverslip, and incubated overnight protected from light in a humid environment. Slides were then washed three times for 10 min each with PBS + 0.05% Tween-20 at room temperature. Secondary antibody was then added to the slides at a final concentration of 1.2 μg/ml, in 5% BSA dissolved in PBS, for 4 hr at room temperature, protected from light in a humid environment. Slides were then washed three times for 10 min each with PBS + 0.05% Tween-20 at room temperature. Slides were mounted with DAPI in Vectashield mounting medium, sealed with nail polish, and stored at 4° protected from light until examined. For data analysis, an experiment was defined as the images taken from one slide, made using cells from one mushroom. Slides made from mushrooms that failed to complete fruiting body development were not used. For each combination of strain and time point examined, sufficient experiments were performed that at least two mushrooms were used and the total number of images combined across experiments was ≥30. Antibodies from separate purifications gave statistically similar results; therefore, data obtained using antibodies from different purifications were pooled. For averages and statistical tests, the number of foci from images generated from different experiments was pooled.
Immunostaining of tissue:
Meiotic spindles were detected in single layers of meiotic tissue with DM1A mouse anti-α-tubulin antibody (Accurate Chemical and Scientific), using the conditions described in Celerin et al. (2000).
Irradiation of spo11-1:
At K + 1 or K + 3 spo11-1 mushrooms were irradiated with 60 krad of gamma radiation, using a 137Cs irradiator (Mark 1 model 68-A, J. L. Shepard and Associates), at 1.2 krad/min. Chromosome spreads were made at K + 2 or K + 4, i.e., 10 min after samples were removed from the irradiator.
Fluorescence in situ hybridization:
Fluorescence in situ hybridization (FISH) was performed as described (Li et al. 1999). Probes 3 and 6 described in Li et al. (1999) were used, representing telomeric and interstitial sites, respectively, of chromosome 13.
Statistical analyses:
For spore viability comparisons and for data from immunostraining of chromosome spreads using anti-Rad51 antibody, comparisons between mean numbers for two groups were performed using Student's t-test, while multiple pairwise comparisons were performed using an ANOVA test and, where appropriate, Tukey's post hoc test. All tests were performed using Minitab or SPSS.
For FISH and electron microscopy (EM) data, statistical comparisons between strains were performed using chi-square tests, using the Excel program (Microsoft). For the analysis of EM data, the categories AE only and SC were pooled.
RESULTS
Cloning of C. cinereus rad50 and its identification as rad12:
We aligned Rad50 protein sequences from humans, mice, S. cerevisiae, and A. thaliana and designed primers to amplify a fragment within the relatively conserved 5′ region of the gene. Using C. cinereus genomic DNA as a template, we amplified and sequenced a 363-bp fragment, which BLAST (Altschul et al. 1997) analysis showed was likely orthologous to Rad50. Previous restriction fragment length polymorphism (RFLP) mapping had shown that the C. cinereus rad12 locus is on chromosome 8 in strain Okayama-7 (Zolan et al. 1993). Therefore, the rad50 PCR fragment was used to probe colony blots of a cosmid library enriched for chromosomes 8 and 9 from the Okayama-7 strain of C. cinereus (Zolan et al. 1992). Three cosmids were isolated, and the Ccrad50 gene was sequenced from subclones of these cosmids. Because Rad50 orthologs are strongly conserved at the N and C termini, start and stop sites were defined by alignment to other Rad50 proteins. The C. cinereus genomic rad50 gene from strain Okayama-7 is 5091 bp in length, and the sequence we determined exactly matches the sequence reported for the same strain by the Broad Institute [Coprinus cinereus Sequencing Project, Broad Institute of MIT and Harvard (http://www.broad.mit.edu/annotation/genome/coprinus_cinereus/Home.html)].
To determine rad50 gene structure, we used RT–PCR to isolate overlapping portions of rad50 cDNA, and we compared cDNA sequence with genomic sequence to identify introns. The Ccrad50 coding region is 3930 bp in length and is interrupted in the genome by 22 introns (Figure 1A; Table 1; GenBank accession no. 1017584). The predicted protein is 1309 amino acids in length, with a molecular weight of 150.7 kDa (Figure 1B).
Figure 1.—
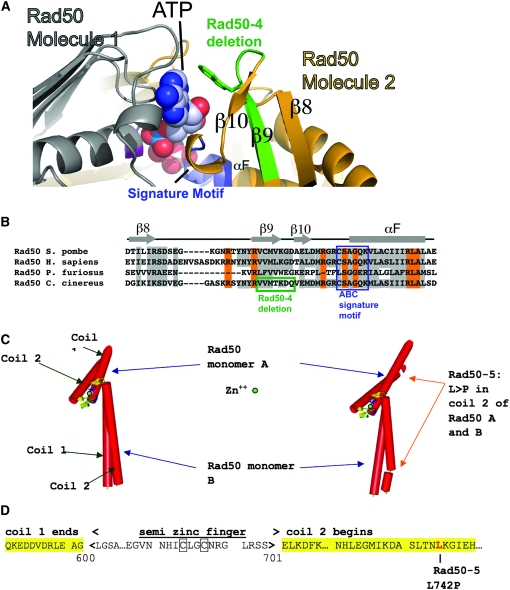
C. cinereus rad50 gene, protein, and mutants. (A) Gene structure. Solid areas represent exons and open areas represent introns. The translational start site is represented by ATG, and the translational stop is represented by TGA. (B) Rad50 protein and mutations. Stars denote the Walker A and Walker B boxes. Asterisks denote nonsense mutations (stop codons). The Rad50-15 insertion is shown above the protein diagram and the Rad50-1, Rad50-6, and Rad50-4 deletions are shown below. Details of the changes in Rad50-1, Rad50-4, and Rad50-15 are given in Table 1, supplemental Figure S1, and the text. (C) Diagrams of predicted Rad50 mutant proteins. Crosses represent sites of single-amino-acid substitution, or, in the case of Rad50-4, its internal deletion and the altered position of the signature motif. Multiple predicted structures of Rad50-4 are summarized as the two classes that encompass the mutant proteins. Also shown is a model of the MRN complex, as in Hopfner et al. (2002b) and de Jager et al. (2004).
TABLE 1.
C. cinereus rad50 mutations
| Mutation
|
Corresponding amino acid changesd | ||||
|---|---|---|---|---|---|
| Allele | Positionb | Codon changec | Predicted mutant protein | Meiotic arrest stage | |
| rad50-1a | Intron 4e | See text | Truncation | Diffuse diplotene | |
| rad50-2 | 644 | TTG > TAG | L215* | Truncation | ND |
| rad50-3 | 1897 | AAG > TAG | K633* | Truncation | Diffuse diplotene |
| rad50-7 | 760 | CAG > TAG | Q254* | Truncation | Diffuse diplotene |
| rad50-11 | 583 | AAA > TAA | K195* | Truncation | Diffuse diplotene |
| rad50-12 | 2164 | AAA > TAA | K722* | Truncation | ND |
| rad50-15a | 2929 | +TAC GCA AGG G | See text | Truncation | Diffuse diplotene |
| rad50-16 | 1783 | CGA > TGA | R595* | Truncation | ND |
| rad50-4a | Intron 20f | See text | Internal deletion/truncation, see text | Diffuse diplotene | |
| rad50-6 | Δ2583–2663 | Δ861–888 | Internal deletion | Diffuse diplotene | |
| rad50-5a | 2279 | CTC > CCC | L742P | Full-length with disrupted coiled coil | Diffuse diplotene |
| rad50-9 | 2279 | CTC > CCC | L742P | Full-length with disrupted coiled coil | Diffuse diplotene |
| rad50-10 | 2279 | CTC > CCC | L742P | Full-length with disrupted coiled coil | Diffuse diplotene |
| rad50-8a | 850 | (aaa > aag) | (K284K) L1234S | Single-site substitution | Metaphase-like |
| 3701 | TTA > TCA | ||||
| rad50-14a | 3703 | GAT > AAT | D1235N | Single-site substitution | Diffuse diplotene |
| rad50-13 | No change found | ND | |||
ND, not determined.
Alleles studied in depth and shown in Figure 1C.
All positions refer to nucleotides of the coding strand of the open reading frame except where indicated.
Nucleotide changes are in boldface type.
Single-letter amino acid designations are used. An asterisk denotes a stop codon.
The end of intron 4's sequence is changed from CAG to CAA in rad50-1.
The end of intron 20's sequence is changed from TAG to TCG in rad50-4.
To test whether the rad50 gene maps to the rad12 genetic locus, we analyzed progeny of a cross between a rad12-4 strain (rad12-4;4-1) and the backcross parent (Okayama-7; Valentine et al. 1995). Strains were assayed for radiation sensitivity, genomic DNA was isolated and cleaved with EcoRI, and fragments were separated on agarose gels. A Southern blot of the digested DNA was probed with a cosmid containing the rad50 sequence. The enzyme digest revealed a polymorphism between the radiation-sensitive (rad12) and radiation-resistant (wild-type) parent. No recombinants were found in 115 isolates examined, indicating that rad50 is tightly linked to rad12.
