Abstract
Like most microorganisms, the yeast Saccharomyces cerevisiae is prototrophic for riboflavin (vitamin B2). Riboflavin auxotrophic mutants with deletions in any of the RIB genes frequently segregate colonies with improved growth. We demonstrate by reporter assays and Western blots that these suppressor mutants overexpress the plasma-membrane riboflavin transporter MCH5. Frequently, this overexpression is mediated by the transcription factor Put3, which also regulates the proline catabolic genes PUT1 and PUT2. The increased expression of MCH5 may increase the concentrations of FAD, which is the coenzyme required for the activity of proline oxidase, encoded by PUT1. Thus, Put3 regulates proline oxidase activity by synchronizing the biosynthesis of the apoenzyme and the coenzyme FAD. Put3 is known to bind to the promoters of PUT1 and PUT2 constitutively, and we demonstrate by gel-shift assays that it also binds to the promoter of MCH5. Put3-mediated transcriptional activation requires proline as an inducer. We find that the increased activity of Put3 in one of the suppressor mutants is caused by increased intracellular levels of proline. Alternative PUT3-dependent and -independent mechanisms might operate in other suppressed strains.
MAMMALS depend on a dietary supply of riboflavin (vitamin B2), which mostly derives from the flavoprotein cofactors FMN and FAD. These are deadenylated or dephosphorylated in the gut followed by the transport of free riboflavin across the mucosal membrane (Foraker et al. 2003). In contrast, although many microorganisms are dependent on various water-soluble vitamins, only few show a riboflavin auxotrophy (Koser 1968). This indicates that most microorganisms are capable of synthesizing riboflavin, a pathway, which starts with GTP and two molecules of ribulose-5-phosphate and is similar but not perfectly conserved in various species (Bacher et al. 2000). In the yeast Saccharomyces cerevisiae, which is an excellent dietary source of riboflavin (Bässler et al. 2002), the enzymes required for riboflavin synthesis are encoded by the genes RIB1, RIB2, RIB3, RIB4, RIB5, and RIB7. Both prokaryotic and eukaryotic microorganisms have been engineered to overproduce riboflavin and are used in industrial processes for riboflavin synthesis (Stahmann et al. 2000).
In addition to being able to synthesize riboflavin, single-celled organisms are also capable of taking up riboflavin from the culture medium. Because the riboflavin transport activities of most wild-type (wt) strains are low, most investigations were performed with riboflavin auxotrophic mutants. At least three different classes of riboflavin transporters exist in bacteria. These have been predicted by phylogenetic footprinting (Vitreschak et al. 2002) and functional data are now available for two proteins. RibU from Lactococcus lactis and Bacillus subtilis appear to work as very high-affinity transporters with five transmembrane domains (Cecchini et al. 1979; Burgess et al. 2006; Vogl et al. 2007). According to our analyses, RibU acts as an active riboflavin transporter in B. subtilis. Proteins of the RibM type are present in Corynebacterium glutamicum and Streptomyces davawensis (Grill et al. 2007; Vogl et al. 2007) and RibM from C. glutamicum acts as a facilitator when expressed in Escherichia coli. The third prototype bacterial riboflavin transporter, ImpX, has not been experimentally studied (Vitreschak et al. 2002). Yet another type of plasma-membrane riboflavin transporter is present in fungi. We used a multicopy suppressor screen of S. cerevisiae riboflavin auxotrophic strains to identify MCH5, the first known eukaryotic riboflavin transporter gene (Reihl and Stolz 2005). Riboflavin transport in yeast is not significantly stimulated by glucose or ethanol and not inhibited by proton ionophors, indicating that Mch5 acts as a facilitator. Strains with a deletion of MCH5 and of a RIB gene show synthetic growth defects and a reduced efficiency in catalyzing FAD-dependent cellular processes. Moreover, the expression of MCH5 is regulated according to the riboflavin supply (Reihl and Stolz 2005). Most recently, a mammalian riboflavin transporter has been characterized, which again is not related to the previously known riboflavin transporters and was earlier mistaken as a G-protein coupled receptor (Yonezawa et al. 2008).
In the course of our experiments on the riboflavin transporter MCH5 we noted that rib deletion strains segregate suppressor mutants with improved growth. A similar phenomenon was observed in a riboflavin auxotrophic strain of Pichia guillermondii (Boretsky et al. 2005). Here, we perform a detailed genetic and biochemical analysis of the S. cerevisiae mutants and find that the suppressor phenotype depends on the transcription factor PUT3. Put3 is hyperactive in the suppressor mutants, which results in increased expression of MCH5 and other Put3 target genes on ammonium-containing media. Since Put3 also regulates the proline catabolic genes PUT1 and PUT2 (Brandriss and Magasanik 1979), this work establishes that riboflavin uptake is a part of the regulatory network that allows S. cerevisiae cells to use proline as the sole source of nitrogen.
MATERIALS AND METHODS
Yeast strains:
Most experiments made use of the haploid strains BY4741, BY4742, or the diploid strain BY4743 (Brachmann et al. 1998). These and the haploid deletion strains rib5Δ, put3Δ, mch1Δ, mch3Δ, mch4Δ, and mch5Δ and the heterozygous diploid rib3Δ/RIB3 and rib7Δ/RIB7 strains were obtained from Euroscarf (Frankfurt/Main, Germany). Haploid rib2Δ, rib3Δ, and rib7Δ strains were generated from heterozygous diploids by sporulation and tetrad dissection on YPD plates with 200 mg/liter added riboflavin. rib4Δ strains were not included in the analysis because they are capable of riboflavin synthesis (Kis et al. 2001; Reihl and Stolz 2005). Alternatively, strains based on W303 (Thomas and Rothstein 1989) or CEN.PK113–13D (Makuc et al. 2001; Reihl and Stolz 2005) were used. The PUT3c strain C74-6D (Mata his4-42 ura3-52 PUT3c-683) and the isogenic wild-type strain MB758-5B (Mata ura3-52) were obtained from Marjorie C. Brandriss (Marczak and Brandriss 1989; Siddiqui and Brandriss 1989).
To generate double-deletion strains, kanMX4 in rib5Δ was replaced with his5+ from Schizosaccharomyces pombe using a restriction fragment from pFA6a-HIS3MX6 (Wach et al. 1994). The rib5Δ∷HIS3MX6 strain was mated to strains with the other desired deletion, followed by sporulation, tetrad dissection, and analysis of nutritional markers. RIB1 was deleted in the BY strain background using a rib1Δ∷LEU2 disruption cassette, which inserted LEU2 as a ScaI/BglII fragment into the natural EcoRV/BamHI sites within RIB1, thus replacing 282 bp of the RIB1 ORF. For one of the strains used in Figure 3E, PUT3 was deleted in rib5Δ∷kanMX using a put3Δ∷LEU2 deletion plasmid. In this construct, a 2602 bp HindIII/BamHI fragment of PUT3 was replaced with the LEU2 marker gene. All genomic modifications were checked by PCR.
