Abstract
A screen for Saccharomyces cerevisiae temperature-sensitive silencing mutants identified a strain with a point mutation in the SIR2 gene. The mutation changed Ser276 to Cys. This amino acid is in the highly conserved NAD+ binding pocket of the Sir2 family of proteins. Haploid strains of either mating type carrying the mutation were severely defective at mating at 37° but normal at 25°. Measurements of RNA from the HMR locus demonstrated that silencing was lost rapidly upon shifting the mutant from the low to the high temperature, but it took >8 hours to reestablish silencing after a shift back to 25°. Silencing at the rDNA locus was also temperature sensitive. On the other hand, telomeric silencing was totally defective at both temperatures. Enzymatic activity of the recombinant wild-type and mutant Sir2 protein was compared by three different assays. The mutant exhibited less deacetylase activity than the wild-type protein at both 37° and 25°. Interestingly, the mutant had much more NAD+–nicotinamide exchange activity than wild type, as did a mutation in the same region of the protein in the Sir2 homolog, Hst2. Thus, mutations in this region of the NAD+ binding pocket of the protein are able to carry out cleavage of NAD+ to nicotinamide but are defective at the subsequent deacetylation step of the reaction.
SACCHAROMYCES cerevisiae Sir2 protein is the founding member of a large family of NAD+-dependent deacetylases, the so-called sirtuins, conserved from bacteria to mammals (Frye 2000). These enzymes deacetylate lysines on proteins in an unusual reaction in which NAD+ is cleaved, if and only if acetyl lysine is in the active site of the enzyme, releasing nicotinamide and subsequently deacetylating the lysine to form a novel compound, 2′-O-acetyl-ADP-ribose (AAR) (Tanner et al. 2000; Tanny and Moazed 2001). Nicotinamide is a noncompetitive inhibitor of these enzymes (Landry et al. 2000a). Several assays for sirtuins are available, including direct measurement of deacetylation, NAD+ hydrolysis during deacetylation, or NAD+–nicotinamide exchange (Landry and Sternglanz 2003). In the latter reaction, nonradioactive NAD+ is incubated with radioactive nicotinamide in the presence of enzyme and an acetylated substrate. Extent of the reaction is determined by the production of radioactive NAD+.
Yeast Sir2 is one of four so-called silent information regulator (SIR) proteins involved in transcriptional silencing of the silent mating-type loci, HML and HMR, as well as genes near telomeres (Gasser and Cockell 2001; Rusche et al. 2003). It facilitates silencing by deacetylating histone H4 lysine 16, thereby forming a binding site for a Sir2, Sir3, Sir4 complex on nucleosomes (Rudner et al. 2005). Sir2 is also present in a different protein complex at the rDNA (Shou et al. 1999; Straight et al. 1999); its role within the rDNA array is to suppress recombination and silence RNA polymerase II transcription (Gottlieb and Esposito 1989; Smith and Boeke 1997), presumably by deacetylating lysines on histones.
Studies on establishment and maintenance of silencing at the HM loci have been aided by the use of a sir3 temperature-sensitive (ts) mutation, sir3-8 (Miller and Nasmyth 1984; Lau et al. 2002). These studies have shown that inactivating Sir3 by raising the temperature to 37° causes an immediate loss of silencing, whereas cells shifted from 37° to the permissive temperature, 25°, must pass through S phase and G2/M of the cell cycle to reestablish silencing. A similar conclusion was reached by workers who used special strains in which silencing was absolutely dependent on Sir1 (Kirchmaier and Rine 2001; Li et al. 2001). In those cases, reestablishment of silencing by turning on expression of Sir1 also required passage through the cell cycle and could not occur in cells held in G1. Interestingly, a recent study using a galactose-regulated promoter to control Sir3 expression suggested that reestablishment of the fully silent state required several generations (Katan-Khaykovich and Struhl 2005). Here we describe the identification of a sir2 temperature-sensitive point mutation that changes a single highly conserved residue in the active site of the enzyme. We characterize its effect on silencing at all four loci where it acts and its enzymatic properties in vitro.
MATERIALS AND METHODS
Strains and plasmids:
Strains and plasmids used in this study are listed in Table 1 and Table 2, respectively. Strains used for the complementation test with strain TS1 were KN207, KN877, RS9, and KN9b. Yeast deletion mutants were constructed using standard methods (Longtine et al. 1998). SIR2 and sir2-276 fragments, containing 300 bp both upstream and downstream of the open reading frame, were amplified from genomic DNA of yeast strains RS1 and TS1, respectively, using primers CLP46 (5′-CCGCTCGAGTTTCTTTGACCCAACGCCTT-3′) and CLP47 (5′-CCGGAATTCATCTAGCACTCCTTCCAACCA-3′). Amplified products were digested with EcoRI and XhoI and cloned into the vector pRS316 (URA3, CEN6) to make pCLW18 and pCLW19, respectively. The same fragments were cloned into the vector pRS313 (HIS3, CEN6) to make pCLW21 and pCLW22, respectively. The entire SIR2 and sir2-276 coding regions were sequenced to confirm that there were no PCR-induced mutations. The GST-tagged SIR2 Escherichia coli expression plasmid, pDM111a, was obtained from D. Moazed (Tanny et al. 1999). A version of pDM111a with the sir2-276 mutation was made by amplifying DNA of yeast strain TS1 with primers CLP44 (5′-CCGGAATTCATGACCATCCCACATATG-3′) and CLP45 (5′-CCGCTCGAGTTAGAGGGTTTTGGGATG-3′). The amplified product was digested with ClaI and XhoI. The ClaI–XhoI fragment, which contains the sir2-276 mutation, was used to replace the ClaI–XhoI fragment of pDM111a to create pCLW20. The HST2 E. coli expression plasmid pJWL03 was constructed as described (Landry et al. 2000b). hst2 R45A, H135A, Q115A, and E128A mutant expression plasmids were made from pJWL03 with a site-directed mutagenesis kit (Stratagene) to create pJWL41, pJWL43, pJWL46, and pJWL47, respectively, as described (Min et al. 2001).