A cosmid containing the complete rad50 gene was used in transformation rescue of the rad12-4 allele. The cosmid complemented both the radiation sensitivity and the spore formation defects of 34/97 (35%) of the transformants, and 6/97 (6.2%) were complemented for one defect but not both.
Previous work had identified 16 mutants in the rad12 complementation group (Valentine et al. 1995; Table 1). Overlapping fragments of the rad50 gene were amplified from DNA isolated from all 16 alleles, and mutations were found within the gene for all but one allele. On the basis of the RFLP mapping, complementation, and DNA sequence data, we renamed the C. cinereus rad12 gene rad50 and henceforth refer to the gene as rad50.
The predicted structures of mutant C. cinereus Rad50 proteins:
Most C. cinereus rad50 mutations (Table 1; Figure 1, B and C) are single-base-pair changes resulting in stop codons that truncate the Rad50 protein in the first half of the molecule. The shortest of these predicted mutant proteins is Rad50-11, with a premature stop after amino acid 195, and the longest is Rad50-12, predicted to be 722 amino acids. We found that two alleles, rad50-1 and rad50-4, have mutations in intron splice acceptor sites. For rad50-1, genomic sequencing revealed a G to A transition mutation in the AG′ splice acceptor site of intron 4 (supplemental Table S1). We performed RT–PCR using RNA isolated from mushroom caps of a heterokaryotic (rad50-1 × wild type) strain, cloned the products, and sequenced multiple clones. We found only two types of clones, either wild-type sequences (28/36 clones sequenced) or sequences representing RNAs that had been spliced at the first available AG (italics in supplemental Table S1) after the mutated splice junction (8/36 sequenced clones). The predicted result of the mutation is a Rad50 protein that is normal for 360 amino acids, followed by a 4-amino-acid sequence in which three of four positions are incorrect and then truncation; the resulting protein would be 364 amino acids in length.
Genomic sequencing of rad50-4 revealed an A to C transversion mutation in the splice acceptor site of intron 20 (Figure 1B and supplemental Figure S1; Table 1 and supplemental Table S1). We sequenced a total of 81 clones from RT–PCR products amplified three separate times from caps of rad50-4 × wild-type crosses and found that in 24 clones intron 20 was correctly spliced; these clones were expected because the heterokaryotic crosses contained one wild-type allele of rad50. For 20 clones, the intron had not been spliced out at all, although upstream and/or downstream introns were spliced in those molecules. The remaining clones revealed three alternatively spliced products. Twenty-one clones had been spliced at the first AG after the normal splice site, 6 had been spliced at the second AG, and 7 had been spliced at the third AG (supplemental Figure S1). Therefore, rad50-4 is predicted to encode a mixture of mutant proteins. If intron 20 is not spliced at all, the resulting protein is identical to wild type for 1189 amino acids and then has 24 incorrect amino acids before a premature stop. If splicing is at the first AG after the wild-type splice site, the resulting protein has a deletion of 5 amino acids (“VVMTK,” aa 1190–1194), if splicing is at the second AG, the protein has a deletion of 7 amino acids (“VVMTKDQ,” aa 1190–1196), and if splicing is at the third AG, the protein has 1190 correct amino acids, 11 incorrect amino acids, and then a stop.
Three rad50 strains have the same mutation, a single T to C transition that results in the substitution of a proline for a leucine at amino acid 742 (Figure 1B; Table 1). The mutants have at least two independent origins; the first, rad50-5, was isolated from a screen that was separate from the mutagenesis that generated the other two (Valentine et al. 1995). However, it is possible that rad50-9 and rad50-10 are actually derived from the same mutagenic event and that the isolate was sampled twice in the screen.
Two other substitution (missense) mutations were found, in rad50-8 and rad50-14, both of which lead to predicted changes within the Walker B box of Rad50 (Figure 1B; Table 1). Rad50-8 has a leucine to serine change at amino acid 1234, and Rad50-14 has an aspartic acid to asparagine change at position 1235. One DNA deletion allele (rad50-6) and one insertion allele (rad50-15) were also found; rad50-6 results in an in-frame deletion of eight amino acids within the second coiled-coil domain of Rad50, and rad50-15 has an insertion of 10 bp (TAC GCA AGG G) at position 2929 of the cDNA (Figure 1B; Table 1); the predicted protein is wild type in sequence until amino acid 976 and then has 25 missense amino acids before it ends after amino acid 1001, thus deleting the entire carboxy-terminal globular domain of the protein.
Molecular modeling:
Diagrams of predicted Rad50 proteins for alleles studied in depth are shown in Figure 1C. To analyze structural repercussions of mutant Rad50 proteins in more detail, we modeled the predicted effects of the seven-amino-acid deletion in Rad50-4 and of the amino acid substitution in Rad50-5 on the basis of crystal structures of P. furiosus Rad50 (Figure 2; Hopfner et al. 2000b). Sequence alignments show the equivalent region encompassing the C. cinereus Rad50-4 deletion maps to P. furiosus Rad50 β-sheet β9 (Figure 2, A and B). In the ATP-bound structure of P. furiosus Rad50, sheets β9 and β10 and the neighboring Rad50 ABC signature motif directly participate in formation of the ATP binding pocket and the nucleotide-mediated Rad50 dimerization interface. When deleted in the rad50-4 mutant, removal of β9 likely causes destabilization of the β8-β9-β10 sheet, disruption of the signature motif, and distortion of the ATP binding pocket (Figure 2, A and B). The signature motif is critical for Rad50 dimerization and for MRN complex DNA binding (Hopfner et al. 2002b). A signature motif mutation in P. furiosus Rad50 protein was found to dramatically lower ATP binding (Moncalian et al. 2004); however, equivalent mutations in the S. cerevisiae and human proteins are defective in adenylate kinase activity but not ATP binding or hydrolysis (Bhaskara et al. 2007).
Figure 2.—
Molecular models of C. cinereus Rad50. (A) The Rad50-4 deletion removes sheet β9, and the β9–β10 connecting loop, thereby disrupting the Rad50 ATP binding and dimerization interaction site. Ribbon models are based upon homology with the Pyrococcus furiosus Rad50 protein. (B) Alignment of the β9 and signature motif regions of P. furiosus, C. cinereus, S. pombe, and H. sapiens Rad50. The green-highlighted amino acids are deleted in one splicing variant of Rad50-4; in another, the five amino acids VVMTK are deleted. The ABC signature motifs are boxed in purple. (C) Rod model of the hook and part of the coiled-coil regions of wild-type Rad50 and Rad50-5. (D) A portion of the Rad50 protein sequence, showing the region mutated in Rad50-5.
The Rad50-5 mutation, a leucine to proline substitution, occurs at the beginning of the second coiled-coil domain of the protein (Table 1; Figures 1B and 2D). This is predicted to result in partial or complete destabilization of the portion of the coil just after the hook domain (Figure 2C).
Meiotic defects in rad50 mutants:
Spore viability of rad50 mutants and heterozygotes:
Because Rad50 functions as a multimer (Hopfner et al. 2000b), the rad50-1, rad50-4, rad50-5, rad50-8, and rad50-15 mutants were assessed for dominance using a spore viability assay of homozygous (homokaryotic) vs. heterozygous (heterokaryotic) strains. Five experiments were performed for each strain, at least 200 spores were scored for each experiment, and spore viabilities were averaged. Spore viability for the wild-type strain, J6;5x4, was 87.8%. Mutant strains rad50-1, rad50-4, rad50-5, rad50-8, and rad50-15 had average spore viabilities of 2.0, 4.4, 3.8, 18.7, and 16.7%, respectively. The heterokaryons of rad50-1, rad50-4, rad50-5, rad50-8, and rad50-15, in each case crossed to wild type, gave spore viabilities of 78.8, 80.4, 79.9, 89.3, and 80.1%, values that exhibited no statistically significant difference from wild-type mushrooms (ANOVA analysis; P-value >0.05).
Diffuse diplotene arrest of rad50 mutants:
Meiosis in C. cinereus is synchronous and takes ∼12 hr from karyogamy through the second meiotic division (Raju and Lu 1970; Pukkila et al. 1984). At 6 hr after karyogamy (K + 6), essentially all meiotic cells in the mushroom cap are in pachytene (Pukkila et al. 1984; Seitz et al. 1996); homologous chromosomes are fully condensed and paired, and the SC is fully developed. At ∼8 hr after karyogamy (K + 8), cells enter diffuse diplotene. At this stage, the SC has dissipated, and chromosomes are diffuse in appearance. Nine to 10 hr after karyogamy (K + 10) chromosomes undergo a second condensation and enter metaphase I, and by 12 hr after karyogamy (K + 12) all basidia have completed both meiotic divisions and contain a tetrad of haploid nuclei (Figure 3A).