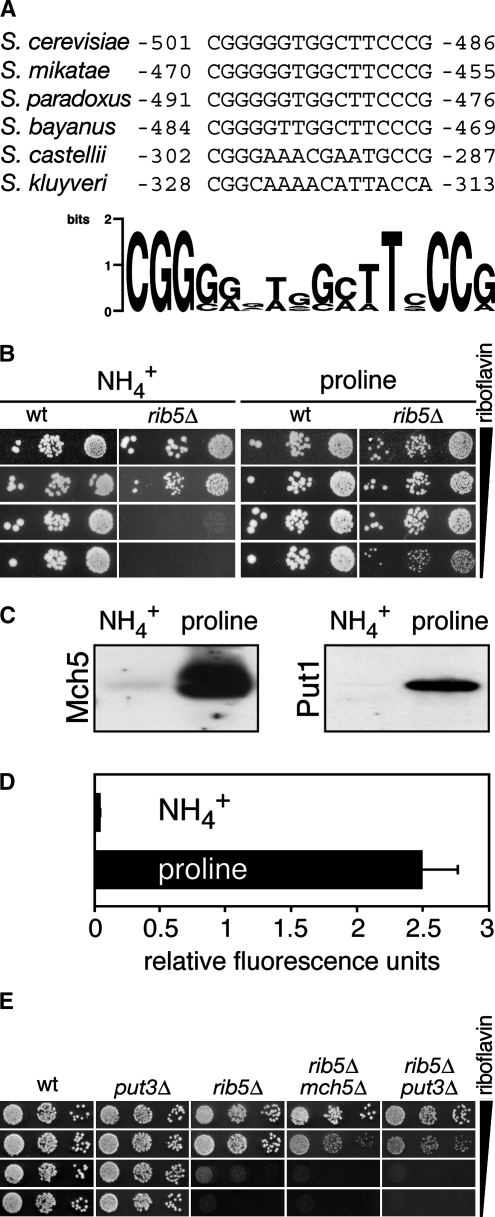
Figure 3.—
Proline induces the expression of MCH5. (A) Alignment of the conserved Put3 binding sites from the MCH5 promoters of various Saccharomyces species. The figure was generated using weblogo (http://weblogo.berkeley.edu/). (B) Growth of rib5Δ and a corresponding wild-type strain on minimal media containing proline or ammonium as sole nitrogen sources and various concentrations of riboflavin (from top to bottom: 200, 20, 2, or 0.2 mg/liter). Serial dilutions of cell suspensions were spotted and grown for 3 days at 30°. (C) Abundance of Mch5 and a Put1 after growth in proline or ammonium. Western blots were performed with the anti-Mch5 antibody or the anti-HA antibody (which binds to the ZZ-tag of Put1). PUT1-ZZ was expressed from a centromeric plasmid under control of its own promoter. Both strains were from the BY genetic background and prototrophic for riboflavin. (D) The Put3 binding site from the MCH5-promoter confers increased reporter activity in proline-grown cells. The GFP reporter construct containing the Put3 binding site from the MCH5 promoter in a MEL1 minimal promoter was transformed into BY4742 wild-type cells. The cells were continuously grown in media containing either ammonium or nitrogen as sole nitrogen source and GFP fluorescence was determined as described in materials and methods. (E) rib5Δ mch5Δ and rib5Δ put3Δ double mutants show an increased requirement for riboflavin. Serial dilutions of cells of the indicated genotype were spotted on YPD plates to which various amounts of riboflavin were added. The additions were (from top to bottom) 200, 20, 2 mg/liter, or no riboflavin as indicated. The rib5Δ mch5Δ double mutant had the genotype rib5Δ∷HIS3MX6 mch5Δ∷KanMX4; the rib5Δ put3Δ double mutant had the genotype rib5Δ∷KanMX4 put3Δ∷LEU2.
Yeast media:
YPD medium (2% d-glucose, 2% bacto peptone and 1% yeast extract) and standard minimal medium (2% d-glucose, 0.67% yeast nitrogen base without amino acids, which contains 0.5% ammonium sulfate as nitrogen source) were prepared from compounds secured from Difco. According to flourimetric measurements (excitation wavelength 449 nm, emission wavelength 526 nm), YPD contains ∼2 mg/liter riboflavin. For some experiments, YPD media were enriched by addition of riboflavin. The concentration of riboflavin in standard minimal medium is 0.2 mg/liter. To create minimal media with lower riboflavin concentrations, vitamin-free yeast nitrogen base without amino acids (BIO101) was supplemented with all vitamins except riboflavin, which was added from a 200 mg/liter stock solution. For media containing l-proline as the sole source of nitrogen, yeast nitrogen base without amino acids without ammonium sulfate (BIO 101) was supplemented with 0.85% l-proline. As a rule, only the required amino acids and nucleobases (adenine, histidin, methionine, tryptophan, and uracil: 20 mg/liter; lysine and leucine: 30 mg/liter) were added to minimal media. Media were solidified with 2% Difco bacto agar.
Isolation of ribΔ* suppressor mutants:
To isolate ribΔ* suppressor mutants and to determine their frequency, 100 μl of a suspension of S. cerevisiae cells (OD600 = 0.1) were plated on YPD and the plate was incubated for 6 days at 30°. To determine the total amount of cells, the cell suspension was further diluted to OD600 = 0.0001 and 100 μl were plated on YPD containing 200 mg/liter riboflavin. The frequency of ribΔ* suppressor mutants was calculated as the number of colonies per 105 cells plated. ribΔ* mutants were purified by streaking on YPD followed by streaking on minimal medium containing 1 mg/liter riboflavin.
Plasmids:
YCplac33 PUT1-ZZ was generated by PCR amplification of the PUT1 ORF including 425 bp of promoter sequence, which included the Put3 binding site (−308 CGGCAATGGCTTTCCG −293) and lacked a stop codon. The product was digested with PstI (partial digest to avoid cleavage of the internal site) and BamHI and ligated into the same sites of the centromeric plasmid YCplac33-ZZ. This vector, based on YCplac33 (Gietz and Sugino 1988), extends the PUT1 ORF with two copies of the IgG binding domain of Staphylococcus aureus protein A, which is followed by a stop codon and the S. cerevisiae ADH1 terminator.
The plasmid to overexpress PUT3 was based on pRS426 (Sikorski and Hieter 1989), into which a PCR fragment of genomic DNA including 236 bp of promoter and 275 bp of terminator sequence was ligated with blunt ends into the SmaI site.
To create a plasmid for the fluorimetric determination of the activity of the MCH5 promoter, the entire promoter (−828 to −1 bp) was amplified from BY4741 genomic DNA, cut at the primer-encoded PstI and BamHI sites, and ligated into the multicopy plasmid YEplac195-GFP, which is a derivative of YEplac195 (Gietz and Sugino 1988). In this vector, the MCH5 promoter drives the expression of a soluble form of GFP, whose translation is initiated at an ATG codon present in the primer to amplify the promoter. GFP starts with the amino acids MGSGRVGAGAGASKGEE (N-terminal extension underlined) and is followed by an ADH1 terminator. The Put3 binding site within the promoter fragment (−501 CGGGGGTGGCTTCCCGA −486) was replaced by the sequence TAATTGAAGCTTCTTCT to produce plasmid YEplac195-MCH5prom mut-GFP.
Alternatively, we used a reporter plasmid that contained only the ORF-proximal Put3 binding site of MCH5 in the UAS-less MEL1 promoter from pMEL-β2 (Melcher et al. 2000). This plasmid was constructed in two steps. First, the MEL1 minimal promoter containing a fragment of the VHT1 promoter (Pirner and Stolz 2006) was ligated into the PstI and BamHI sites of YEplac195-GFP. Next, the VHT1 fragment was replaced with a PCR product containing the Put3 binding site (−501 to −486 bp relative to the start ATG of MCH5) along with 24 bp of upstream and 23 bp of downstream sequence. In the final product, the Put3 binding site lies in position −281 to −266 relative to the start ATG of GFP. A similar fragment amplified from YEplac195-MCH5prom mut-GFP was also used to replace the fragment containing VHT1 sequences.
For sequencing, genomic DNA from rib5Δ*1 and rib5Δ*3 was used to amplify PUT3 in two independent PCR reactions with high-fidelity Phusion DNA polymerase (New England Biolabs). The 3896-bp PCR products, which extended into the coding regions of the neighboring genes ATP7 and URB1, were ligated with blunt ends into the SmaI site of pBluescript and sequenced.
Mch5 antiserum and Western blots:
For immunization of rabbits, amino acids 3–106 from the hydrophilic N terminus of Mch5 were fused to the maltose-binding protein and produced as a soluble protein from vector pMAL-c2X in E. coli BL21(DE3) cells. The fusion protein was purified using amylose columns (New England Biolabs) and injected in rabbits in 2-week intervals until the serum detected specific signals in yeast cell extracts (Pineda Antikörper Service, Berlin). The serum was purified using a Mch5-GST fusion protein, which was produced from vector pGEX-2TK and contained the same N-terminal amino acids of Mch5. The Mch5-GST fusion was soluble after expression in E. coli and was affinity purified using glutathione sepharose 4B (GE Healthcare). Following purification it was immobilized on cyanogen bromide activated CH-sepharose 4B (GE Healthcare). Rabbit serum was diluted with PBS, loaded on the affinity column, and Mch5-specific antibodies were eluted with 200 mm glycine/HCl pH 2.9, followed by neutralization and dialysis against PBS.