TABLE 1.
Strains
| Strain | Genotype |
|---|---|
| KN207 | MATasir1-1 ade6 arg4 leu2 trp1 |
| KN877 | HMLaMATaHMRasir2-275 leu2-1 can1-100 trp1-1 his3 his4 ade2-1 Met− |
| RS9 | HMLaMATaHMRasir3∷LEU2 leu2-1 can1-100 trp1-1 his3 his4 ade2-1 Met− |
| KN9b | HMLaMATaHMRasir4 ade2 his4 leu2 tyr can1 trp1 ura3 |
| W303-1a | MATaade2-1 his3-11,15 leu2-3,112 can1-100 ura3-1 trp1-1 |
| W303-1b | MATα ade2-1 his3-11,15 leu2-3,112 can1-100 ura3-1 trp1-1 |
| DC16 | MATahis1 |
| DC17 | MATα his1 |
| RS869 | W303-1a sir2∷URA3 |
| RS1717 | W303-1b sir2Δ∷his5+ |
| RS1 | W303-1b HMR/TRP1 |
| TS1 | W303-1b HMR/TRP1 sir2-276 |
| AMR1 | W303-1a nat1-5∷LEU2 |
| RS2291 | W303-1b sir2-276 (segregant of TS1 × AMR1) |
| RS2292 | W303-1a sir2-276 (segregant of TS1 × AMR1) |
| CCF100 | W303-1a HMRΔE∷TRP1 rDNA∷ADE2-CAN1 TELVR∷URA3 |
| RS2350 | MATα HMRΔE∷TRP1 rDNA∷ADE2-CAN1 TELVR∷URA3 sir2-276 (segregant of CCF100 × RS2291) |
| RS2351 | MATα HMRΔE∷TRP1 rDNA∷ADE2-CAN1 TELVR∷URA3 sir2-276 (segregant of CCF100 × RS2291) |
| DS2804 | W303-1a NTS1∷mURA3-LEU2 |
| DS2805 | W303-1b NTS1∷mURA3-LEU2 |
| RS2364 | DS2804 sir2Δ∷kanMX6 |
| RS2365 | DS2805 sir2Δ∷kanMX6 |
TABLE 2.
Plasmids
| Name | Description |
|---|---|
| pCLW18 | pRS316 SIR2 (URA3, CEN6, its own promoter) |
| pCLW19 | pRS316 sir2-276 (URA3, CEN6, its own promoter) |
| pCLW21 | pRS313 SIR2 (HIS3, CEN6, its own promoter) |
| pCLW22 | pRS316 sir2-276 (HIS3, CEN6, its own promoter) |
| pDM111a | pGEX-5X-1 SIR2 (GST-SIR2) |
| pCLW20 | pGEX-5X-1 sir2-276 (GST-sir2-276) |
| pJWL03 | pET28b HST2 (His6-HST2) |
| pJWL41 | pET28b hst2 R45A (His6-hst2 R45A) |
| pJWL43 | pET28b hst2 H135A (His6-hst2 H135A) |
| pJWL46 | pET28b hst2 Q115A (His6-hst2 Q115A) |
| pJWL47 | pET28b hst2 E128A (His6-hst2 E128A) |
Mating assays:
For patch mating assays, cells transformed with the indicated plasmids were patched onto the appropriate synthetic complete selective medium and grown at 25° or 37° for 1 day. Patches were transferred onto yeast–peptone–dextrose (YPD) medium by replica plating along with the appropriate mating tester strain (DC16 or DC17). After 6 hr, these cells were transferred by replica plating onto synthetic dextrose (SD) plates to select for diploids. Diploids were allowed to grow at 25° or 37° for 2 days. For quantitative mating assays, cells were grown in liquid YPD medium at 25° or 37° to exponential phase. Cells were diluted in YPD and plated onto YPD to count the total number of cells. Appropriate dilutions were also plated onto SD with 107 cells of an exponentially growing mating tester strain (DC16 or DC17) at 25° or 37°. Mating efficiency was calculated as the fraction of cells that mated. All values are the means of data from three independent assays.