Previous analysis of chromosome spreads of rad50-1, rad50-4, and rad50-15 (Ramesh and Zolan 1995) showed that they undergo karyogamy 30–60 min later than wild-type strains but are similar to wild-type strains in the timing of the formation and dissolution of AEs and SC during meiosis. That is, although rad50 mutants do not achieve complete SC formation (i.e., pachytene), the timing of maximal AE and SC formation is comparable to that of wild type, and the AEs and SCs that do form then disperse at times similar to that of wild type. However, we also found that rad50 mutants arrest in diffuse diplotene. We extended this study within our collection of rad50 mutants and examined single-layer gill segments taken from mushrooms at K + 12 (Table 1). We found that all rad50 mutants examined apparently arrest at diffuse diplotene except for rad50-8 (Figure 3A). The majority of basidia in rad50-8 arrest at metaphase I in a spindle-like shape, which implies that this mutant progresses farther through meiosis. To examine rad50 arrest in more detail, we stained arrested tissue from rad50-4 with both DAPI and an antibody against α-tubulin (Figure 3B). We found that 43% of the cells arrested with larger discs of chromatin and that these were without organized spindles, while 57% of cells had more compact DAPI-staining discs and spindle-shaped microtubule organization (n = 60). Therefore, the rad50-4 arrest phenotype is actually a mixture of two distinct, arrested cell types, probably corresponding to diffuse diplotene and condensed chromatin entering metaphase I. The rad50-8 allele allows all cells to progress to the metaphase I-like state.
The spo11-1 mutation is epistatic to rad50 mutations and mre11-1 for meiotic arrest:
In S. cerevisiae, most rad50 mutants are phenotypically similar to spo11 null mutants, in that meiotic DSBs are not formed. However, in rad50S mutants (Alani et al. 1990), DSBs are made but not processed; Spo11 protein remains bound to the 5′ ends of the DSB (Keeney et al. 1997). In C. cinereus, the spo11-1 mutant is likely defective at DSB formation, because gamma irradiation of mushrooms partially suppresses the mutant's meiotic defects (Celerin et al. 2000); in addition, Rad51 foci are not induced during meiosis in spo11-1 (Figure 4F; supplemental Figures S3 and S4). In spo11-1, meiotic cells progress into an aberrant, metaphase I-like stage and then undergo programmed cell death (PCD) (Figure 3C; Celerin et al. 2000). In contrast, rad50 mutants remain arrested in either diffuse diplotene or a metaphase I-like state for several hours (Ramesh and Zolan 1995; Figure 3A).
We thought it possible that the C. cinereus rad50 mutants were all permissive for meiotic DSB formation but defective in DSB processing. If this were the case, then spo11-1, which does not make meiotic DSBs, would be epistatic to rad50 mutations. To test this prediction, we constructed rad50;spo11-1 double mutants and found that they underwent signature PCD chromatin changes characteristic of the spo11-1 single mutant (data for the rad50-4 and rad50-8 mutants are shown in Figure 3C; similar results were obtained for rad50-5). This result indicated that Spo11 has activity in rad50 mutants and that this activity is required for the rad50 mutant arrest. In a previous study, we found that mre11-1 arrests variably at diffuse diplotene or in a metaphase I-like state (Gerecke and Zolan 2000). We constructed an mre11-1;spo11-1 double mutant and found that it also exhibited a spo11-1 PCD phenotype, indicating that spo11-1 PCD is epistatic to the mre11-1 arrest phenotype (Figure 3D). Therefore, Spo11 likely functions before both Mre11 and Rad50 in C. cinereus meiosis; this observation also implies that Spo11-catalyzed DSB formation may be MRN complex independent in C. cinereus.
Rad51 foci are induced during meiosis for some rad50 alleles:
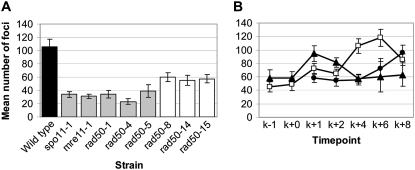
We reasoned that if Spo11 forms DSBs in C. cinereus MRN complex mutants, then the array of Rad50 mutant proteins in our alleles might vary in their ability to process those breaks and succeed with recombination-dependent events of prophase I. As shown in S. cerevisiae (reviewed in Keeney 2008), break formation is followed by MRN complex-dependent processing, which removes Spo11 from the 5′ ends of the breaks and asymmetrically forms single-strand ends that bind Rad51 and Dmc1 and invade a nonsister chromatid. To investigate recombination events in C. cinereus, we raised and purified antibody against full-length Rad51 protein (supplemental Figures S2–S4) and used it to examine the time course of Rad51 focus formation during meiosis in a wild-type dikaryon (Figure 4). We found that Rad51 foci are induced after karyogamy and peak in number at K + 4 and K + 6, corresponding to the zygotene and pachytene stages of prophase I, and our results are similar to those obtained previously using immunofluorescence staining of C. cinereus tissue sections with an independently derived anti-Rad51 antibody (Nara et al. 2001). In contrast, Rad51 foci are not induced during meiosis in spo11-1 (Figure 4F; supplemental Figures S3 and S4), in which the number of foci was not significantly different from that seen in wild type before and during karyogamy (in two-sample t-tests: K − 1, P = 0.830; K + 0, P = 0.258), and remained essentially constant throughout prophase I. Our results with spo11-1 are similar to those reported for Dmc1 by Holzen et al. (2006), who found a low, residual number of foci in an S. cerevisiae spo11 mutant.
To compare wild-type and mutant strains, we examined Rad51 foci at K + 4 in mre11-1 and an array of rad50 mutants. We found that mutant strains fell into two clear classes (Figure 5A). The first, which includes mre11-1, rad50-1, rad50-4, and rad50-5, formed numbers of foci that were not significantly different from that found in spo11-1. The second class of mutants, rad50-8, rad50-14, and rad50-15, formed significantly greater numbers of Rad51 foci than spo11-1, although significantly fewer foci than wild type. These three strains were examined further for the timing of Rad51 focus formation. In rad50-14, the number of foci increased at later meiotic stages, to levels greater than that of wild type by K + 8 (Figure 5B). For both rad50-8 and rad50-15, early meiotic time points showed higher than wild-type levels of foci at K + 1 and then a decrease in foci to an intermediate level by K + 4 (data for rad50-15 are shown in Figure 5B). For rad50-15, we further examined its time course by staining samples taken before karyogamy (K − 1 and K + 0), to examine the initial appearance of foci, and at K + 6 and K + 8, to determine whether the number of foci remains constant at these later time points (Figure 5B). At K − 1 and K + 0, the numbers of Rad51 foci in a rad50-15 strain were not significantly different from those in either spo11-1 or wild type (ANOVA, P = 0.107 at K − 1 and P = 0.330 at K + 0). This indicates that the increase in foci seen in rad50-15 at K + 1 is occurring in response to breaks formed during meiotic prophase and not during the karyogamy or premeiotic S phases. Examination of the later time points showed a persistent level of foci, likely indicating that unresolved recombination events remained.
Figure 5.—
Mutants fall into two classes for Rad51 focus formation. (A) Mean number of Rad51 foci observed at K + 4. (B) Time course of Rad51 foci in rad50-14 and rad50-15. Wild-type data (□) are the same as in Figure 4 and shown for reference. •, rad50-14; ▴, rad50-15. Error bars show 95% confidence intervals.
If Rad51 foci in rad50 mutants are meiosis specific, they should be Spo11 dependent. As predicted, a rad50-8;spo11-1 double mutant examined at K + 6 had significantly fewer foci (22.9 ± 3.0) than that observed in rad50-8, rad50-14, or rad50-15 at any time point (P < 0.001).
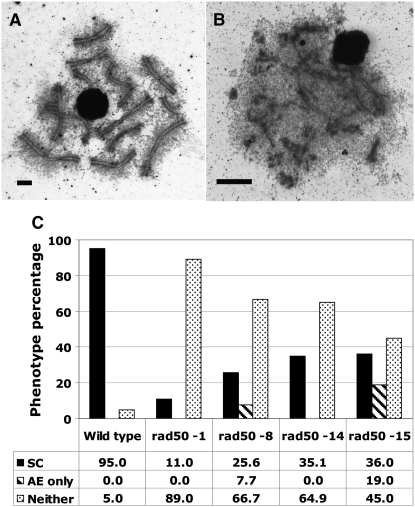
Synaptonemal complex formation in rad50 mutants:
To characterize SC formation in rad50 mutants, we examined electron micrographs of silver-stained surface-spread nuclei. The three rad50 alleles found to be defective for Rad51 focus formation, rad50-1, rad50-4, and rad50-5 (Figure 5A), were also severely deficient for SC formation (Figures 6 and 7); the SC data for rad50-1 and rad50-4 are similar to those we previously reported (Ramesh and Zolan 1995). In contrast, the three rad50 alleles with significantly more Rad51 foci than spo11-1, rad50-8, rad50-14, and rad50-15 formed more AEs and SCs than the severely defective alleles (Figure 6). The allele rad50-15 had previously been examined after one outcross generation (Ramesh and Zolan 1995), and our results were similar to those previously obtained.