Western blots were performed by transfer of gel-separated proteins to a nitrocellulose membrane, which was blocked and incubated with the primary and secondary antibodies as required. Put1-ZZ was detected in total cell extracts prepared by shaking 5 OD600 units of cells with glass beads in 100 μl SDS sample buffer in a FastPrep instrument (BIO 101). After heating the samples (2 min, 95°) and a brief centrifugation, 10 μl were loaded per lane. Mch5 was detected in total membrane preparations generated from 35 OD600 units of cells by centrifugation for 20 min at 20,000 × g. The membrane pellet was resuspended in 50 μl of 25 mm Tris/HCl, 5 mm EDTA, pH 7.5, diluted with SDS sample buffer, heated (2 min, 42°), and 10 μl were used per lane. After transfer, the nitrocellulose membrane was sequentially incubated with primary [rabbit polyclonal anti-Mch5 or rabbit polyclonal anti-HA (Santa Cruz, sc-805)] and peroxidase-linked secondary antibodies (Sigma A-6154) and developed with chemiluminescence reagents.
GFP as a reporter of promoter activity:
S. cerevisiae cells containing reporter plasmids were grown in SD media containing 20 mg/liter riboflavin, diluted in media with 0.2 mg/liter riboflavin, and grown to log phase. The cells were washed twice with water and resuspended in water for OD600 ≈ 0.1 in a 2.5 ml plastic cuvette. The fluorescence of the cells was measured in a Spex Industries FluoroMax-2 fluorescence spectrophotometer (excitation wavelength 488 nm, emission wavelength 512 nm) and corrected by subtracting the signal of cells lacking a GFP reporter plasmid. In parallel, the OD600 of the cell suspension was determined. Relative fluorescence units were calculated by dividing the corrected emission signal by the number of cells present in the cuvette assuming that a cell suspension of OD600 = 0.1 contains 106 cells/ml. Most measurements were performed in triplicate with three independent yeast cultures for calculation of means (represented by bars) and the standard deviation (represented by error bars). Values from single measurements are shown as columns lacking error bars.
Determination of proline:
S. cerevisiae cells were grown to exponential phase, washed with water and 50 OD600 units of cells were frozen and stored at −80°. After thawing, the cells were suspended in 100 μl water and lysed with glass beads. The lysate was transferred to a fresh tube, the glass beads were washed with 2 × 100 μl water, and the fractions were pooled. After centrifugation to separate soluble from insoluble material, 40 μl of the supernatant were labeled with iTRAQ reagents (AA 45/32 kit, Applied Biosystems) as recommended by the manufacturer and analyzed on an Applied Biosystems 3200 Q TRAP LC/MS/MS system equipped with a RP-C18-column (150 mm length, 4.6 mm diameter, 5 μm particle size). The pellets containing insoluble cell components were dried at 60° and weighed. Proline concentrations are reported in micromole/gram insoluble cell material.
Gel-shift assays:
Oligonucleotide Mch5-1 (ctcgagACAACGCACCTGTTATTATGATCTCGGGGGTGGCTTCCCGAACATCGTCGGTTGAATACTGCGgtcgag), putative Put3 binding site underlined, residues identical to the MCH5 promoter [(nt −525 to nt −463) in uppercase] was purchased in a Cy5 labeled and unlabeled version and annealed to a complementary oligonucleotide. The annealed DNA was separated from single-stranded oligonucleotides by 12% polyacrylamide gel electrophoresis at 4°, excised from the gel, extracted in 10 mm EDTA 100 mm Tris/HCl, pH 7.5 overnight at 37°, and adjusted to 1–2 ng/μl with the same buffer. For protein extracts, 100 OD600 units of cells were resuspended in 25 mm Tris/HCl, pH 7.5, 5 mm EDTA, 1 mm PMSF and lysed with glass beads in a FastPrep instrument. Insoluble material was removed by centrifugation and the supernatant was stored in liquid nitrogen. Binding assays were performed by incubating 5 μl protein extract and 2 μl 5× gel binding buffer [20% glycerol, 5 mm MgCl2, 2.5 mm EDTA, 2.5 mm DTT, 250 mm NaCl, 0.25 mg/mg poly(dI-dC), and 50 mm Tris/HCl (pH 7.5)] for 10 min on ice. Then, 1 μl of Cy5-labeled ds-DNA (1 ng/μl) and 2 μl H2O were added and incubated for 20 min at 25°. Competition experiments contained 2 μl of unlabeled ds-DNA (2 ng/μl) for a fourfold excess instead of H2O. Binding reactions were separated on a 5.3% acrylamide gel at 4°. The gel was scanned with a Typhoon Trio+ imager (GE Healthcare) and data were loaded into Adobe Photoshop software for contrast enhancement and noise reduction.
RESULTS
Suppressor mutants do not bypass individual steps of riboflavin biosynthesis:
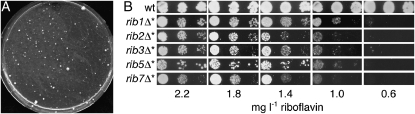
S. cerevisiae strains with deletions that disable riboflavin biosynthesis (ribΔ strains) frequently segregate colonies with improved growth (Lesuisse et al. 2005; Reihl and Stolz 2005). These extragenic suppressor mutants, which we will refer to as ribΔ* mutants, appeared as colonies of variable sizes on YPD plates, where the majority of the cells were unable to grow (Figure 1A). These suppressed mutants occurred at an exceptionally high frequency in various strain backgrounds (∼10−3–10−4; Table 1).
Figure 1.—
Isolation and analysis of suppressor mutants. (A) Approximately 105 cells of the BY4742 rib5Δ deletion strain were plated on YPD and incubated for 6 days at 30°. Whereas the majority of the cells are unable to grow, some spontaneous suppressor mutants form colonies of various sizes. Similar analyses were performed for other strains and used to calculate the frequencies presented in Table 1. (B) Growth of purified rib5Δ* suppressor mutants on minimal medium containing the indicated concentrations of riboflavin. A riboflavin prototrophic wild-type strain (BY4742) was used as control. Nonsuppressed rib5Δ deletion strains require 20 mg/liter riboflavin (see Figure 3B).
TABLE 1.
Frequency of ribΔ* mutants in different strains
| Relevant genotype | Suppressor mutants per 105 cells |
|---|---|
| BY4741 rib1Δa | 32 ± 6 |
| BY4742 rib1Δa | 11 ± 4 |
| rib2Δ | 28 ± 9 |
| rib3Δ | 17 ± 7 |
| BY4741 rib5Δ | 22 ± 9 |
| BY4742 rib5Δ | 31 ± 4 |
| rib7Δ | 187 ± 75 |
| rib1Δ rib5Δ | 61 ± 16 |
| rib2Δ rib5Δ | 32 ± 5 |
| rib3Δ rib5Δ | 129 ± 16 |
| rib7Δ rib5Δ | 62 ± 23 |
| rib5Δ/rib5Δ diploid | 140 ± 26 |
| W303-1A rib5Δ | 102 ± 21 |
| W303-1B rib5Δ | 144 ± 33 |
| BY4742 rib5Δ mch1Δ | 16 ± 5 |
| BY4742 rib5Δ mch3Δ | 66 ± 7 |
| BY4742 rib5Δ mch4Δ | 173 ± 33 |
| BY4742 rib5Δ mch5Δ | 0b |
| CEN.PK rib5Δ | 112 ± 21 |
| CEN.PK rib5Δ mch5Δ | 0b |
| CEN.PK rib5Δ (mch1-5)Δ | 0b |
| BY4742 rib5Δ put3Δ | 14 ± 8 |
| BY4742 rib5Δ*1/BY4741 rib5Δ diploidc | 620 ± 72 |
| BY4742 rib5Δ*3/BY4741 rib5Δ diploidc | 67 ± 11 |
A suspension containing ∼500,000 S. cerevisiae cells of the given genotype were plated on five YPD plates without additional riboflavin. All colonies visible after 6 days at 30° were scored. Total cell counts were determined on YPD plates with added 200 mg/liter riboflavin. The values are means ± SD from five plates.
Single or double mutants denoted BY4741 or BY4742 were derived from the respective parental strains by transformation with knockout cassettes. Double mutants lacking this designation were derived from BY4743 heterozygous diploids or from mating of deletion mutants in BY4741 and BY4742, followed by isolation of haploid progeny.
The frequency of suppressor mutations is <5 × 10−5.
Obtained by crossing.