Quantitative RNA measurements:
Cells were grown overnight in YPD and used to innoculate 30 ml of fresh YPD. Cells were then grown to an A600 ∼0.8, harvested, washed with diethyl pyrocarbonate-treated water, and total RNA was purified according to the protocol described (Ambion). Before cDNA synthesis, PCR controls were performed to confirm the absence of chromosomal DNA. To generate cDNA, 2 μg of total RNA were used in a 20-μl reaction with reverse transcriptase (Invitrogen). Two microliters of each cDNA reaction were amplified with a thermal cycler set of 94° for 4 min and then 23 cycles (for detection of a1 or PMA1 transcript in MATα strains) or 33 cycles (for detection of α1 or PMA1 transcript in MATa strains). a1 transcript was detected from cDNA using primers as described previously (Smeal et al. 1996). α1 transcript was detected using CLP52 (5′-TCCAGATTCCTGTTCCTTCCT-3′) and CLP53 (5′- CATTCTTCAGCGAGCAGAGAA-3′). PMA1 transcript was detected using PMA1-2018 (5′-CTATTATTGATGCTT TGAAGACCTCCAG -3′) and PMA1-2290 (5′-TTATGGGGTATGTCTATTATTTTG GGCA-3′). PCR products were resolved on a 2% agarose gel stained with ethidium bromide. For radioactive PCR, the same amounts of cDNAs were analyzed by standard reaction mixtures to which 1 μCi of [32P]dATP (3 Ci/μmol) had been added. Products were resolved on a 2% agarose gel and quantified using a Storm840 scanner and ImageQuant software (Molecular Dynamics).
For temperature-shift experiments overnight cultures were grown at 25° or 37° for 2 hr and then shifted to 37° or 25°, respectively. Cells were collected at different time points in 3 (25°–37° shift) to 12 hr (37°–25° shift) and total RNA isolated for RT–PCR by using primers that can specifically amplify HMR a1 transcripts. Primers that can amplify PMA1 transcripts were also used as a control. PCR products were resolved on a 2% agarose gel stained with ethidium bromide.
Silencing assays:
Strains CCF100, RS2350, and RS2351 were used to measure telomeric silencing. Strains were grown in YPD for 1 day at 25° or 37° and then plated in fivefold serial dilutions on YPD medium and synthetic complete (SC) medium containing 0.1% 5-fluoroorotic acid (5-FOA). For rDNA silencing assays, strains RS2364 and RS2365 transformed with plasmids pCLW21 (SIR2), pCLW22 (sir2-276), or pRS313 (vector) were grown in SC −His −Leu medium at 25° or 37° for 1 day. Cells were spotted in fivefold serial dilutions on SC −His and SC −His −Leu + 5-FOA medium. Plates were incubated at 25° and 37°, respectively for 2–4 days, depending on medium and temperatures.
Protein expression in E. coli:
Sir2 and Sir2 S276C were expressed in E. coli BL21(DE3) from pDM111a and pCLW20 after a 3-hr induction with 0.5 mm isopropyl β-d-thiogalactoside (IPTG) at room temperature. Proteins were purified using glutathione sepharose 4 fast flow affinity resin (Amersham) according to the manufacturer's instructions. Hst2 and mutants were expressed from pJWL03 (wild type), pJWL41 (R45A), pJWL43 (H135A), pJWL46 (Q115A), and pJWL47 (E128A) in E. coli BL21(DE3) and purified as described (Landry et al. 2000b). Purified proteins were dialyzed against 50 mm sodium phosphate (pH 7.2) and frozen at −80° in 50 mm sodium phosphate (pH 7.2), 0.5 mm DTT, and 10% glycerol. Protein concentrations were estimated by comparing Coomassie brilliant blue staining of samples with BSA standards, analyzed by SDS/PAGE.
NAD+ hydrolysis assays:
Reactions were carried out as described previously (Landry and Sternglanz 2003), using 50 μm H4 peptide or H4 lysine 16-acetylated peptide, 1 μCi [4-3H]NAD (0.37 Ci/mmol), and 50–150 ng purified Sir2 or Sir2 S276C. Reactions were incubated at 25° or 37° for 1 hr and terminated by adding 13.5 μl 0.5 m sodium borate (pH 8.0) and quenching on ice. Released nicotinamide was extracted by the addition of 0.5 ml of water-saturated ethyl acetate. After vortexing, samples were centrifuged at 16,000 × g for 2 min. A fraction of ethyl acetate (0.35 ml) was transferred to 3 ml of Ecoscint scintillation fluid and the released nicotinamide quantified in a liquid scintillation counter.
Deacetylation assays:
Reactions were carried out as previously described (Landry and Sternglanz 2003), using 50 μm H4 peptide or H4 lysine 16-acetylated peptide, 125 μm NAD, and 100 ng Sir2 or Sir2 S276C. Reactions were incubated at 25° or 37° for 1 hr and terminated by adding 13.5 μl 0.5 m sodium borate (pH 8.0) and quenching on ice. One microliter of each reaction was spotted to a nitrocellulose membrane and histone deacetylation was detected by Western blotting with an anti-histone H4 acetyl K16 antibody (Serotec).
NAD+–nicotinamide exchange assays:
Assays were performed as described previously (Landry and Sternglanz 2003) at 30° for 1 hr in 20 μl with 0.5 mm NAD+ (Sigma N-1511), enzymes (200–600 ng for Sir2 or Sir2 S276C; 1.3 ng for Hst2 mutants), 0.1 mm [14C]nicotinamide (Sigma N-2142, 53.1 mCi/mmol), 50 mm sodium phosphate (pH 7.2), 0.5 mm DTT, and 0.2 mg/ml chicken erythrocyte histones. After incubation, 8 μl of each reaction were spotted to Whatman HPKF silica gel 60 A thin-layer chromatography plates. The plates were developed in a preequilibrated chamber with 80:20 ethanol:2.5 m ammonium acetate. After chromatography, the plates were air dried, sprayed with EN3HANCE (NEN NEF981), and exposed to film (Kodak X-Omat AR) at −80° for 24 hr.