Figure 6.—
SC formation in wild type, rad50-8, rad50-14, and rad50-15. Surface spreads of wild-type (A) and rad50-14 (B) nuclei from the K + 6 time point were stained with silver nitrate and photographed using transmission electron microscopy. Bars, 2 μm for all images. (C) Percentage of SC and AE formation for each strain. The number of images categorized was as follows: wild type, 40; rad50-8, 78; rad50-14, 57; rad50-15, 100.
Figure 7.—
The coiled coil of Rad50 proximal to the hook is required for AE formation. (A and C) Samples were taken at 6 hr after karyogamy, fixed, spread, and stained and photographed using EM as described (Seitz et al. 1996). (B and D) Data from nuclear spreads from a single mushroom were plotted according to increasing SC length. Each graph corresponds to the genotype shown in the image above it. Images were chosen to represent the major class of nuclear spreads observed for each genotype. Bars, 2 μm.
Homolog pairing:
The concordance between elevated (relative to spo11-1) numbers of Rad51 foci and elevated SC formation for rad50-8, rad50-14, and rad50-15 led us to ask whether stable homolog pairing correlates with these indicators of recombination activity. We used FISH (Li et al. 1999) to examine the extent of meiotic pairing on two sites on chromosome 13, one telomeric and one interstitial, for rad50-1, rad50-5, rad50-8, and rad50-14 (Table 2). The data for rad50-1 were similar to those obtained previously for mre11-1 (Gerecke and Zolan 2000) and those we obtained for spo11-1; ∼25% of nuclei showed pairing for both probes at K + 6. In contrast, rad50-5 had less pairing than any other strain; only 13% of nuclei had both loci paired at K + 1, and only 6% had both loci paired at K + 6.
TABLE 2.
Homolog pairing
| % pairing
|
||||||
|---|---|---|---|---|---|---|
| Strain | Time point | N | Interstitial probe | Telomeric probe | Both probes | Either probe |
| Wild type | K + 1 | 82 | 78 | 72 | 60 | 90 |
| Wild type | K + 6 | 39 | 90 | 92 | 82 | 100 |
| spo11-1 | K + 6 | 39 | 33 | 49 | 26 | 56 |
| rad50-1 | K + 1 | 40 | 70 | 33 | 23 | 80 |
| rad50-1 | K + 6 | 37 | 51 | 43 | 24 | 70 |
| rad50-5 | K + 1 | 45 | 27 | 20 | 13 | 33 |
| rad50-5 | K + 6 | 68 | 24 | 15 | 6 | 32 |
| rad50-8 | K + 6 | 83 | 43 | 43 | 36 | 51 |
| rad50-14 | K + 6 | 93 | 52 | 49 | 40 | 61 |
Data are arrayed by time point and strain. For each combination of time point and strain at least two experiments, examining two different mushrooms, were performed. Data shown combine images from all experiments on a given time point and strain.
The rad50 strains that had some partial recombination activity exhibited a higher amount of pairing at K + 6 (rad50-8, 36%; rad50-14, 40%) than rad50-1 (24%), although with our data set this difference is not statistically significant (P = 0.202 for the rad50-8 comparison with rad50-1 and P = 0.096 for the rad50-14 comparison).
Because we used both a telomere-proximal probe (Li et al. 1999) and an interstitial probe, we were able to examine differences in pairing on the same chromosome. For the rad50-1 strain we observed a greater amount of pairing with the interstitial probe as compared to the telomeric probe. This difference was significant at K + 1 (70% compared to 33%; chi-square test, P < 0.001), but was not significant at K + 6 (P = 0.342). In addition, none of the other rad50 mutants examined showed a significant difference in percentage of pairing between the two probes at any time point.
Analysis of AE formation in rad50 mutants:
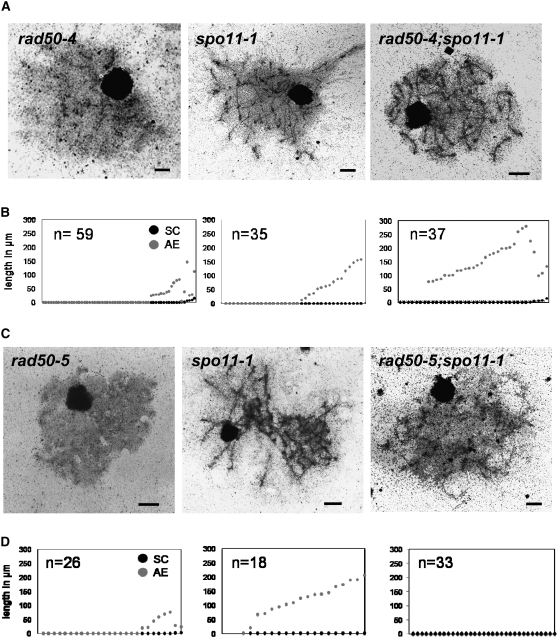
Most studies of Rad50 have focused on mutations that affect the catalytic activities of the MRN complex. In addition, the hook portion of the Rad50 protein has been shown to be of critical importance in both intracomplex and intercomplex interactions (Hopfner et al. 2002a) and in the in vivo functions of the complex (Wiltzius et al. 2005). We were interested in separating the functions of Rad50 in meiotic DSB formation and processing from any structural role the protein may have in meiosis. Therefore, we investigated AE formation by rad50 mutants in the spo11-1 background, in which DSBs are not made.
Using rad50;spo11-1 double mutants and sibling progeny that were single mutants for each gene or wild type, we generated electron micrographs of silver-stained, surface-spread meiotic nuclei and scored each strain for AE and SC formation (Figure 7; Table 3). AE and SC lengths were measured from nuclear spreads taken from one mushroom of each strain (data for rad50-4 and rad50-5 are shown in Figure 7, B and D).
TABLE 3.
Effect of hook-proximal coiled-coil structure on AE and SC formation
| Straina | % SCb | % AE | % no structure | Total (N) |
|---|---|---|---|---|
| Cross A: rad50-4 × spo11-1 | ||||
| rad50-4 11 × 30 | 6 | 15 | 79 | 79 |
| spo11-1 93 × 44 | 0 | 43 | 57 | 62 |
| rad50-4;spo11-1 18 × 12 | 7 | 49 | 44 | 120 |
| Cross B: rad50-1 × spo11-1 | ||||
| rad50-1 96 × 13 | 11 | 0 | 89 | 75 |
| spo11-1 195 × 176 | 0 | 45 | 55 | 55 |
| rad50-1;spo11-1 47 × 59 | 0 | 21 | 79 | 34 |
| Cross C: rad50-5 × spo11-1 | ||||
| rad50-5 19 × 45 | 0 | 4 | 96 | 137 |
| spo11-1 7 × 29 | 7 | 48 | 44 | 81 |
| rad50-5;spo11-1 59 × 21 | 0 | 7 | 93 | 75 |
| Cross D: rad50-5 × spo11-1 | ||||
| rad50-5 60 × 69 | 2 | 8 | 90 | 137 |
| spo11-1 35 × 50 | 0 | 30 | 70 | 169 |
| rad50-5;spo11-1 48 × 6 | 1 | 8 | 91 | 248 |
| rad50-5;spo11-1 40 × 70 | 7 | 14 | 78 | 222 |
Strains used were dikaryons formed by crosses between the indicated, numbered, sibling monokaryons.
For some crosses, percentages do not sum to 100 due to rounding.
As expected and as previously reported for other spo11-1 strains (Celerin et al. 2000), nuclei from the spo11-1 single mutants showed either no structure or AE formation only (Figure 7; Table 3). The number of AE segments and their lengths in those spo11-1 nuclei that formed AEs were variable, with 5 to 42 AE segments forming per nucleus, individual AE lengths ranging from 0.3 to 20 μm, per nucleus lengths ranging from 12 to 173 μm, and overall AE length per nucleus ranging from 12 to 206 μm. The average AE lengths per nucleus for the two spo11-1 strains shown in Figure 7, A and C, were 73 and 126 μm, respectively. For comparison, a congenic wild-type strain consistently formed 26 AEs, with an average AE length per nucleus of 124 μm. Although the rad50-1, rad50-4, and rad50-5 single mutants were similar to one another, the rad50-4;spo11-1 double mutant was strikingly different from the rad50-1;spo11-1 and rad50-5;spo11-1 double mutants (Figure 7; Table 3). We found that the presence of Rad50-4 was sufficient for the formation of long AE's in a spo11-1 mutant background (Figure 7, A and B). For those nuclei that formed AE, the number of AE segments ranged from 17 to 40, and the average AE length per nucleus was 153 μm. However, in both the rad50-1;spo11-1 and the rad50-5;spo11-1 double mutants, little AEs was found.