To get the first idea of the molecular basis of this suppression, we analyzed a panel of different riboflavin biosynthetic mutants from the BY strain background. With the exception of rib4Δ, which is prototrophic for riboflavin (Kis et al. 2001; Reihl and Stolz 2005), all available ribΔ mutants were assayed. Most of the ribΔ strains segregated mutants with a frequency of 1–3 × 10−4 and the suppressors appeared with even higher frequency in rib7Δ (Table 1). This similar incidence of mutants in strains with different blocks in riboflavin biosynthesis made it unlikely that suppression was caused by reactions that bypass individual steps of riboflavin biosynthesis. This conclusion was supported by the analysis of double mutants that combined a deletion of RIB5, disabling the last step of riboflavin biosynthesis, with deletions abrogating earlier steps. The double knockouts segregated suppressed strains at similar rates as the single knockouts (Table 1). Taken together, a bypass of the riboflavin biosynthetic pathway is unlikely to account for the better growth of suppressed strains.
We also analyzed the ribΔ* mutants for growth on minimal media containing defined amounts of riboflavin. Whereas wild-type cells grew on all plates, the suppressed mutants isolated from different ribΔ backgrounds showed no growth unless 1.0 mg/liter riboflavin were present (Figure 1B). Nonsuppressed rib5Δ cells required a riboflavin supplement of 20 mg/liter for growth (Reihl and Stolz 2005), (also see Figure 3B). Thus, the ribΔ* mutants were riboflavin auxotrophs like their parents, but they possessed a reduced riboflavin requirement. We additionally found that suppressed strains were recovered with similar frequencies from haploid and homozygous diploid rib5Δ mutants (Table 1). At first sight, this indicated that suppression was caused by a dominant mutation. However, when we crossed two individual strains (rib5Δ*1 and rib5Δ*3) to a nonsuppressed rib5Δ mutant, we found that the resulting diploids produced suppressed colonies at largely different rates (Table 1). In the diploid deriving from rib5Δ*3, the frequency was similar to the spontaneous rate of suppressor mutations, indicating that rib5Δ*3 carried a recessive mutation. In the diploid deriving from rib5Δ*1, the incidence of suppressed colonies was 10-fold higher but much lower than expected for a dominant mutation. The reason for this is not clear but it may indicate that the suppressor mutations in rib5Δ*1 and rib5Δ*3 are nonallelic. Thus, multiple genes may exist that when mutated give rise to the same phenotype and this may also account for the high frequency with which ribΔ* mutants occur.
Suppressor mutants overexpress the riboflavin transporter MCH5:
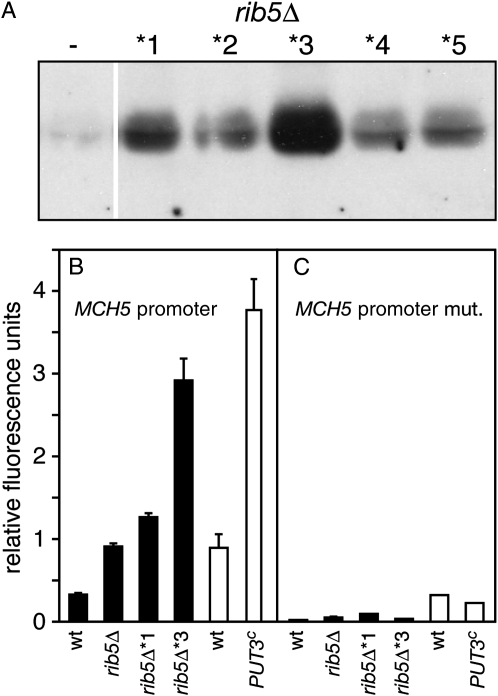
The results presented above indicated that suppression of ribΔ mutants was not caused by a mechanism that created an alternative pathway for riboflavin biosynthesis. We thus turned our attention to the plasma-membrane riboflavin transporter encoded by MCH5, which provides a second route to secure intracellular riboflavin. To analyze if the amount of Mch5 was changed, we prepared membrane protein extracts from rib5Δ and from five independently isolated suppressor mutants and analyzed them by Western blotting (Figure 2A). This showed that although the abundance of Mch5 was variable between individual rib5Δ* mutants, all had elevated levels of Mch5 when compared to a nonsuppressed rib5Δ strain (Figure 2A).
Figure 2.—
Suppressor mutants overexpress MCH5 and have increased amounts of Mch5. (A) Five independently isolated ribΔ* suppressor mutants were analyzed by Western blotting for the relative abundance of Mch5. A nonsuppressed rib5Δ strain was used as a control. (B) A reporter construct containing the full-length MCH5 promoter fused to GFP was transformed into BY4742-based strains (solid bars) or into MB758-5B (wt) or an isogenic PUT3c strain (open bars). Prior to the measurement, the cells were grown in media containing 0.2 mg/liter riboflavin for 6 hr and relative fluorescence units were determined as described in materials and methods. (C) The same strains used in B carrying a similar plasmid lacking the Put3 binding site were subjected to GFP reporter assays.
To analyze if this increased amount of Mch5 was caused by an increased expression of the MCH5 gene, we generated a reporter plasmid in which the promoter of MCH5 was fused to GFP. In intact cells carrying this plasmid, the activity of the MCH5 promoter could be determined by fluorescence measurements. Using lacZ as a reporter we previously demonstrated that MCH5 is more strongly expressed in rib5Δ cells (Reihl and Stolz 2005). Measurements with a rib5Δ strain carrying the GFP reporter plasmid indicated a 2.8-fold induction relative to wild-type levels (Figure 2B), demonstrating that the GFP reporter is functional.
Fluorescence measurements were repeated with rib5Δ*1 and rib5Δ*3 that possessed the highest levels of Mch5 in the Western analysis (Figure 2A). Both mutants also showed increased levels of GFP relative to rib5Δ and, similar to the Western analysis, rib5Δ*1 had lower levels of activity than rib5Δ*3 (Figure 2B). Thus, the rib5Δ* mutants appear to overexpress MCH5. To check if MCH5 is directly involved in the development of the suppressor phenotype, we analyzed if suppressor mutants can be derived from rib5Δ mch5Δ. Contrary to preliminary findings (Reihl and Stolz 2005), we were not able to isolate suppressor mutants from this strain. These experiments were performed in the BY and CEN.PK genetic backgrounds (Table 1) but neither led to the identification of a single suppressed mutant. However, suppressed mutants appeared at normal rates when other MCH genes, that do not encode riboflavin transporters (Reihl and Stolz 2005), were deleted in addition to RIB5. Moreover, a strain lacking all MCH genes and RIB5, similar to a strain lacking only MCH5 and RIB5, did not segregate suppressed colonies (Table 1). Thus, MCH5 is necessary for the development of the suppressor phenotype.
Proline induces the expression of MCH5:
The promoter of MCH5 contains two binding sites for the transcription factor Put3. Whereas the ORF-proximal site (−501 to −486) is well conserved in different Saccharomyces species (Figure 3A), the distant site (−1060 to −1045) is less conserved. A genomewide screen found Put3 bound to both sites in vivo (Harbison et al. 2004). Put3 also binds to the promoters of PUT1 and PUT2, the structural genes encoding proline oxidase and Δ1-pyrroline-5-carboxylate dehydrogenase, respectively, and increases their expression when the inducer proline is present (des Etages et al. 1996). Together, Put1 and Put2 convert proline to glutamate and are essential for growth when proline is the sole source of nitrogen. Because PUT1 and PUT2 are not sufficiently expressed in its absence, PUT3 is also essential for growth when proline is the sole nitrogen source (Brandriss 1987).
To test if the Put3 binding site is functional in regulating MCH5, we made use of the fact that rib5Δ mutants show better growth on riboflavin-limited plates when MCH5 is overexpressed (Reihl and Stolz 2005). Indeed, growth of rib5Δ cells required 20 mg/liter riboflavin on ammonium plates but only 0.2 mg/liter riboflavin on proline (Figure 3B), indicating that the expression of MCH5 is induced by proline. Similar to these observations, also rib2Δ, rib3Δ, rib4Δ, and rib7Δ mutants had a drastically reduced riboflavin requirement when the plates contained proline instead of ammonium (data not shown). These results were confirmed by Western blots that detected much more Mch5 in membrane protein extracts from proline-grown wild-type cells (Figure 3C). As expected, Put1 was also more abundant when cells were grown in proline rather than ammonium (Figure 3C).