RESULTS
Identification and characterization of a sir2 ts mutant:
To identify ts silencing mutants, a yeast strain, RS1, with a TRP1 reporter gene at HMR was used (Brand et al. 1985). This strain could not grow without tryptophan in the medium due to silencing of the TRP1 gene (a Trp− phenotype). Spontaneous mutants were selected that were Trp+ at 37° but Trp− at 25°. One such mutant, TS1, was analyzed further by crossing it with sir1, sir2, sir3, or sir4 mutants of opposite mating type (see materials and methods and Table 1 for a description of these strains). Analysis of the Trp phenotype of the resultant diploids demonstrated that the mutation in strain TS1 failed to complement the sir2 mutation and thus suggested the silencing defect in TS1 was due to a mutation in the SIR2 gene. Further evidence that the mutation was in the SIR2 gene came from analysis of a cross of strain TS1 with a nat1∷LEU2 mutant (Mullen et al. 1989). NAT1 and SIR2 are adjacent genes and therefore recombination between them should be extremely rare. Eleven tetrads resulting from this cross were analyzed. All were parental, i.e., all the Leu− segregants exhibited ts mating and/or a ts Trp phenotype while none of the Leu+ segregants did. This demonstrated that the ts silencing mutation was tightly linked to NAT1 and thus consistent with a sir2 mutation.
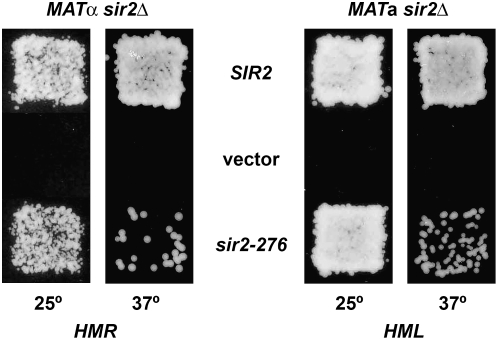
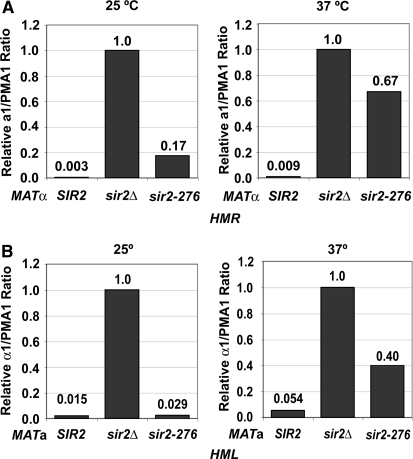
Therefore the SIR2 gene was amplified from strain TS1 by PCR and sequenced. A single nucleotide change was found when the sequence was compared to that of the parental wild-type strain. It was predicted to change Ser276 to Cys. The amplified SIR2 fragment from both the mutant and the wild-type parent was cloned into a yeast shuttle vector. The two resulting plasmids were transformed into sir2Δ strains and mating of the transformants tested. As seen in Figure 1A, mating is normal at both 25° and 37° for wild-type SIR2 but is ts for the mutant. This demonstrates conclusively that the mutation causing the ts silencing defect is in SIR2. We named this allele sir2-276. The mutation was also crossed into strains of both a and α mating types. Again mating of these strains was seen to be ts, as judged by both patch mating (not shown) and quantitative mating (Table 3). Note that mating was >10-fold worse at 37° in the MATα strain than in the MATa strain, suggesting that silencing was more defective at HMR than at HML. Quantitative RT–PCR was used to measure a1 RNA from HMR in the MATα strain and α1 RNA from HML in the MATa strain (Figure 2, A and B). This confirmed that the mutation conferred a silencing defect and it was greater at HMR than at HML. The data also showed that some RNA from HMR was detected even at 25°, showing that the Sir2 protein from the mutant is somewhat defective even at a lower temperature.
Figure 1.—
The sir2-276 mutation confers a temperature-sensitive mating defect. The indicated strains were transformed with a plasmid expressing SIR2, a vector or a plasmid expressing sir2-276 (pCLW18, pRS316, and pCLW19 for the MATα sir2 strain, RS1717; pCLW20, pRS313, and pCLW21 for the MATa sir2 strain, RS869). Mating was measured at 25° or 37° as indicated.
TABLE 3.
Quantitative mating of SIR2 and sir2-276 strains
|
MATα
|
MATa
|
|||
|---|---|---|---|---|
| 25° | 37° | 25° | 37° | |
| SIR2 | 1 | 1 | 1 | 1 |
| sir2-276 | 7.0 × 10−1 | 3.9 × 10−5 | 1.3 | 1.6 × 10−4 |
Figure 2.—
The sir2-276 mutation has a temperature-sensitive silencing defect at the HM loci. Expression from the HM loci of SIR2, sir2Δ, or sir2-276 strains was determined by RT–PCR. (A) cDNA derived from RNA from MATα strains (W303-1b, RS1717, and RS2291) grown at either 25° or 37° was amplified with primers to a1 and PMA1. The a1/PMA1 ratios were calculated and shown relative to the value for the sir2Δ strain, which was set to 1. (B) cDNA from MATa strains (W303-1a, RS869, and RS2292) cultured at either 25° or 37° was amplified with primers to α1 and PMA1. The α1/PMA1 ratios were calculated and shown as in A.