Chi-square analysis showed that for rad50-4;spo11-1, the distribution of nuclei into classes with chromosome structures (AE and/or SC) and without structure was the same as for spo11-1 (P = 0.116). In contrast, for both rad50-1;spo11-1 and rad50-5;spo11-1, the distributions of nuclei in the two categories were significantly different from that of the sibling spo11-1 strain and in all but one case not significantly different from that of the sibling rad50 single mutant. For rad50-1, progeny from one cross to spo11-1 were examined (Table 3, cross B), and the rad50-1;spo11-1 double mutant was different from spo11-1 (P = 0.018) and the same as rad50-1 (P = 0.322). For rad50-5, two different crosses were examined (Table 3, crosses C and D). For progeny from the first cross, the rad50-5;spo11-1 double mutant was significantly different from spo11-1 (P < 0.001) and not different from the rad50-5 single mutant (P = 0.322). For cross D, two different rad50-5;spo11-1 double mutants were examined. The first (48 × 6) was significantly different from spo11-1 (P < 0.001) and the same as rad50-5 (P = 0.754). The second (40 × 70) was significantly different from spo11-1 (P = 0.047) but also significantly different from the sibling rad50-5 strain examined (P = 0.007). Overall, our data show that the disruption (in Rad50-5) or absence (in Rad50-1) of the Rad50 hook-proximal coiled coil makes the formation of stable AE unlikely. Our results indicate that there is a structural role for Rad50, separate from the DSB-processing activity of the MRN complex, which is required for the formation of meiotic chromosome structures.
DISCUSSION
C. cinereus rad50 mutations are recessive:
For proteins that normally function as multimers, mutants that can assemble into complexes often exhibit dominant-negative phenotypes in heterozygotes (Herskowitz 1987). Because of the known structure of the Mre11/Rad50 complex, we expected that at least some rad50 alleles would behave as dominant negatives. However, heterokaryon spore viabilities did not show statistically significant differences from wild-type spore viabilities, indicating that the rad50 mutations examined (rad50-1, rad50-4, rad50-5, rad50-8, and rad50-15) are not dominant. A simple explanation for this would be instability of all mutant Rad50 proteins. However, the phenotypes of a number of rad50 mutants are distinct (e.g., Figure 4; Ramesh and Zolan 1995); therefore, at least some of them make stable protein. In addition, recent studies have shown that Rad50-5 forms foci on meiotic chromosomes (A. Many, C. Melki, D. Maillet, S. Acharya and M. Zolan, unpublished results). Therefore, although it is possible or even likely that some alleles (e.g., rad50-1) encode unstable protein, the recessive nature of all rad50 mutations examined cannot be explained by a simple absence of stable protein. It is possible that none of the mutant proteins can form sufficiently stable dimers with wild-type Rad50 to become incorporated into complete MRN complexes or to diminish the pool of wild-type complexes. In any case, the pool of fully functional MRN is sufficient in the heterokaryons for the production of viable meiotic products, although subtle effects on meiosis (e.g., on crossover frequency or distribution) would not have been detected by our analysis.
C. cinereus rad50 mutants form meiotic DSBs:
In rad50;spo11-1 and mre11-1;spo11-1 double mutants, the spo11-1 PCD phenotype is epistatic to the rad50 or mre11-1 phenotype, in all allele combinations examined (Figure 3 and data not shown). Therefore, our data imply that Spo11 has activity in rad50 and mre11 mutants. Although Spo11 has meiotic functions that are independent of its DSB-forming activity (e.g., Cha et al. 2000), the spo11-1 single mutant forms essentially no SC (Celerin et al. 2000), whereas rad50 and mre11 mutants form SC in variable amounts (Ramesh and Zolan 1995; Gerecke and Zolan 2000; this work). As discussed recently by Borde (2007), the partial SC formation in C. cinereus rad50 and mre11 mutants is similar to that seen in those S. cerevisiae MRN mutants (such as rad50S; Alani et al. 1990) in which Spo11 forms but does not process meiotic DSBs (Keeney et al. 1997). In addition, like S. cerevisiae rad50S (Gasior et al. 1998), some of the C. cinereus rad50 alleles form Rad51 foci at levels lower than that observed in wild-type strains but significantly higher than that found for spo11-1 (Figure 5). Because the Rad51 foci correlate in the rad50 alleles with enhanced levels of stable homolog pairing and SC formation (relative to spo11-1; Figure 6 and Table 2), and because they are Spo11-dependent, at least a portion of them must represent progression through a normal meiotic program of Spo11-dependent DSB processing, leading to synapsis (Kleckner 2006).
For the rad50 alleles that formed Rad51 foci, the time courses of focus formation and loss were different from that of the wild-type strain examined (Figure 5B). For rad50-14, the number of Rad51 foci increased at time points corresponding to late prophase in wild-type strains. The predicted Rad50-14 protein has an aspartate to asparagine substitution in the Walker B domain (Figure 1B; Table 1), which might result in DSB processing that is stalled or delayed compared to that in wild-type cells. An alternative possibility is that these late events may not be related to Spo11-induced DSBs but may be repair events resulting from the accumulation of DNA damage as the cells attempt to proceed through meiotic prophase. For rad50-8 and rad50-15, we found that a large number of foci are observed at K + 1 and decrease thereafter. One simple interpretation of this is that these foci mark recombination events in which an aberrant processing step occurs soon after the formation of meiotic DSBs, followed by disassociation of the MRN complex.
Because all of the rad50 mutants, and mre11-1 arrest in meiosis, because this arrest is Spo11 dependent (Figure 3), and because some of the alleles form inducible, Spo11-dependent Rad51 foci (Figure 5) and therefore must be forming DSBs, we think it likely that arrest in both mre11-1 and the collection of rad50 mutants is the result of unrepaired or improperly processed DSBs in the genome and that Rad50 and Mre11 are dispensable for DSB formation in C. cinereus, but required for appropriate DSB processing. This interpretation assumes that truncation alleles (e.g., rad50-1 and mre11-1) in fact encode unstable protein. Alternatively, the Walker A box alone of Rad50 and the N-terminal 40% of Mre11 are sufficient to allow formation, but not complete and/or proper processing, of meiotic DSBs. Although S. cerevisiae rad50 and mre11 null or deletion mutants show no meiotic DSB formation, as assayed by pulsed-field gel analysis of genomic DNA (Cao et al. 1990; Nairz and Klein 1997), rad50 deletion mutants of S. pombe do form meiotic DSBs (Young et al. 2004). In addition, Spo11 has DSB-forming activity in mre11 mutants of S. pombe (Young et al. 2004) and A. thaliana (Puizina et al. 2004). In C. elegans, Rad50 facilitates DSB formation by Spo11, but Rad50 is apparently not strictly required for Spo11 activity, because a defect in a cohesin protein, Rec8, allows Spo11-dependent DSB formation in the absence of Rad50 function (Hayashi et al. 2007). Thus, S. cerevisiae may be the exception, rather than the rule, in its requirement for the MRN complex in meiotic DSB formation.
Rad50's structural roles in meiosis:
The C. cinereus rad50 mutants undergo varying amounts of AE and SC formation, and a comparison of the predicted Rad50 structures (Figures 1 and 2) with the meiotic phenotypes of the mutants (Ramesh and Zolan 1995; Figure 4) is informative. Three alleles, rad50-1, rad50-4, and rad50-5, are comparably severe as single mutants (except in homolog pairing, discussed below). Because rad50-1 encodes a severely truncated protein, it is not surprising that little function is retained, and this allele is most likely effectively null. For Rad50-4, modeling revealed that the loss of a β-strand (Figure 2A) likely distorts the ATP binding pocket and neighboring signature motif. The signature motif (in the C-terminal globular domain) of Rad50-4 may not be in the correct orientation for contact with the ATP bound to the N-terminal domain of another Rad50-4. This may result in a loss of Rad50 head domain dimerization (Moncalian et al. 2004). In addition, the signature motif is essential for the recently reported and essential adenylate kinase activity of S. cerevisiae Rad50 (Bhaskara et al. 2007) and may be abrogated in Rad50-4 molecules. Also, because the deletion is at the junction where the Rad50 globular ATPase domain adjoins the coiled coil, it may affect coil stability in this region and therefore Mre11 binding (Hopfner et al. 2001). An unspliced rad50-4 transcript is predicted to make a protein similar to Rad50-15, and transcripts containing an unspliced intron composed about one-third of the mutant cDNAs examined. Given that there is a mix of aberrantly spliced transcripts in the rad50-4 mutant, it is perhaps surprising that only the severe phenotype prevails. It may be that there is insufficient truncated, Rad50-15-like protein to allow SC formation in rad50-4.