Since PUT3 is required for the growth on proline, we could not use put3Δ strains to demonstrate that Put3 is necessary for the proline-induced expression of MCH5. However, we constructed a plasmid containing the ORF-proximal Put3 binding site of the MCH5 promoter to validate that the proline-induced expression of MCH5 requires Put3. This construct is based on the minimal promoter of MEL1 and is followed by the GFP reporter. Cells containing this plasmid had a 60-fold increased expression of GFP in proline relative to ammonium (Figure 3D). The strong induction of MCH5 by proline is comparable to PUT1, which is induced 50-fold when proline is present (Wang and Brandriss 1987) and proves that the Put3 binding site mediates proline regulation of MCH5. These findings are also supported by transcriptome analyses in which increased expression of MCH5 was found in proline-grown cells (Boer et al. 2007; Godard et al. 2007).
To further support these data, we constructed strains with deletions of the PUT3 gene and assayed their growth on YPD plates to which different amounts of riboflavin were added. On all plates, put3Δ was indistinguishable from wild type. rib5Δ possessed an increased requirement for riboflavin but showed full growth with riboflavin additions ≥20 mg/liter. In contrast, rib5Δ put3Δ was similar to rib5Δ mch5Δ. Both produced only small colonies with 20 mg/liter added riboflavin and required 200 mg/liter added riboflavin for full growth (Figure 3E). Together, these experiments confirm that Put3 is necessary for the expression of MCH5 and that loss of either protein reduces growth on limiting riboflavin concentrations. In summary, the expression of the MCH5 gene is strongly increased by proline and this depends on the transcription factor Put3.
Put3 binds to the promoter of MCH5 in vitro and the binding site is necessary and sufficient for the increased expression in ribΔ* mutants:
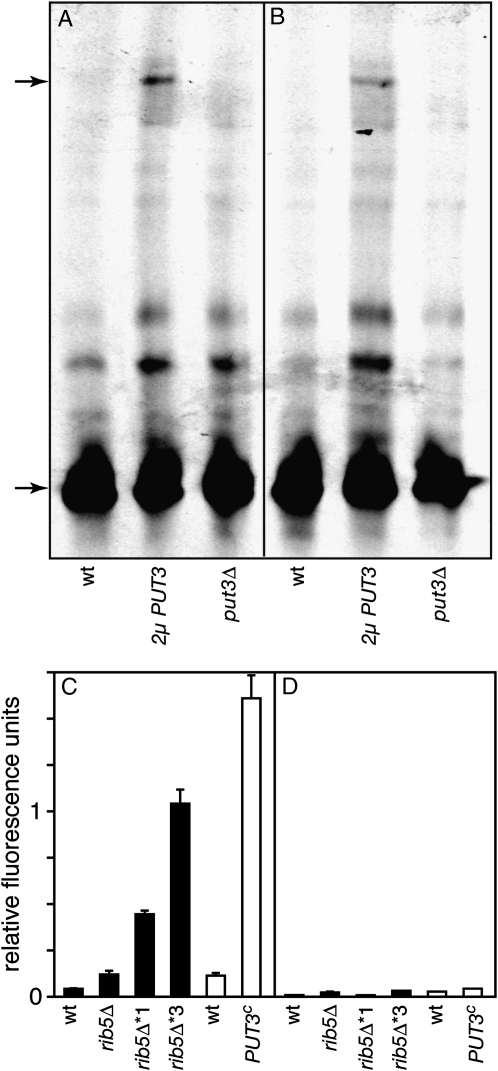
We next addressed if the increased expression of MCH5 in ribΔ* strains is mediated by Put3. To this end, we performed gel-shift assays with a fluorescently labeled DNA probe containing the Put3 binding site from the MCH5 promoter. The DNA fragment was incubated with extracts from ammonium-grown wild-type cells, put3Δ mutants or from wild-type cells that carried PUT3 on a multicopy plasmid. The extracts from PUT3 overexpressing cells led to the formation of a protein-DNA complex with drastically reduced gel mobility that was undetectable in the other extracts (Figure 4A). Moreover, the intensity of this band decreased in the presence of a fourfold excess of the unlabeled probe (Figure 4B). Visualization of this binding event required neither cultivation of the cells in proline nor the addition of proline to the binding assay. However, overexpression of PUT3 was required, indicating that wild-type cells contain only little free Put3. In conclusion, Put3 binds to a fragment of the MCH5 promoter containing the conserved Put3 binding site in vitro.
Figure 4.—
Put3 binds to the promoter of MCH5 and deletion of the binding site reduces its expression. (A and B) A double-stranded, Cy5-labeled DNA probe covering the Put3-binding site of the MCH5 promoter was incubated with protein extracts from ammonium-grown cells. As indicated, these cells were either wild type, overexpressing PUT3 from a multicopy plasmid (2μ PUT3), or put3Δ mutants. The binding reactions were analyzed by nondenaturing polyacrylamid gel electrophoresis, followed by fluorescence scanning. In B, a fourfold excess of the unlabeled probe was present during the binding assay. The positions of the free probes and of the bands appearing in cells overexpressing PUT3 are marked with arrows. (C) A GFP reporter construct containing the Put3 binding site from the MCH5 promoter in a MEL1 minimal promoter was transformed into BY4742-based strains (solid bars), or into MB758-5B (wt), or an isogenic PUT3c strain (open bars). The cells were grown in media containing 0.2 mg/liter riboflavin for 6 hr and GFP fluorescence was determined as described in materials and methods. (D) The same strains used in C carrying a similar plasmid lacking the Put3 binding site were subjected to GFP reporter assays.
To analyze the effect of Put3 binding on the transcriptional activity of MCH5, we deleted the Put3 binding site from the full-length MCH5 promoter and repeated the GFP reporter assays. Upon deletion, the increased activity observed in rib5Δ and in the two rib5Δ* mutants was nullified (Figure 2C). Deletion of the Put3 binding site also reduced the promoter activity in wild-type cells. Thus, similar to PUT1 and PUT2 (Siddiqui and Brandriss 1989), Put3 appears to be necessary for the basal expression of MCH5. Similar results were obtained with the MEL1 minimal promoter containing the conserved Put3 site from the MCH5 promoter (Figure 4, C and D). In conclusion, Put3 binds to the conserved site in the MCH5 promoter and this appears to be required for the improved growth of ribΔ* mutants.
The suppressor phenotype is caused by mutations that activate Put3:
Consistent with the above results, we found that a rib5Δ put3Δ strain produced fewer suppressor mutants than any of the other rib5Δ strains analyzed except rib5Δ mch5Δ (Table 1). This demonstrates that while many suppressor mutations operate by activating Put3, other pathways exist that lead to suppression of rib5Δ.
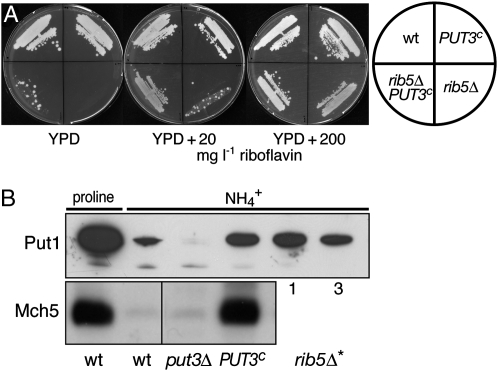
Put3 is constitutively bound to DNA and becomes activated in the presence of proline (Axelrod et al. 1991; Sellick and Reece 2003). Binding of proline leads to a conformational change that is thought to increase the ability of Put3 to function as a transcriptional activator. Several dominant mutations in PUT3 that cause constitutive activation of Put3 and increased expression of its target genes PUT1 and PUT2 in noninducing ammonium-containing media have been isolated and designated PUT3c (Brandriss and Magasanik 1979; Marczak and Brandriss 1989, 1991). We deleted RIB5 in a PUT3c strain to compare the growth properties of the double mutant to rib5Δ and PUT3c single mutants. As expected, PUT3c was similar to wild type and was able to grow without added riboflavin. The rib5Δ deletion strain was unable to grow on YPD, displayed numerous faster growing colonies (likely spontaneous suppressor mutants) on YPD plus 20 mg/liter riboflavin, and showed full growth when more riboflavin was present (Figure 5A). On all plates used, rib5Δ PUT3c showed improved growth relative to rib5Δ cells. Thus, the growth properties of rib5Δ PUT3c are similar to rib5Δ*.