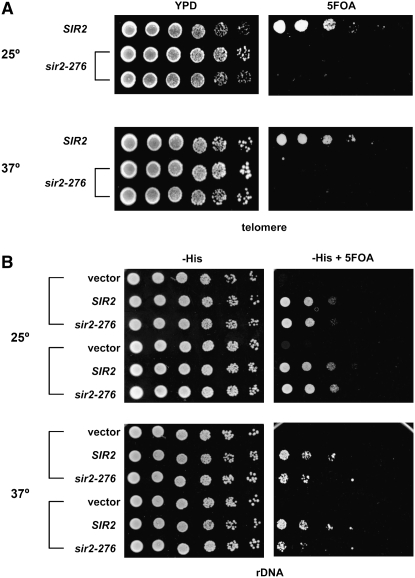
The sir2-276 mutation was also introduced into a strain with a URA3 reporter gene near a telomere. As can be seen in Figure 3A, the mutation led to a complete loss of telomeric silencing at both 25° and 37°. To assess rDNA silencing, plasmids containing either wild-type SIR2 or sir2-276 were transformed into two sir2Δ strains with a URA3 reporter gene in the rDNA array. The rDNA silencing of the transformants was then tested. As shown in Figure 3B, silencing at rDNA was normal at both 25° and 37° for wild-type SIR2 but was somewhat defective in the mutant at 37° for each of the two strains tested. In summary, sir2-276 caused a temperature-sensitive silencing defect at both HM loci and rDNA and a complete silencing defect at telomeres.
Figure 3.—
The sir2-276 mutation confers a complete silencing defect at telomeres and a temperature-sensitive silencing defect at rDNA. (A) Telomeric silencing. Fivefold serial dilutions of overnight cultures grown in YPD medium of SIR2 (CCF100) and sir2-276 strains (RS2350 and RS2351) containing a URA3 reporter gene integrated near telomere VR were spotted onto YPD plates and 5-FOA plates at either 25° or 37°. Good growth on 5-FOA plates means good silencing. (B) rDNA silencing. Strains RS2364 and RS2365, both with a sir2Δ mutation and containing a URA3 reporter gene within the rDNA, were transformed with a SIR2 plasmid, a plasmid expressing sir2-276, or vector. Transformed strains were grown overnight in SC −His −Leu medium at 25° or 37° and fivefold serial dilutions spotted on selective medium without and with 5-FOA.
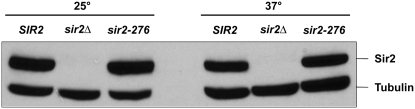
To determine if the mutant Sir2 protein was degraded at 37°, Westerns were performed on extracts from cells grown at 25° and 37°. As seen in Figure 4, the mutant protein is present in amounts similar to that of the wild-type strain at both temperatures. Thus, the observed silencing defect of the mutant is due to loss of function of the mutant protein, not to its degradation.
Figure 4.—
The Sir2-276 mutant protein is stable at 37°. A Western with an antibody to Sir2 showed that similar amounts of Sir2 were present in wild-type cells and the sir2-276 mutant grown at 25° or at 37°. No signal was seen for a sir2Δ strain. An antibody to tubulin was used to show that similar amounts of total protein were present in all lanes.
Temperature shifts:
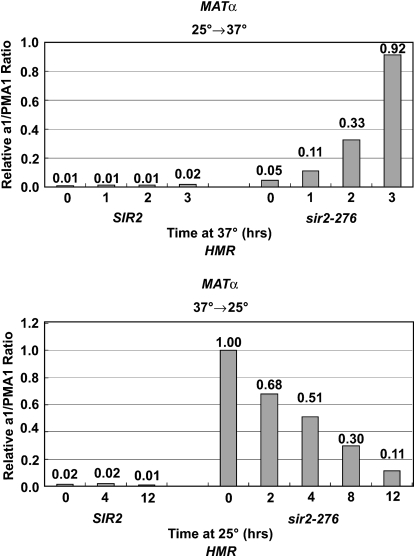
Previous experiments with a sir3 ts mutant showed that mutant cells that shifted from a low to a high temperature lost silencing rapidly while cells that shifted from a high temperature to a low temperature required many hours and passage through the cell cycle to regain full silencing (Miller and Nasmyth 1984; Lau et al. 2002). We wanted to determine whether the same behavior would be seen with the sir2-276 mutant. As seen in Figure 5, when cells were shifted from 25° to 37° (top), silencing at HMR was lost rapidly. On the other hand, when cells were shifted from 37° to 25° (Figure 5, bottom), it took >8 hr to see significant silencing of HMR. These results indicate that sir2-276, similar to sir3-8, also needs to pass through the cell cycle to reestablish complete silencing.
Figure 5.—
Measurements of a1 mRNA from HMR after temperature shifts of MATα SIR2 and sir2-276 strains. Cells were grown at 25° for 2 hr and raised to 37° for 0, 1, 2, and 3 hr (top), or grown at 37° for 2 hr and dropped to 25° for 0, 2, 4, 8, and 12 hr (bottom). cDNAs collected at different time points were amplified with primers to a1 and PMA1 and RNA was quantified as in Figure 2.