The rad50-5 mutation causes a leucine to proline change at the beginning of the second coil (Figures 1 and 2, C and D). From modeling, it is predicted to result in either an interruption or a distortion of the second coil. The stability of coiled-coil structures and their segmented structure (M. Oakley, personal communication) predicts that only the beginning of the coiled-coil region, and probably the hook region, is affected in Rad50-5. Therefore, it is striking that the meiotic defects in rad50-5 strains are as severe as those caused by the rad50-1 allele and, in fact, that homolog pairing is worse in rad50-5 than in rad50-1 or spo11-1 (Table 2). The analysis of Wiltzius et al. (2005) showed that the hook region of S. cerevisiae Rad50 is essential for DNA repair and meiotic DSB formation. Our data show that this region is also essential for the formation of stable AEs and SCs.
The requirement for the hook-proximal coiled coil of Rad50 in stable AE formation is shown dramatically by our analysis of AE formation in rad50;spo11-1 double mutants (Figure 7). For rad50-4, the spo11-1 mutation seems to be fully epistatic; the rad50-4;spo11-1 double mutant is the same as spo11-1 for both PCD (Figure 3) and AE formation (Figure 7; Table 3). In contrast, the spo11-1 phenotype of variable AE formation is masked, in rad50-1 and rad50-5 strains, by the inability of those mutants, even in the spo11-1 background, to form stable AEs. Therefore, Rad50 has a structural role in AE formation that must be independent of its role in processing Spo11-catalyzed DSBs.
In contrast with rad50-1, rad50-4, and rad50-5, the rad50-15 mutant makes extensive AEs and SCs. This allele contains an insertion that leads to the production of a protein with 25 incorrect amino acids after position 976, followed by a stop (Table 1 and Figure 1, B and C). The Walker A box is intact and the coiled domains presumably fold normally, but the globular domain containing the Walker B motif is missing. Since the Walker B motif is absent, and this mutant is radiation sensitive, it is unlikely that Rad50-15 is catalytically active. Similarly, rad50-8 and rad50-14 have mutations within the Walker B region, which should abrogate or at least diminish function. The phenotypes of these mutants imply that AE and SC formation are to a large extent independent of Rad50's ATPase or adenylate kinase functions. Instead, the main role of Rad50 in meiotic DSB processing may be via its interaction with Mre11. Recent biochemical data from S. cerevisiae and S. pombe implicate Mre11 as the nuclease that removes the covalently bound Spo11 from meiotic DSBs (Neale et al. 2005). On the basis of their predicted structures (Figures 1C and 2), we expect that Rad50-8 and Rad50-14 retain the ability to interact with Mre11. Although Rad50-15 is missing the N-terminal portion of the Mre11 binding site, its phenotype predicts that Rad50-15 does interact with Mre11 as well.
Although our data show that catalytic activity by the Rad50 molecules themselves cannot be essential for AE formation or meiotic DSB processing, none of the Rad50 mutant proteins allows the formation of wild-type levels of SC, and all mutant strains arrest in meiosis. Therefore, enzymatic activity conferred by intact C- and N-terminal globular domains is necessary for complete synapsis and meiotic progression.
What is the role of Rad50 in AE formation? AE formation results from structural changes in meiotic chromatin that may require Rad50 or require Rad50 for stability. In comparison to wild-type S. cerevisiae meiotic cells, rad50Δ strains show increased sensitivity of DNA to micrococcal nuclease treatment (Ohta et al. 1998). It is possible that in the absence of Rad50, or of its coiled-coil regions, structural changes of the chromatin do not occur, and AE cannot form. It is additionally possible that Rad50 has a structural role either in the AE core itself or in mediating appropriate sister-chromatid interactions that are necessary for AE formation. In S. pombe rad50Δ mutants, sister-chromatid recombination is reduced and homologous recombination frequencies are increased (Hartsuiker et al. 2001). The same is true of homologous recombination frequencies in S. cerevisiae rad50Δ strains (Malone et al. 1990). Haploid rad50Δ G2 strains are more sensitive to radiation than diploid rad50Δ G1 strains, suggesting that Rad50 is associated with sister-chromatid interactions (Fabre et al. 1984; Ivanov et al. 1992; Paques and Haber 1999). Additionally, a C. cinereus rad50-4;msh5-22 double mutant, which enters meiosis with unreplicated chromosomes, is rescued for SC formation (Merino et al. 2000). Therefore, the role of Rad50 in AE formation could also be due to its requirement in sister-chromatid interactions. Although a recent study found no loss of mitotic sister-chromatid cohesion in rad50 mutants of S. cerevisiae (Wiltzius et al. 2005), there are several lines of study that argue that meiotic recombination imposes additional requirements for sister-chromatid cohesion. First, DSB repair requires the recruitment of additional cohesin to sites of DNA damage (Strom et al. 2004; Unal et al. 2004). Cohesin recruitment to DSB sites requires Mre11, and although it is unknown whether the entire MRN complex is necessary, Mre11 does not localize to chromatin in the absence of Nbs1 (Tauchi et al. 2001), arguing that complex formation is necessary. Second, a spo11 mutation partially suppresses the need for Rec8 and Spo76 in Sordaria macrospora (Storlazzi et al. 2008), meaning that chiasma formation imposes an increased requirement for cohesion. Finally, the rad9-1 mutant of C. cinereus has no obvious mitotic growth defect but is almost completely defective in meiosis (Zolan et al. 1988). Rad9 is the C. cinereus ortholog of S. cerevisiae Scc2 (Michaelis et al. 1997), S. pombe Mis4 (Furuya et al. 1998), D. melanogaster NippedB (Rollins et al. 1999), and the human delangin protein (Krantz et al. 2004; Tonkin et al. 2004) and is required for cohesin localization to chromatin (Ciosk et al. 2000). The difference in mitotic and meiotic phenotypes for rad9-1 argues for an enhanced role of sister-chromatid cohesion in meiosis. Because Rad50 is an SMC-like protein, structurally similar to members of condensin and cohesin complexes (Losada and Hirano 2005; Nasmyth and Haering 2005), its role in meiotic sister-chromatid interactions may be as a member of the MRN complex, required for cohesin recruitment (Strom et al. 2004; Unal et al. 2004), or the Rad50 protein may be structurally required for appropriate sister-chromatid interactions. These two roles are not mutually exclusive, and it is also possible that the modification of chromatin structure by Rad50 may be a key factor in cohesin recruitment to sites of recombination.
Acknowledgments
We thank Claire Walczak and Joel Ybe for assistance in developing antibody purification techniques, Ellen Quardokus for immunofluorescence assistance and advice, and Jim Bever for statistics advice. We also thank F. Rudolph Turner for preparation of electron micrographs, Katy Van Hook for preliminary antibody purification work, Martha Oakley for helpful discussion, Pat Pukkila and Elizabeth A. Sierra for helpful discussion and critical reading of the manuscript, and Megan Kingslover for clerical support. In addition, we thank Kengo Sakaguchi and his laboratory for their generous gift of the Dmc1 expression construct. This work was supported by National Institutes of Health grants GM43930 (to M.E.Z.), CA117638 and CA092584 (to J.A.T.), and (training grant) T32 GMOO7757 (to S.N.A.).
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession no. 1017584.