Figure 5.—
Similarity of PUT3c and rib5Δ* mutants. (A) Cells of the indicated genotype were streaked on YPD plates containing various amounts of added riboflavin. Growth was recorded after 5 days at 30°. (B) Cells of the indicated genotype were grown in media containing proline or ammonium as the sole nitrogen source. Put1 was detected in total cell extracts using the C-terminally fused ZZ tag and Mch5 was detected with the specific antiserum. For details, see legend to Figure 3C; for expression of MCH5 in rib5Δ*1 and rib5Δ*3, see Figure 2A.
When transformed with reporter plasmids containing either the full-length MCH5 promoter (Figure 2B) or only the Put3 binding site from MCH5 (Figure 4A), the PUT3c strain displayed much higher levels of GFP fluorescence than an isogenic strain with a normal PUT3 allele. For both types of reporter plasmids, the increased fluorescence in PUT3c was abolished after mutation of the Put3 binding site (Figures 2C and 4B).
To find independent proof that the Put3 transcription factor is hyperactive in rib5Δ* mutants we analyzed the expression of the Put3 target PUT1 in Western blots. Whereas no signal was obtained in put3Δ strains, PUT1 was weakly expressed in wild-type strains grown in ammonium media and highly expressed when proline was the nitrogen source. The expression of PUT1 in ammonium media was increased in the PUT3c strain and a similar increase was also seen in rib5Δ*1 and rib5Δ*3 suppressor mutants (Figure 5B). Membrane protein extracts from the same cells were also analyzed for Mch5, whose abundance was increased in proline-grown wild-type cells and PUT3c cells grown in ammonium (Figure 5B). We have shown before that rib5Δ*1 and rib5Δ*3 cells possessed increased levels of Mch5 relative to wild-type cells (Figure 2A). Taken together, rib5Δ* strains are similar to strains that carry a hyperactive allele of PUT3. Both overexpress MCH5, which leads to improved growth of riboflavin auxotrophic mutants.
Reasons for the increased activity of Put3:
Many reasons could account for the increased activity of Put3 in rib5Δ* suppressor mutants. The phenotypic similarity of rib5Δ* suppressor mutants to PUT3c mutants prompted us to first look for mutations in the PUT3 gene from rib5Δ*1 and rib5Δ*3. However, the sequence reads were fully identical with the published sequence of this region.
A priori, overproduction of Put3 alone could also lead to a suppressor phenotype. However, this explanation was unlikely because we did not find PUT3 as a multicopy suppressor of rib4Δ and rib5Δ mutants in the genetic screen in which 14 MCH5-containing plasmids were isolated (Reihl and Stolz 2005). Indeed, Western blots to detect Put3 expressed from a genomically integrated tagged allele (PUT3-3HA) demonstrated equal levels of Put3 in wild-type cells, rib5Δ, rib5Δ*1, and rib5Δ*3 (data not shown).
Since the activity of Put3 requires proline as an inducer, we also analyzed if the cellular proline content was increased. This experiment was only performed with rib5Δ*3 because the rib5Δ*1 suppressor mutant used in the above experiments was genetically unstable and had lost some of its characteristics. While still able to grow on YPD without added riboflavin, it had lost the elevated expression of MCH5 and of GFP reporter plasmids. This likely originated from the accumulation of additional unknown mutations. The phenotype of rib5Δ*3, in contrast, was stable and we found that these cells contained more than twofold increased proline levels compared to wild-type cells. Relative to rib5Δ, the proline content was >3.5-fold higher in rib5Δ*3 (Table 2). Thus, the higher activity of Put3 in rib5Δ*3 was likely due to its higher proline content, which led to an increased expression of MCH5. The finding that a proline-accumulating S. cerevisiae mutant (put1-54 pro1-D154N, a gift from Hiroshi Takagi) also displayed elevated levels of Mch5 in Western blots (data not shown) makes this hypothesis all the more appealing.
TABLE 2.
Intracellular proline concentrations in different strains
| Genotype | Proline concentration (μmol/gram) | Proline concentration (%) |
|---|---|---|
| Wild type | 22.4 ± 8.8 | 100 |
| rib5Δ | 14.7 ± 1.5 | 65.6 |
| rib5Δ*3 | 52.0 ± 8.8 | 232a |
S. cerevisiae strains were grown in media containing 20 mg/liter riboflavin and 100 OD600 units of cells were used for the determination of the cellular proline concentrations. Values are reported as micromole proline per gram of insoluble cell material, which remained after lysis and centrifugation of the cells and mostly consists of cell walls.
Significant (P < 0.05, t-test) increase relative to the proline content of rib5Δ cells.
The analysis of suppressor mutants that spontaneously arise in yeast strains with defects in riboflavin biosynthesis thus led to the surprising finding that the expression of the riboflavin transporter gene MCH5 is regulated by proline via the transcription factor Put3. Whereas MCH5 is normally not highly expressed in ammonium-containing media, its expression is increased by unknown mutations that activate the transcription factor Put3. These mutations do not lie within PUT3 but at least some of them seem to raise the intracellular concentration of proline, which is the inducer of Put3. Further experiments will be necessary to identify the mechanism of proline accumulation in rib5Δ*3 and to resolve the basis of suppression in other mutants.
DISCUSSION
The S. cerevisiae Put3 protein is a dimeric Zn2 Cys6 transcription factor that is required both for the basal as well as the proline-induced transcription of PUT1 and PUT2 (Marczak and Brandriss 1991; Todd and Andrianopoulos 1997; Huang and Brandriss 2000; Sellick and Reece 2005). The N-terminal domain of Put3 binds to the DNA motif CGGNANGCNANNNCCGA in vivo and in vitro and independent of the presence or absence of proline (Todd and Andrianopoulos 1997). This domain is followed by a coiled-coil domain that mediates dimer formation and a C-terminal acidic domain that acts as a transcriptional transactivator (des Etages et al. 1996; Swaminathan et al. 1997; Walters et al. 1997). Binding of proline to Put3 leads to a conformational change that is thought to expose the transactivation domain (Axelrod et al. 1991; des Etages et al. 1996, 2001). Binding sites for Put3 are found in the promoters of PUT1 and PUT2, encoding enzymes involved in proline degradation, but also in MCH5, the S. cerevisiae gene encoding the plasma-membrane transporter for riboflavin. Riboflavin-auxotrophic S. cervisiae mutants frequently segregate suppressed strains with improved growth on ammonium-containing media. The frequency of suppressor mutants is independent of the underlying defect in the riboflavin biosynthetic pathway, reduced in cells lacking PUT3, and further reduced in strains lacking MCH5. On ammonium-containing media, Mch5 and Put1 were more abundant in suppressed strains and in a mutant carrying a constitutively active (proline-independent) allele of PUT3. These assays were confirmed by reporter assays using the full-length MCH5 promoter or fragments thereof harboring the Put3 binding site. Moreover, we demonstrate that Put3 binds to a fragment of the MCH5 promoter in vitro. In summary, this led to the finding that the riboflavin transporter gene MCH5 is regulated by Put3.