Enzymatic assays:
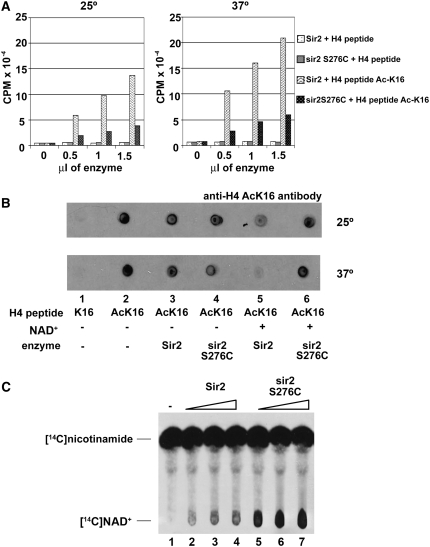
The Ser276C mutation was introduced into a Sir2 plasmid used to express the recombinant protein in E. coli. Both the wild-type and mutant recombinant proteins were purified using glutathione beads and then assayed for enzymatic activity. First, an H4 peptide acetylated on Lys16 was assayed by an NAD+ hydrolysis assay, commonly used to quantitate deacetylation by sirtuins (Landry and Sternglanz 2003). As seen in Figure 6A, the mutant protein had less activity than the wild-type protein at both 25° and 37° but retained some activity at both temperatures. Note that no activity for either protein was seen with an unacetylated H4 peptide, as expected. In another assay, deacetylation of the acetylated H4 peptide was measured directly by use of an antibody specific to H4 acetyl-Lys16. A Western blot of the reactions demonstrated good NAD+-dependent deacetylation activity for the wild-type protein but little or no activity for the mutant at both temperatures (Figure 6B). Next we assayed both proteins using a NAD+–nicotinamide exchange assay (Landry and Sternglanz 2003). A surprising result was obtained. The mutant reproducibly showed greater exchange activity than wild type (Figure 6C). The same protein preparations were used in the two deacetylase assays described above as in this exchange assay. Since the mutant protein had less activity in both deacetylase assays, the greater NAD+–nicotinamide exchange activity seen for the mutant is a real effect and cannot be due to a higher concentration of functional mutant protein.
Figure 6.—
Enzymatic assays with recombinant Sir2 and sir2 S276C. (A) NAD+ hydrolysis. Assays were performed by incubating enzyme (0–1.5 μl of Sir2 or sir2 S276C, 100 ng/μl), radioactive NAD+ labeled on the nicotinamide moiety, and H4 peptide (unacetylated or acetyl-K16) at 25° or 37° for 1 hr. Activity was determined by measuring the amount of [3H]nicotinamide released. The sir2 S276C mutant protein shows defective NAD+ hydrolysis activity at both temperatures. (B) Deacetylase assays. The assays were performed by incubating enzyme (100 ng of Sir2 or sir2 S276C), +/− NAD+, and H4 peptide (unacetylated or acetyl-K16) at 25° or 37° for 1 hr. The reaction mixture was spotted onto paper and activity was determined by probing with an anti-H4 acetyl-K16 antibody. Deacetylase activity is shown by a weaker response to the H4 acetyl-K16 antibody. As in A, sir2 S276C shows defective deacetylase activity at both temperatures. (C) NAD+–nicotinamide exchange assays. The assays were performed by incubating enzyme (2–6 μl of Sir2 or sir2 S276C, 100 ng/μl), NAD+, radioactive nicotinamide and chicken histones at 30° for 1 hr. Activity was determined by the amount of [14C]NAD+ formed. sir2 S276C protein shows enhanced NAD+–nicotinamide exchange activity compared with Sir2.
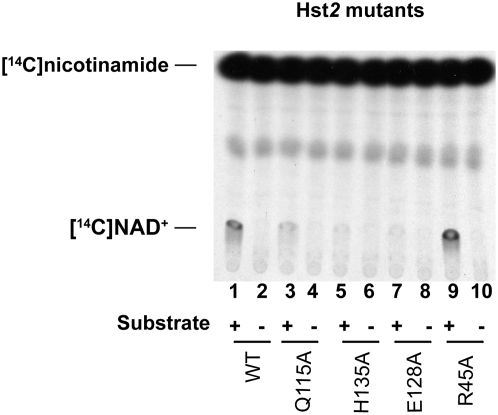
This result was reminiscent of an unpublished result we had obtained previously with a particular Hst2 mutant. As shown in Figure 7, an Hst2 Arg45Ala mutant also had greater exchange activity than wild type, whereas three other point mutants in conserved residues had lower activity than wild type. All four mutants tested had lower-than-wild-type deacetylase activity (data not shown). The Arg45 residue of Hst2 corresponds to Arg275 of Sir2 and thus is right next to Ser276. Why mutations in this conserved region of Sir2 have greater NAD+–nicotinamide exchange activity but less deacetylation activity will be discussed below.
Figure 7.—
NAD+–nicotinamide exchange assays with Hst2 mutants. Recombinant wild-type and mutant Hst2 proteins were assayed by NAD+–nicotinamide exchange assays. The assays were done by incubating 1.3 ng of enzyme, NAD+, radioactive nicotinamide, and chicken histones (substrate, with or without) at 30° for 1 hr. The Q115A, H135A, and E128A mutants show reduced exchange activity while the R45A mutant shows enhanced activity.