References
- Ajimura, M., S. H. Leem and H. Ogawa, 1993. Identification of new genes required for meiotic recombination in Saccharomyces cerevisiae. Genetics 133 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani, E., R. Padmore and N. Kleckner, 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61 419–436. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, C. F., M. L. Sikes, R. Sullivan, L. E. Huye, M. M. Le Beau et al., 2002. Cancer predisposition and hematopoietic failure in Rad50(S/S) mice. Genes Dev. 16 2237–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara, V., A. Dupre, B. Lengsfeld, B. B. Hopkins, A. Chan et al., 2007. Rad50 adenylate kinase activity regulates DNA tethering by Mre11/Rad50 complexes. Mol. Cell 25 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binninger, D. M., C. Skrzynia, P. J. Pukkila and L. A. Casselton, 1987. DNA-mediated transformation of the basidiomycete Coprinus cinereus. EMBO J. 6 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde, V., 2007. The multiple roles of the Mre11 complex for meiotic recombination. Chromosome Res. 15 551–563. [DOI] [PubMed] [Google Scholar]
- Cao, L., E. Alani and N. Kleckner, 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61 1089–1101. [DOI] [PubMed] [Google Scholar]
- Carney, J. P., R. S. Maser, H. Olivares, E. M. Davis, M. Le Beau et al., 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93 477–486. [DOI] [PubMed] [Google Scholar]
- Celerin, M., S. T. Merino, J. E. Stone, A. M. Menzie and M. E. Zolan, 2000. Multiple roles of Spo11 in meiotic chromosome behavior. EMBO J. 19 2739–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha, R. S., B. M. Weiner, S. Keeney, J. Dekker and N. Kleckner, 2000. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 14 493–503. [PMC free article] [PubMed] [Google Scholar]
- Chamankhah, M., Y. F. Wei and W. Xiao, 1998. Isolation of hMRE11B: failure to complement yeast mre11 defects due to species-specific protein interactions. Gene 225 107–116. [DOI] [PubMed] [Google Scholar]
- Cherry, S. M., C. A. Adelman, J. W. Theunissen, T. J. Hassold, P. A. Hunt et al., 2007. The Mre11 complex influences DNA repair, synapsis, and crossing over in murine meiosis. Curr. Biol. 17 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk, R., M. Shirayama, A. Shevchenko, T. Tanaka, A. Toth et al., 2000. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5 243–254. [DOI] [PubMed] [Google Scholar]
- Cox, B. S., and J. M. Parry, 1968. The isolation, genetics and survival characteristics of ultraviolet light-sensitive mutants in yeast. Mutat. Res. 6 37–55. [DOI] [PubMed] [Google Scholar]
- Cummings, W. J., M. Celerin, J. Crodian, L. K. Brunick and M. E. Zolan, 1999. Insertional mutagenesis in Coprinus cinereus: use of a dominant selectable marker to generate tagged, sporulation-defective mutants. Curr. Genet. 36 371–382. [DOI] [PubMed] [Google Scholar]
- Cummings, W. J., S. T. Merino, K. G. Young, L. Li, C. W. Johnson et al., 2002. The Coprinus cinereus adherin Rad9 functions in Mre11-dependent DNA repair, meiotic sister-chromatid cohesion, and meiotic homolog pairing. Proc. Natl. Acad. Sci. USA 99 14958–14963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jager, M., K. M. Trujillo, P. Sung, K. P. Hopfner, J. P. Carney et al., 2004. Differential arrangements of conserved building blocks among homologs of the Rad50/Mre11 DNA repair protein complex. J. Mol. Biol. 339 937–949. [DOI] [PubMed] [Google Scholar]
- Dolganov, G. M., R. S. Maser, A. Novikov, L. Tosto, S. Chong et al., 1996. Human Rad50 is physically associated with human Mre11: identification of a conserved multiprotein complex implicated in recombinational DNA repair. Mol. Cell. Biol. 16 4832–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre, F., A. Boulet and H. Roman, 1984. Gene conversion at different points in the mitotic cycle of Saccharomyces cerevisiae. Mol. Gen. Genet. 195 139–143. [DOI] [PubMed] [Google Scholar]
- Furuya, K., K. Takahashi and M. Yanagida, 1998. Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes Dev. 12 3408–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego, M. E., M. Jeanneau, F. Granier, D. Bouchez, N. Bechtold et al., 2001. Disruption of the Arabidopsis RAD50 gene leads to plant sterility and MMS sensitivity. Plant J. 25 31–41. [DOI] [PubMed] [Google Scholar]
- Game, J. C., T. J. Zamb, R. J. Braun, M. Resnick and R. M. Roth, 1980. The role of radiation (rad) genes in meiotic recombination in yeast. Genetics 94 51–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior, S. L., A. K. Wong, Y. Kora, A. Shinohara and D. K. Bishop, 1998. Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes. Genes Dev. 12 2208–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerecke, E. E., and M. E. Zolan, 2000. An mre11 mutant of Coprinus cinereus has defects in meiotic chromosome pairing, condensation and synapsis. Genetics 154 1125–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsuiker, E., E. Vaessen, A. M. Carr and J. Kohli, 2001. Fission yeast Rad50 stimulates sister chromatid recombination and links cohesion with repair. EMBO J. 20 6660–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, M., G. M. Chin and A. M. Villeneuve, 2007. C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet. 3 e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz, I., 1987. Functional inactivation of genes by dominant negative mutations. Nature 329 219–222. [DOI] [PubMed] [Google Scholar]
- Holzen, T. M., P. P. Shah, H. A. Olivares and D. K. Bishop, 2006. Tid1/Rdh54 promotes dissociation of Dmc1 from nonrecombinogenic sites on meiotic chromatin. Genes Dev. 20 2593–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner, K. P., and J. A. Tainer, 2003. Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr. Opin. Struct. Biol. 13 249–255. [DOI] [PubMed] [Google Scholar]
- Hopfner, K. P., A. Karcher, D. Shin, C. Fairley, J. A. Tainer et al., 2000. a Mre11 and Rad50 from Pyrococcus furiosus: cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J. Bacteriol. 182 6036–6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner, K. P., A. Karcher, D. S. Shin, L. Craig, L. M. Arthur et al., 2000. b Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101 789–800. [DOI] [PubMed] [Google Scholar]
- Hopfner, K. P., A. Karcher, L. Craig, T. T. Woo, J. P. Carney et al., 2001. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105 473–485. [DOI] [PubMed] [Google Scholar]
- Hopfner, K. P., L. Craig, G. Moncalian, R. A. Zinkel, T. Usui et al., 2002. a The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418 562–566. [DOI] [PubMed] [Google Scholar]
- Hopfner, K. P., C. D. Putnam and J. A. Tainer, 2002. b DNA double-strand break repair from head to tail. Curr. Opin. Struct. Biol. 12 115–122. [DOI] [PubMed] [Google Scholar]
- Ivanov, E. L., V. G. Korolev and F. Fabre, 1992. XRS2, a DNA repair gene of Saccharomyces cerevisiae, is needed for meiotic recombination. Genetics 132 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka, K., and H. Ogawa, 1995. Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics 139 1521–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney, S., 2008. Spo11 and the formation of DNA double-strand breaks in meiosis, pp. 81–123 in Recombination and Meiosis, edited by R. Egel and D.-H. Lankenau. Springer, Berlin/Heidelberg, Germany. [DOI] [PMC free article] [PubMed]
- Keeney, S., C. N. Giroux and N. Kleckner, 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 375–384. [DOI] [PubMed] [Google Scholar]
- Kleckner, N., 2006. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115 175–194. [DOI] [PubMed] [Google Scholar]
- Krantz, I. D., J. McCallum, C. DeScipio, M. Kaur, L. A. Gillis et al., 2004. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat. Genet. 36 631–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis, P. J., 1991. Molscript—a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Lee, J. H., and T. T. Paull, 2004. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science 304 93–96. [DOI] [PubMed] [Google Scholar]
- Lee, J. H., and T. T. Paull, 2005. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308 551–554. [DOI] [PubMed] [Google Scholar]
- Li, L., E. E. Gerecke and M. E. Zolan, 1999. Homolog pairing and meiotic progression in Coprinus cinereus. Chromosoma 108 384–392. [DOI] [PubMed] [Google Scholar]
- Losada, A., and T. Hirano, 2005. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 19 1269–1287. [DOI] [PubMed] [Google Scholar]
- Luo, G., M. S. Yao, C. F. Bender, M. Mills, A. R. Bladl et al., 1999. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl. Acad. Sci. USA 96 7376–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone, R. E., 1983. Multiple mutant analysis of recombination in yeast. Mol. Gen. Genet. 189 405–412. [DOI] [PubMed] [Google Scholar]
- Malone, R. E., and R. E. Esposito, 1981. Recombinationless meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1 891–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone, R. E., T. Ward, S. Lin and J. Waring, 1990. The RAD50 gene, a member of the double strand break repair epistasis group, is not required for spontaneous mitotic recombination in yeast. Curr. Genet. 18 111–116. [DOI] [PubMed] [Google Scholar]
- Matsuura, S., H. Tauchi, A. Nakamura, N. Kondo, S. Sakamoto et al., 1998. Positional cloning of the gene for Nijmegen breakage syndrome. Nat. Genet. 19 179–181. [DOI] [PubMed] [Google Scholar]
- Mehrotra, S., and K. S. McKim, 2006. Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet. 2 e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino, S. T., W. J. Cummings, S. N. Acharya and M. E. Zolan, 2000. Replication-dependent early meiotic requirement for Spo11 and Rad50. Proc. Natl. Acad. Sci. USA 97 10477–10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis, C., R. Ciosk and K. Nasmyth, 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91 35–45. [DOI] [PubMed] [Google Scholar]