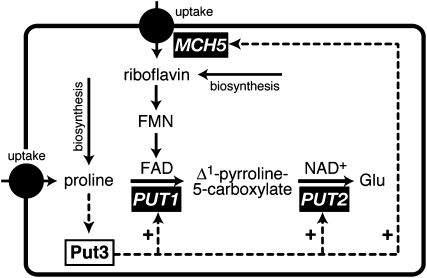
Most yeast media formulations contain ammonium ions as these are the preferred nitrogen source of S. cerevisiae (Godard et al. 2007). In their natural habitats (e.g., on grapes), yeasts mostly encounter the amino acids arginine and proline, which are poorer sources of nitrogen (des Etages et al. 1996; Boer et al. 2007; Godard et al. 2007). We have demonstrated above that proline strongly increases the expression of the riboflavin transporter gene MCH5 and that this effect is mediated by Put3. In batch cultures, no other genes besides PUT1, PUT2, and MCH5 are significantly induced by proline, whereas in chemostat cultures, a group of 26 genes that also includes MCH5 is induced (Boer et al. 2007; Godard et al. 2007). Why are riboflavin transport and proline degradation coregulated processes? PUT1 encodes proline oxidase, an enzyme that creates Δ1-pyrroline 5-carboxylate, which is further metabolized by Put2 to produce glutamate (Figure 6). Proline oxidase is localized to the inner mitochondrial membrane and contains the cofactor FAD, which derives from riboflavin. To increase proline oxidase activity, the cells have to increase the production of the Put1 apoprotein and also provide more FAD to generate the enzymatically active holoform of Put1. The data presented here suggest that the extra riboflavin required for the activity of Put1 derives from uptake via Mch5 and not from biosynthesis. Possibly, proline and riboflavin occur together in the natural habitats of yeast, making it highly economical to use external sources of riboflavin to facilitate proline degradation (Figure 6). Thus, the increase in proline oxidase activity is coordinated by Put3, which synchronizes the formation of apo-proline oxidase with the formation of the coenzyme FAD. However, even when proline is used as the sole source of nitrogen, phenotypes associated with a lack of MCH5 can only be observed in rib mutants. The likely reason for this is that, in laboratory growth conditions, intracellular riboflavin largely derives from biosynthesis. We speculate, however, that the Put3-mediated control of MCH5, PUT1, and PUT2 may have biological significance for wild-type cells in natural habitats. The fact that regulation of MCH5 by proline can be observed in wild-type cells (Figures 3C and 5B) and that the Put3 binding site in the MCH5 promoter is evolutionary conserved (Figure 3A) suggests that it is more than an esoteric twist of nature.
Figure 6.—
Regulation of proline degradation in Saccharomyces cerevisiae. The scheme illustrates the pathway of proline catabolism in S. cerevisiae. Intracellular proline activates the transcription factor Put3, which acts as a positive regulator (dashed arrows) of the proline catabolic genes PUT1 (encoding proline oxidase) and PUT2 (encoding Δ1-pyrroline-5-carboxylate dehydrogenase). This work demonstrates that Put3 also regulates MCH5, the plasma-membrane riboflavin transporter. Intracellular riboflavin is necessary for the generation of FAD, the catalytic cofactor required by Put1. For clarity, details of the subcellular compartmentation of these pathways were omitted, as were the pathways of proline and riboflavin biosynthesis. At least four amino acid permeases contribute to proline uptake in S. cerevisiae (Andreasson et al. 2004).
In the absence of MCH5, no suppressed strains could be isolated. Thus, activation of MCH5 appears to be the basis for the suppressor mutants that develop in riboflavin auxotrophic strains. The frequency of suppressor mutants was reduced in the absence of Put3, indicating that many suppression events were caused by the aberrant activity of Put3 in ammonium-containing media. For one of the suppressor mutants we found that the cells contained increased levels of proline, a known inducer of Put3. Accumulation of proline, for example, can be caused by dominant alleles of PRO1. This gene encodes the first enzyme in the proline biosynthetic pathway and the mutated form of the protein is insensitive to feedback inhibition by proline (Sekine et al. 2007). In other suppressed strains, however, other mutations may activate Put3. These mutations may cause proline-independent activity of Put3 (similar to PUT3c), alter the phosphorylation of Put3, or lead to the accumulation of other metabolites that activate Put3 (Sellick and Reece 2003). Put3 is hyperphosphorylated in the presence of poor nitrogen sources and full activation requires both the presence of the inducer proline and the absence of good nitrogen sources (Huang and Brandriss 2000). Consistent with this, PUT1 and PUT2 are much higher expressed in nitrogen starvation (Godard et al. 2007), or in the presence of rapamycin (Saxena et al. 2003), conditions that do not affect the expression of PUT3 (Axelrod et al. 1991; des Etages et al. 2001). The pathways that govern the phosphorylation of Put3 are not resolved, but mutations in these might lead to overexpression of MCH5.
Whatever the reason for the suppression in individual strains might be, further work with the suppressor mutants is difficult and hampered by their genetic instability. This fact, as well as the initial observation that suppressor mutants arise in all ribΔ strains with an extraordinary high frequency, might be related to an additional function served by the riboflavin biosynthetic pathway. It is known for E. coli that riboflavin biosynthesis contributes to the removal of 8-oxo-GTP, a spontaneous oxidation product of GTP, which is a mutagenic substrate for DNA synthesis, and that the expression level of ribA (encoding GTP cyclohydrolase II, the first enzyme of the riboflavin biosynthetic pathway), correlates with the rate of spontaneous mutations (Kobayashi et al. 1998). Although yeasts, like other organisms, have multiple pathways to sanitize oxidized nucleotides (Nunoshiba et al. 2004; Ishchenko et al. 2005) the high mutation rate in all ribΔ mutants suggests that riboflavin biosysnthesis also serves this purpose. The rare incidence of riboflavin auxotrophic microorganisms (Koser 1968) might also reflect this additional function of riboflavin biosynthesis, which in contrast to the generation of the flavin cofactors, cannot be satisfied by exogeneous riboflavin. Because the rib5Δ* suppressor mutants characterized here are still defective in riboflavin biosynthesis, they are also likely to have increased amounts of 8-oxo-GTP, which results in continued genetic instability. In support of this possible function of the riboflavin biosynthetic pathway, P. guilliermondii rib1 mutants also display increased rates of spontaneous mutations and develop more canavanine and 5-fluoro orotic acid resistant clones than wild-type strains (Boretsky et al. 2005).
In summary, our analysis of spontaneous suppressor mutants arising in riboflavin auxotrophic strains has led to the provocative finding that the riboflavin transporter gene MCH5 is regulated by proline via the transcription factor Put3. We provide strong evidence that the transcriptional activation of MCH5 by Put3 forms the molecular basis of the suppression mechanism that improves the growth of rib mutants on ammonium-containing media.
Acknowledgments
We thank Martin Loibl, Johannes Schönberger, and Stefan Ringlstetter for technical support; Marjorie C. Brandriss and Hiroshi Takagi for strains; Klaus-Jürgen Tiefenbach for help with the fluorescence spectrophotometer; Gabriele Schmidt for the proline determination; and members of the group of Martin Klingenspor for help with the band-shift assay. This work was supported by the Deutsche Forschungsgemeinschaft (STO434/2-1).
References
- Andreasson, C., E. P. Neve and P. O. Ljungdahl, 2004. Four permeases import proline and the toxic proline analogue azetidine-2-carboxylate into yeast. Yeast 21 193–199. [DOI] [PubMed] [Google Scholar]
- Axelrod, J. D., J. Majors and M. C. Brandriss, 1991. Proline-independent binding of PUT3 transcriptional activator protein detected by footprinting in vivo. Mol. Cell. Biol. 11 564–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher, A., S. Eberhardt, M. Fischer, K. Kis and G. Richter, 2000. Biosynthesis of vitamin b2 (riboflavin). Annu. Rev. Nutr. 20 153–167. [DOI] [PubMed] [Google Scholar]
- Boer, V. M., S. L. Tai, Z. Vuralhan, Y. Arifin, M. C. Walsh et al., 2007. Transcriptional responses of Saccharomyces cerevisiae to preferred and nonpreferred nitrogen sources in glucose-limited chemostat cultures. FEMS Yeast Res. 7 604–620. [DOI] [PubMed] [Google Scholar]
- Boretsky, Y. R., K. Y. Kapustyak, L. R. Fayura, O. V. Stasyk, M. M. Stenchuk et al., 2005. Positive selection of mutants defective in transcriptional repression of riboflavin synthesis by iron in the flavinogenic yeast Pichia guilliermondii. FEMS Yeast Res. 5 829–837. [DOI] [PubMed] [Google Scholar]
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li et al., 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14 115–132. [DOI] [PubMed] [Google Scholar]
- Brandriss, M. C., 1987. Evidence for positive regulation of the proline utilization pathway in Saccharomyces cerevisiae. Genetics 117 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss, M. C., and B. Magasanik, 1979. Genetics and physiology of proline utilization in Saccharomyces cerevisiae: mutation causing constitutive enzyme expression. J. Bacteriol. 140 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, C. M., D. J. Slotboom, E. R. Geertsma, R. H. Duurkens, B. Poolman et al., 2006. The riboflavin transporter RibU in Lactococcus lactis: molecular characterization of gene expression and the transport mechanism. J. Bacteriol. 188 2752–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bässler, K.-H., I. Golly, D. Loew and K. Pietrzuk, 2002. Vitamin-Lexikon. Urban & Fischer, München, Jena, Germany.