DISCUSSION
As mentioned, sir2-276 was identified as a spontaneous mutation that led to a temperature-sensitive silencing defect at HMR. This mutation changes only one residue in Sir2: Ser276 to Cys. Interestingly, such a minor change of a side chain from -CH2-OH to -CH2-SH caused a significant enzymatic defect in vitro and significant silencing defects in vivo, showing the structural and enzymatic importance of Ser276 in Sir2.
This amino acid is in a conserved disordered loop of the NAD+ binding pocket of the enzyme, termed the C site in the Sir2 superfamily (Min et al. 2001). The cocrystal structure of the yeast Sir2 homolog Hst2 with the product of NAD hydrolysis, 2′-O-acetyl-ADP-ribose, has been solved (Zhao et al. 2003). The structure indicates that Ser46, which is the homologous residue of Sir2 Ser276, is adjacent to Phe44 and Arg45 that form hydrogen bonds with the phosphate group of AAR. These results support the idea that Ser276 in Sir2 is important for its enzymatic activity as discussed below.
The Sir2 family of NAD+-dependent deacetylases acts in two steps (Landry et al. 2000a; Jackson et al. 2003; Sauve and Schramm 2003). In the first step, NAD is cleaved, releasing nicotinamide. This step is reversible in the presence of excess nicotinamide and makes the exchange reaction possible in vitro. In the second step, which is irreversible, the protein substrate is deacetylated, releasing AAR. In our enzymatic assays, we observed that Sir2 S276C protein is defective in deacetylation as determined by two assays, NAD hydrolysis and Western blotting. In these two assays the activities were measured by detecting the final products of the whole two-step reaction, nicotinamide or unacetylated lysine, respectively. In the NAD+–nicotinamide exchange assay, which measures the reversible first step, both Sir2 S276C and Hst2 R45A mutant proteins showed more activity than wild-type protein. We think this is because the S276C mutation and the adjacent mutation in Hst2 inhibit the second step but not the first. This increases the lifetime of an acetyl-lysine-ADP ribose 1′-O-alkylamidate intermediate and thus leads to increased NAD+–nicotinamide exchange (Denu 2005). Recently, Khan and Lewis (2006) determined kinetic parameters for several different hst2 mutants, including the R45A mutant we have used. They found that the R45A mutant had a very similar binding constant (Km) as wild type for either NAD+ or an acetylated lysine substrate but the rate constant (kcat) was about six times slower than wild type. They therefore proposed, as we do, that the function of Arg45 in Hst2 was to stabilize the acetyl-lysine-ADP ribose intermediate formed between step one and step two of the reaction, supporting our hypothesis that the adjacent Arg and Ser residues in the conserved loop play an important role in the second step of the deacetylation reaction.
Two silencing assays, mating (Figure 1 and Table 3) and quantitative RNA measurements (Figure 2), both showed that the sir2-276 mutation had a greater silencing defect at HMR than at HML. This result was somewhat surprising. Generally mutations that do not completely abolish silencing, such as nat1/ard1, sas2, or leaky sir3 mutations, abolish silencing at telomeres, weaken it at HML, and hardly affect HMR. In other words, silencing is considered strongest at HMR, perhaps because the HMR-E silencer is the only one of the four HM silencers with binding sites for three proteins, ORC, Rap1, and Abf1. Thus, it is not clear why the sir2-276 mutant affects HMR more than HML.
Our in vitro enzymatic assays have shown that Sir2 S276C is equally defective at both 25° and 37° (Figures 6 and 7). On the other hand, the mutant exhibited a temperature-sensitive mating and silencing defect (Figures 1 and 2) and an rDNA silencing defect (Figure 3B). However, the mutant had a complete loss of telomeric silencing at both temperatures (Figure 3A). Moreover, in the quantitative RT–PCR assays, some RNA from HMR was detected even at 25° (Figure 2), showing that the mutant is somewhat defective at a lower temperature. It is not clear why the sir2-276 strain has a strong temperature-sensitive silencing defect at the HM loci and rDNA when the mutant protein is enzymatically defective even at a lower temperature. Apparently, more Sir2 activity is required in vivo at 37° than at 25°.
Holmes and colleagues recently described several sir2 ts mutants generated by error-prone mutagenesis (Matecic et al. 2006; Hickman et al. 2007). All but one of these mutants had multiple amino acid changes. Their mutants fell into two classes. One group had defects in deacetylase activity and the other had defects in binding to Sir4 (Matecic et al. 2006). Similar to the sir2-276 mutant described here, they found that the deacetylase-defective mutants lost silencing rapidly when raised to the nonpermissive temperature. Another study in which silencing was reestablished by galactose induction of Sir3 showed that restoration of complete silencing of HMR took more than three generations (Katan-Khaykovich and Struhl 2005). Similarly, we found that it took >8 hr after lowering the temperature of the sir2 ts mutant to observe significant silencing of HMR (Figure 5), clearly more than one generation. It will be interesting to study the cell cycle dependence of silencing reestablishment at both silent loci with this mutant and compare it with previous studies in which the controlled production of Sir3 or Sir1 was used to examine reestablishment of silencing as a function of the cell cycle.
Acknowledgments
R.S. thanks Kim Nasmyth in whose lab this work was initiated many years ago. We thank Danesh Moazed for the Sir2 expression plasmid. This work was supported by National Institutes of Health grant GM28220.