- Moncalian, G., B. Lengsfeld, V. Bhaskara, K. P. Hopfner, A. Karcher et al., 2004. The rad50 signature motif: essential to ATP binding and biological function. J. Mol. Biol. 335 937–951. [DOI] [PubMed] [Google Scholar]
- Moreno-Herrero, F., M. de Jager, N. H. Dekker, R. Kanaar, C. Wyman et al., 2005. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature 437 440–443. [DOI] [PubMed] [Google Scholar]
- Nairz, K., and F. Klein, 1997. mre11S–a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 11 2272–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nara, T., F. Hamada, S. Namekawa and K. Sakaguchi, 2001. Strand exchange reaction in vitro and DNA-dependent ATPase activity of recombinant LIM15/DMC1 and RAD51 proteins from Coprinus cinereus. Biochem. Biophys. Res. Commun. 285 92–97. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K., and C. H. Haering, 2005. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 74 595–648. [DOI] [PubMed] [Google Scholar]
- Neale, M. J., J. Pan and S. Keeney, 2005. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436 1053–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, K., A. Nicolas, M. Furuse, A. Nabetani, H. Ogawa et al., 1998. Mutations in the MRE11, RAD50, XRS2, and MRE2 genes alter chromatin configuration at meiotic DNA double-stranded break sites in premeiotic and meiotic cells. Proc. Natl. Acad. Sci. USA 95 646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques, F., and J. E. Haber, 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull, T. T., and M. Gellert, 1998. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell 1 969–979. [DOI] [PubMed] [Google Scholar]
- Paull, T. T., and M. Gellert, 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrini, J. H., M. E. Walsh, C. DiMare, X. N. Chen, J. R. Korenberg et al., 1995. Isolation and characterization of the human MRE11 homologue. Genomics 29 80–86. [DOI] [PubMed] [Google Scholar]
- Pitts, S. A., H. S. Kullar, T. Stankovic, G. S. Stewart, J. I. Last et al., 2001. hMRE11: genomic structure and a null mutation identified in a transcript protected from nonsense-mediated mRNA decay. Hum. Mol. Genet. 10 1155–1162. [DOI] [PubMed] [Google Scholar]
- Puizina, J., J. Siroky, P. Mokros, D. Schweizer and K. Riha, 2004. Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16 1968–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila, P. J., and B. C. Lu, 1985. Silver staining of meiotic chromosomes in the fungus, Coprinus cinereus. Chromosoma 91 108–112. [DOI] [PubMed] [Google Scholar]
- Pukkila, P. J., B. M. Yashar and D. M. Binninger, 1984. Analysis of meiotic development in Coprinus cinereus, pp. 177–194 in Controlling Events in Meiosis, edited by C. W. Evans and H. G. Dickinson. Society for Experimental Biology, Cambridge, UK. [PubMed]
- Pukkila, P. J., C. Skrzynia and B. C. Lu, 1992. The rad3-1 mutant is defective in axial core assembly and homologous chromosome pairing during meiosis in the basidiomycete Coprinus cinereus. Dev. Genet. 13 403–410. [Google Scholar]
- Raju, N. B., and B. C. Lu, 1970. Meiosis in Coprinus. III. Timing of meiotic events in C. lagopus (sensu Buller). Can. J. Bot. 48 2183–2186. [Google Scholar]
- Ramesh, M. A., and M. E. Zolan, 1995. Chromosome dynamics in rad12 mutants of Coprinus cinereus. Chromosoma 104 189–202. [DOI] [PubMed] [Google Scholar]
- Raymond, W. E., and N. Kleckner, 1993. RAD50 protein of S. cerevisiae exhibits ATP-dependent DNA binding. Nucleic Acids Res. 21 3851–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins, R. A., P. Morcillo and D. Dorsett, 1999. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics 152 577–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz, L. C., K. Tang, W. J. Cummings and M. E. Zolan, 1996. The rad9 gene of Coprinus cinereus encodes a proline-rich protein required for meiotic chromosome condensation and synapsis. Genetics 142 1105–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharples, G. J., and D. R. Leach, 1995. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol. Microbiol. 17 1215–1217. [DOI] [PubMed] [Google Scholar]
- Stajich, J. E., B. Birren, C. Burns, L. A. Casselton, F. Dietrich et al., 2006. Genomic analysis of Coprinus cinereus. Proceedings of the International Symposium on Mushroom Science, pp. 59–66. Akita Prefectural University, Akita, Japan.
- Stassen, N. Y., J. M. Logsdon, Jr., G. J. Vora, H. H. Offenberg, J. D. Palmer et al., 1997. Isolation and characterization of rad51 orthologs from Coprinus cinereus and Lycopersicon esculentum, and phylogenetic analysis of eukaryotic recA homologs. Curr. Genet. 31 144–157. [DOI] [PubMed] [Google Scholar]
- Stewart, G. S., R. S. Maser, T. Stankovic, D. A. Bressan, M. I. Kaplan et al., 1999. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99 577–587. [DOI] [PubMed] [Google Scholar]
- Storlazzi, A., S. Tesse, G. Ruprich-Robert, S. Gargano, S. Poggeler et al., 2008. Coupling meiotic chromosome axis integrity to recombination. Genes Dev. 22 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom, L., H. B. Lindroos, K. Shirahige and C. Sjogren, 2004. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16 1003–1015. [DOI] [PubMed] [Google Scholar]
- Sym, M., J. A. Engebrecht and G. S. Roeder, 1993. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72 365–378. [DOI] [PubMed] [Google Scholar]
- Tauchi, H., J. Kobayashi, K. Morishima, S. Matsuura, A. Nakamura et al., 2001. The forkhead-associated domain of NBS1 is essential for nuclear foci formation after irradiation but not essential for hRAD50·hMRE11·NBS1 complex DNA repair activity. J. Biol. Chem. 276 12–15. [DOI] [PubMed] [Google Scholar]
- Tavassoli, M., M. Shayeghi, A. Nasim and F. Z. Watts, 1995. Cloning and characterisation of the Schizosaccharomyces pombe rad32 gene: a gene required for repair of double strand breaks and recombination. Nucleic Acids Res. 23 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin, E. T., T. J. Wang, S. Lisgo, M. J. Bamshad and T. Strachan, 2004. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat. Genet. 36 636–641. [DOI] [PubMed] [Google Scholar]
- Trujillo, K. M., S. S. Yuan, E. Y. Lee and P. Sung, 1998. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 273 21447–21450. [DOI] [PubMed] [Google Scholar]
- Unal, E., A. Arbel-Eden, U. Sattler, R. Shroff, M. Lichten et al., 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16 991–1002. [DOI] [PubMed] [Google Scholar]
- Valentine, G., Y. J. Wallace, F. R. Turner and M. E. Zolan, 1995. Pathway analysis of radiation-sensitive meiotic mutants of Coprinus cinereus. Mol. Gen. Genet. 247 169–179. [DOI] [PubMed] [Google Scholar]
- Varon, R., C. Vissinga, M. Platzer, K. M. Cerosaletti, K. H. Chrzanowska et al., 1998. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 93 467–476. [DOI] [PubMed] [Google Scholar]
- Williams, R. S., and J. A. Tainer, 2005. A nanomachine for making ends meet: MRN is a flexing scaffold for the repair of DNA double-strand breaks. Mol. Cell 19 724–726. [DOI] [PubMed] [Google Scholar]
- Williams, R. S., J. S. Williams and J. A. Tainer, 2007. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 85 509–520. [DOI] [PubMed] [Google Scholar]
- Williams, R. S., G. Moncalian, J. S. Williams, Y. Yamada, O. Limbo et al., 2008. Mre11 dimers coordinate DNA end bridging and nuclease processing in double strand break repair. Cell 135 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltzius, J., M. Hohl, J. Fleming and J. Petrini, 2005. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat. Struct. Mol. Biol. 12 403–407. [DOI] [PubMed] [Google Scholar]
- Wolf, E., P. S. Kim and B. Berger, 1997. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 6 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, M. M. J., J. R. Cassidy and P. J. Pukkila, 1983. Polymorphisms in DNA of Coprinus cinereus. Curr. Genet. 7 385–392. [DOI] [PubMed] [Google Scholar]
- Xiao, Y., and D. T. Weaver, 1997. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 25 2985–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai, Y., E. Sonoda, M. S. Sasaki, C. Morrison, T. Haraguchi et al., 1999. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 18 6619–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J. A., R. W. Hyppa and G. R. Smith, 2004. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics 167 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., S. Petersen, L. Tessarollo and A. Nussenzweig, 2001. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol. 11 105–109. [DOI] [PubMed] [Google Scholar]
- Zolan, M. E., and P. J. Pukkila, 1986. Inheritance of DNA methylation in Coprinus cinereus. Mol. Cell. Biol. 6 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolan, M. E., C. J. Tremel and P. J. Pukkila, 1988. Production and characterization of radiation-sensitive meiotic mutants of Coprinus cinereus. Genetics 120 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolan, M. E., J. R. Crittenden, N. K. Heyler and L. C. Seitz, 1992. Efficient isolation and mapping of rad genes of the fungus Coprinus cinereus using chromosome-specific libraries. Nucleic Acids Res. 20 3993–3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolan, M. E., N. K. Heyler and M. A. Ramesh, 1993. Gene mapping using marker chromosomes in Coprinus cinereus, pp. 31–35 in Industrial Microorganisms: Basic and Applied Molecular Genetics, edited by R. H. Baltz, G. D. Hegeman and P. L. Skatrud. American Society for Microbiology, Washington, DC.
- Zolan, M. E., N. Yeager, Y. Stassen, M. A. Ramesh, B. C. Lu et al., 1995. Meiotic mutants and DNA repair genes of Coprinus cinereus. Can. J. Bot. Rev. Can. Bot. 73 S226–S233. [Google Scholar]