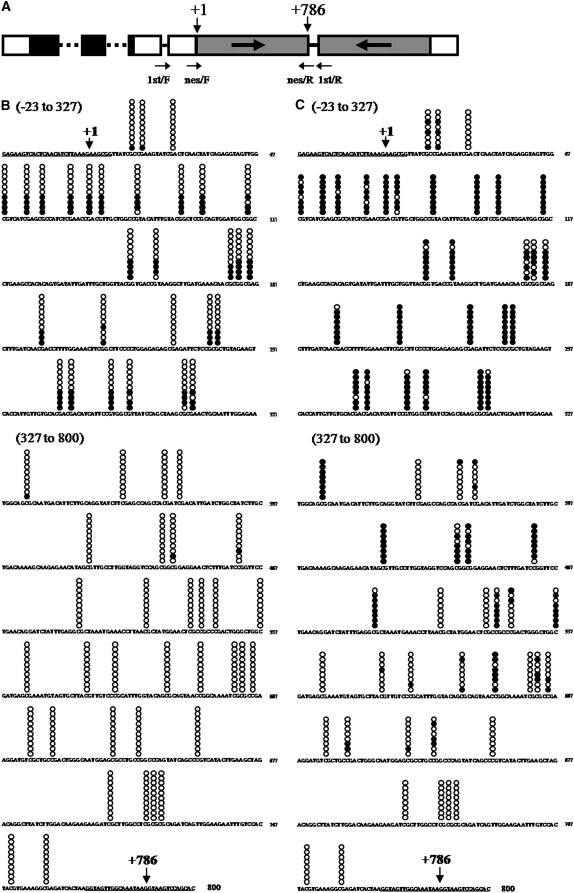
Figure 5.—
Analyses of methylated cytosine in the inverted repeat region of the silencer. (A) A schematic map of the silencer DNA construct. Annealing sites for the degenerate primers are indicated by arrows. (B and C) The CpG methylation patterns of a strongly silenced subclone and a weakly silenced subclone, respectively. Methylated cytosines were detected by the bisulfite sequencing method. Promoter region, −23 to −1; 5′-truncated aadA ORF region, +1 to +786; loop constructing intron, +787 to +800. Annealing sites for nested primers (see supplemental Table 5) are underlined. CpG dinucleotide positions are marked by stacked circles on the sequence. Each horizontal set of circles represents the methylation pattern of a cloned PCR product. Ten PCR clones obtained from a strongly silenced RNAi-37 subclone and 8 PCR clones obtained from a weakly silenced RNAi-37 subclone were sequenced. Solid circles indicate methylated cytosines and open circles indicate unmethylated cytosines. Each horizontal set of circles represents the methylation patterns of a single cloned PCR product.