References
- Brand, A. H., L. Breeden, J. Abraham, R. Sternglanz and K. Nasmyth, 1985. Characterization of a “silencer” in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell 41 41–48. [DOI] [PubMed] [Google Scholar]
- Denu, J. M., 2005. The Sir2 family of protein deacetylases. Curr. Opin. Chem. Biol. 9 431–440. [DOI] [PubMed] [Google Scholar]
- Frye, R. A., 2000. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273 793–798. [DOI] [PubMed] [Google Scholar]
- Gasser, S. M., and M. M. Cockell, 2001. The molecular biology of the SIR proteins. Gene 279 1–16. [DOI] [PubMed] [Google Scholar]
- Gottlieb, S., and R. E. Esposito, 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56 771–776. [DOI] [PubMed] [Google Scholar]
- Hickman, M., K. McCullough, A. Woike, L. Raducha-Grace, T. Rozario et al., 2007. Isolation and characterization of conditional alleles of the yeast SIR2 gene. J. Mol. Biol. 367 1246–1257. [DOI] [PubMed] [Google Scholar]
- Jackson, M. D., M. T. Schmidt, N. J. Oppenheimer and J. M. Denu, 2003. Mechanism of nicotinamide inhibition and transglycosidation by Sir2 histone/protein deacetylases. J. Biol. Chem. 278 50985–50998. [DOI] [PubMed] [Google Scholar]
- Katan-Khaykovich, Y., and K. Struhl, 2005. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J. 24 2138–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, A. N., and P. N. Lewis, 2006. Use of substrate analogs and mutagenesis to study substrate binding and catalysis in the Sir2 family of NAD-dependent protein deacetylases. J. Biol. Chem. 281 11702–11711. [DOI] [PubMed] [Google Scholar]
- Kirchmaier, A. L., and J. Rine, 2001. DNA replication-independent silencing in S. cerevisiae. Science 291 646–650. [DOI] [PubMed] [Google Scholar]
- Landry, J., and R. Sternglanz, 2003. Enzymatic assays for NAD-dependent deacetylase activities. Methods 31 33–39. [DOI] [PubMed] [Google Scholar]
- Landry, J., J. T. Slama and R. Sternglanz, 2000. a Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem. Biophys. Res. Commun. 278 685–690. [DOI] [PubMed] [Google Scholar]
- Landry, J., A. Sutton, S. T. Tafrov, R. C. Heller, J. Stebbins et al., 2000. b The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci.USA 97 5807–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, A., H. Blitzblau and S. P. Bell, 2002. Cell-cycle control of the establishment of mating-type silencing in S. cerevisiae. Genes Dev. 16 2935–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. C., T. H. Cheng and M. R. Gartenberg, 2001. Establishment of transcriptional silencing in the absence of DNA replication. Science 291 650–653. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, 3rd, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953–961. [DOI] [PubMed] [Google Scholar]
- Matecic, M., K. Martins-Taylor, M. Hickman, J. Tanny, D. Moazed et al., 2006. New alleles of SIR2 define cell-cycle-specific silencing functions. Genetics 173 1939–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, A. M., and K. A. Nasmyth, 1984. Role of DNA replication in the repression of silent mating type loci in yeast. Nature 312 247–251. [DOI] [PubMed] [Google Scholar]
- Min, J., J. Landry, R. Sternglanz and R. M. Xu, 2001. Crystal structure of a SIR2 homolog-NAD complex. Cell 105 269–279. [DOI] [PubMed] [Google Scholar]
- Mullen, J. R., P. S. Kayne, R. P. Moerschell, S. Tsunasawa, M. Gribskov et al., 1989. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 8 2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner, A. D., B. E. Hall, T. Ellenberger and D. Moazed, 2005. A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin. Mol. Cell. Biol. 25 4514–4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche, L. N., A. L. Kirchmaier and J. Rine, 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72 481–516. [DOI] [PubMed] [Google Scholar]
- Sauve, A. A., and V. L. Schramm, 2003. Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Biochemistry 42 9249–9256. [DOI] [PubMed] [Google Scholar]
- Shou, W., J. H. Seol, A. Shevchenko, C. Baskerville, D. Moazed et al., 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97 233–244. [DOI] [PubMed] [Google Scholar]
- Smeal, T., J. Claus, B. Kennedy, F. Cole and L. Guarente, 1996. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell 84 633–642. [DOI] [PubMed] [Google Scholar]
- Smith, J. S., and J. D. Boeke, 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11 241–254. [DOI] [PubMed] [Google Scholar]
- Straight, A. F., W. Shou, G. J. Dowd, C. W. Turck, R. J. Deshaies et al., 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97 245–256. [DOI] [PubMed] [Google Scholar]
- Tanner, K. G., J. Landry, R. Sternglanz and J. M. Denu, 2000. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA 97 14178–14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny, J. C., G. J. Dowd, J. Huang, H. Hilz and D. Moazed, 1999. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99 735–745. [DOI] [PubMed] [Google Scholar]
- Tanny, J. C., and D. Moazed, 2001. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. USA 98 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, K., X. Chai and R. Marmorstein, 2003. Structure of the yeast Hst2 protein deacetylase in ternary complex with 2′-O-acetyl ADP ribose and histone peptide. Structure 11 1403–1411. [DOI] [PubMed] [Google Scholar]