- Cecchini, G., M. Perl, J. Lipsick, T. P. Singer and E. B. Kearney, 1979. Transport and binding of riboflavin by Bacillus subtilis. J. Biol. Chem. 254 7295–7301. [PubMed] [Google Scholar]
- des Etages, S. A., D. A. Falvey, R. J. Reece and M. C. Brandriss, 1996. Functional analysis of the PUT3 transcriptional activator of the proline utilization pathway in Saccharomyces cerevisiae. Genetics 142 1069–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- des Etages, S. A., D. Saxena, H. L. Huang, D. A. Falvey, D. Barber et al., 2001. Conformational changes play a role in regulating the activity of the proline utilization pathway-specific regulator in Saccharomyces cerevisiae. Mol. Microbiol. 40 890–899. [DOI] [PubMed] [Google Scholar]
- Foraker, A. B., C. M. Khantwal and P. W. Swaan, 2003. Current perspectives on the cellular uptake and trafficking of riboflavin. Adv. Drug Deliv. Rev. 55 1467–1483. [DOI] [PubMed] [Google Scholar]
- Gietz, R. D., and A. Sugino, 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74 527–534. [DOI] [PubMed] [Google Scholar]
- Godard, P., A. Urrestarazu, S. Vissers, K. Kontos, G. Bontempi et al., 2007. Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 27 3065–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill, S., H. Yamaguchi, H. Wagner, L. Zwahlen, U. Kusch et al., 2007. Identification and characterization of two Streptomyces davawensis riboflavin biosynthesis gene clusters. Arch. Microbiol. 188 377–387. [DOI] [PubMed] [Google Scholar]
- Harbison, C. T., D. B. Gordon, T. I. Lee, N. J. Rinaldi, K. D. Macisaac et al., 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H. L., and M. C. Brandriss, 2000. The regulator of the yeast proline utilization pathway is differentially phosphorylated in response to the quality of the nitrogen source. Mol. Cell. Biol. 20 892–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishchenko, A. A., X. Yang, D. Ramotar and M. Saparbaev, 2005. The 3′→5′ exonuclease of Apn1 provides an alternative pathway to repair 7,8-dihydro-8-oxodeoxyguanosine in Saccharomyces cerevisiae. Mol. Cell. Biol. 25 6380–6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kis, K., K. Kugelbrey and A. Bacher, 2001. Biosynthesis of riboflavin. The reaction catalyzed by 6,7-dimethyl-8-ribityllumazine synthase can proceed without enzymatic catalysis under physiological conditions. J. Org. Chem. 66 2555–2559. [DOI] [PubMed] [Google Scholar]
- Kobayashi, M., Y. Ohara-Nemoto, M. Kaneko, H. Hayakawa, M. Sekiguchi et al., 1998. Potential of Escherichia coli GTP cyclohydrolase II for hydrolyzing 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J. Biol. Chem. 273 26394–26399. [DOI] [PubMed] [Google Scholar]
- Koser, S. A., 1968. Vitamin Requirements of Bacteria and Yeasts. Charles C. Thomas, Springfield, IL.
- Lesuisse, E., S. A. Knight, M. Courel, R. Santos, J. M. Camadro et al., 2005. Genome-wide screen for genes with effects on distinct iron uptake activities in Saccharomyces cerevisiae. Genetics 169 107–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makuc, J., S. Paiva, M. Schauen, R. Kramer, B. Andre et al., 2001. The putative monocarboxylate permeases of the yeast Saccharomyces cerevisiae do not transport monocarboxylic acids across the plasma membrane. Yeast 18 1131–1143. [DOI] [PubMed] [Google Scholar]
- Marczak, J. E., and M. C. Brandriss, 1989. Isolation of constitutive mutations affecting the proline utilization pathway in Saccharomyces cerevisiae and molecular analysis of the PUT3 transcriptional activator. Mol. Cell. Biol. 9 4696–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczak, J. E., and M. C. Brandriss, 1991. Analysis of constitutive and noninducible mutations of the PUT3 transcriptional activator. Mol. Cell. Biol. 11 2609–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher, K., B. Sharma, W. V. Ding and M. Nolden, 2000. Zero background yeast reporter plasmids. Gene 247 53–61. [DOI] [PubMed] [Google Scholar]
- Nunoshiba, T., R. Ishida, M. Sasaki, S. Iwai, Y. Nakabeppu et al., 2004. A novel Nudix hydrolase for oxidized purine nucleoside triphosphates encoded by ORFYLR151c (PCD1 gene) in Saccharomyces cerevisiae. Nucleic Acids Res. 32 5339–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirner, H. M., and J. Stolz, 2006. Biotin sensing in Saccharomyces cerevisiae is mediated by a conserved DNA element and requires the activity of biotin-protein ligase. J. Biol. Chem. 281 12381–12389. [DOI] [PubMed] [Google Scholar]
- Reihl, P., and J. Stolz, 2005. The monocarboxylate transporter homolog Mch5p catalyzes riboflavin (vitamin B2) uptake in Saccharomyces cerevisiae. J. Biol. Chem. 280 39809–39817. [DOI] [PubMed] [Google Scholar]
- Saxena, D., K. B. Kannan and M. C. Brandriss, 2003. Rapamycin treatment results in GATA factor-independent hyperphosphorylation of the proline utilization pathway activator in Saccharomyces cerevisiae. Eukaryot. Cell 2 552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine, T., A. Kawaguchi, Y. Hamano and H. Takagi, 2007. Desensitization of feedback inhibition of the Saccharomyces cerevisiae gamma-glutamyl kinase enhances proline accumulation and freezing tolerance. Appl. Environ. Microbiol. 73 4011–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick, C. A., and R. J. Reece, 2003. Modulation of transcription factor function by an amino acid: activation of Put3p by proline. EMBO J. 22 5147–5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick, C. A., and R. J. Reece, 2005. Eukaryotic transcription factors as direct nutrient sensors. Trends Biochem. Sci. 30 405–412. [DOI] [PubMed] [Google Scholar]
- Siddiqui, A. H., and M. C. Brandriss, 1989. The Saccharomyces cerevisiae PUT3 activator protein associates with proline-specific upstream activation sequences. Mol. Cell. Biol. 9 4706–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahmann, K. P., J. L. Revuelta and H. Seulberger, 2000. Three biotechnical processes using Ashbya gossypii, Candida famata, or Bacillus subtilis compete with chemical riboflavin production. Appl. Microbiol. Biotechnol. 53 509–516. [DOI] [PubMed] [Google Scholar]
- Swaminathan, K., P. Flynn, R. J. Reece and R. Marmorstein, 1997. Crystal structure of a PUT3-DNA complex reveals a novel mechanism for DNA recognition by a protein containing a Zn2Cys6 binuclear cluster. Nat. Struct. Biol. 4 751–759. [DOI] [PubMed] [Google Scholar]
- Thomas, B. J., and R. Rothstein, 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56 619–630. [DOI] [PubMed] [Google Scholar]
- Todd, R. B., and A. Andrianopoulos, 1997. Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet. Biol. 21 388–405. [DOI] [PubMed] [Google Scholar]
- Vitreschak, A. G., D. A. Rodionov, A. A. Mironov and M. S. Gelfand, 2002. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 30 3141–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl, C., S. Grill, O. Schilling, J. Stülke, M. Mack et al., 2007. Characterization of riboflavin (vitamin B2) transport proteins from Bacillus subtilis and Corynebacterium glutamicum. J. Bacteriol. 189 7367–7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 1793–1808. [DOI] [PubMed] [Google Scholar]
- Walters, K. J., K. T. Dayie, R. J. Reece, M. Ptashne and G. Wagner, 1997. Structure and mobility of the PUT3 dimer. Nat. Struct. Biol. 4 744–750. [DOI] [PubMed] [Google Scholar]
- Wang, S. S., and M. C. Brandriss, 1987. Proline utilization in Saccharomyces cerevisiae: sequence, regulation, and mitochondrial localization of the PUT1 gene product. Mol. Cell. Biol. 7 4431–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa, A., S. Masuda, T. Katsura and K. Inui, 2008. Identification and functional characterization of a novel human and rat riboflavin transporter, RFT1. Am. J. Physiol. Cell Physiol. 295 C632–C641. [DOI] [PubMed] [Google Scholar]