Abstract
Auxin controls numerous plant growth processes by directing cell division and expansion. Auxin-response mutants, including iba response5 (ibr5), exhibit a long root and decreased lateral root production in response to exogenous auxins. ibr5 also displays resistance to the phytohormone abscisic acid (ABA). We found that the sar3 suppressor of auxin resistant1 (axr1) mutant does not suppress ibr5 auxin-response defects, suggesting that screening for ibr5 suppressors might reveal new components important for phytohormone responsiveness. We identified two classes of Arabidopsis thaliana mutants that suppressed ibr5 resistance to indole-3-butyric acid (IBA): those with restored responses to both the auxin precursor IBA and the active auxin indole-3-acetic acid (IAA) and those with restored response to IBA but not IAA. Restored IAA sensitivity was accompanied by restored ABA responsiveness, whereas suppressors that remained IAA resistant also remained ABA resistant. Some suppressors restored sensitivity to both natural and synthetic auxins; others restored responsiveness only to auxin precursors. We used positional information to determine that one ibr5 suppressor carried a mutation in PLEIOTROPIC DRUG RESISTANCE9 (PDR9/ABCG37/At3g53480), which encodes an ATP-binding cassette transporter previously implicated in cellular efflux of the synthetic auxin 2,4-dichlorophenoxyacetic acid.
AUXIN is an essential plant hormone controlling root elongation, lateral root initiation, stem elongation, embryo patterning, and leaf expansion through its effects on cell division and expansion (reviewed in Davies 2004; Woodward and Bartel 2005). Auxin signaling requires auxin recognition by TIR1/ABF receptor proteins, which are components of SCFTIR1/ABF ubiquitin-protein ligases that promote degradation of Aux/IAA transcriptional repressors by the 26S proteasome (reviewed in Parry and Estelle 2006). Aux/IAA protein degradation is thought to allow auxin-responsive transcription by relieving repression of the activating class of AUXIN RESPONSE FACTOR (ARF) proteins. Loss-of-function mutations in genes encoding or modulating the SCFTIR1/ABF complex (reviewed in Woodward and Bartel 2005) and gain-of-function stabilizing mutations in certain Aux/IAA proteins (reviewed in Reed 2001) can confer resistance to applied and endogenous auxin.
The phytohormone abscisic acid (ABA) controls diverse processes including shoot and root growth, stomatal closure, seed storage protein synthesis, and seed dormancy (reviewed in Davies 2004). Although responses to auxin and ABA are distinct, sensitivity to auxin appears to correlate with ABA sensitivity. For example, mutations in AUX1, AXR1, AXR2, IBR5, and TIR1, which were all isolated in mutant screens for reduced auxin sensitivity (Lincoln et al. 1990; Wilson et al. 1990; Bennett et al. 1996; Ruegger et al. 1998; Monroe-Augustus et al. 2003), also confer decreased ABA sensitivity (Wilson et al. 1990; Tiryaki and Staswick 2002; Monroe-Augustus et al. 2003; Strader et al. 2008). Although connections have been made between auxin and ABA signaling, the molecular nature of the relationship between these two phytohormones remains largely undefined.
The Arabidopsis iba response5 (ibr5) mutant was originally isolated in a screen for resistance to the auxin indole-3-butyric acid (IBA; Zolman et al. 2000) and is defective in a putative dual-specificity protein phosphatase (Monroe-Augustus et al. 2003). In addition to IBA resistance, loss-of-function ibr5 mutants are resistant to natural and synthetic auxins as well as to ABA (Monroe-Augustus et al. 2003). ibr5 exhibits decreased basal and auxin-induced expression of the auxin-responsive transcriptional reporter DR5-GUS (Monroe-Augustus et al. 2003; Strader et al. 2008), but, unlike other characterized auxin-response mutants, Aux/IAA proteins are not stabilized in ibr5 (Strader et al. 2008). These results suggest that ARF functions can be regulated by means in addition to modulation of Aux/IAA repressor protein stability.
Genetic modifiers can be useful for uncovering additional components in signaling pathways. Previous screens for suppressors of auxin-resistant mutants have identified pax1, which partially suppresses axr3-1 (Tanimoto et al. 2007), and sar1 and sar3, which partially restore auxin response to axr1 (Cernac et al. 1997; Parry et al. 2006). SAR1 and SAR3 encode nucleoporins; these mutants may suppress axr1 by altering Aux/IAA protein transport into the nucleus (Parry et al. 2006). We found that sar3 fails to suppress tir1 or ibr5 auxin resistance. To better understand IBR5 function, we isolated extragenic suppressors that restored ibr5 responsiveness to IBA. We found that these suppressors fell into two classes: those that restored ibr5 sensitivity to both IBA and indole-3-acetic acid (IAA) (class 1) and those that restored sensitivity to IBA but not to IAA (class 2). Suppressors that restored ibr5 IAA sensitivity also restored ABA sensitivity, whereas those that remained IAA resistant retained ABA resistance. We mapped four ibr5-suppressing mutations to four distinct loci and used recombination mapping to clone the gene defective in one class 2 suppressor. This suppressor restored ibr5 responses to a subset of auxins, but not to ABA, and carries a mutation in PDR9/ABCG37, which encodes an ATP-binding cassette (ABC) transporter previously reported to transport the auxinic compound 2,4-dichlorophenoxyacetic acid (2,4-D) out of cells (Ito and Gray 2006). Our results suggest that PDR9 may also facilitate IBA efflux.
MATERIALS AND METHODS
Plant materials and growth conditions:
Arabidopsis thaliana accession Colombia (Col-0) was used as wild type for all experiments. Surface-sterilized (Last and Fink 1988) seeds were plated on plant nutrient medium (PN) Haughn and Somerville 1986) supplemented with 0.5% (w/v) sucrose (PNS), solidified with 0.6% (w/v) agar. Hormone stocks were dissolved in ethanol at 0.1, 1.0, or 100 mm and ethanol-supplemented media were used as controls with all treatments normalized to the same ethanol content (<0.2 μl ethanol/ml medium). Seedlings were grown at 22° under continuous illumination through yellow long-pass filters to slow indolic compound breakdown (Stasinopoulos and Hangarter 1990) unless otherwise indicated.
Mutant isolation and nomenclature:
ibr5-1 seeds (Monroe-Augustus et al. 2003) were mutagenized with ethyl methanesulfonate (EMS; Normanly et al. 1997). M2 seeds were surface sterilized (Last and Fink 1988) and plated on PNS supplemented with 8 μm IBA at ∼1000 seeds/150-mm plate. After 8 days, putative modifier mutants with short roots were selected, transferred to unsupplemented medium to recover for several days, moved to soil, genotyped for the ibr5-1 mutation (Monroe-Augustus et al. 2003), and allowed to self-fertilize. M3 progeny lines were retested by comparing root lengths of seedlings grown on mock- and 8 μm IBA-supplemented media. Lines displaying IBA-responsive root elongation inhibition similar to wild type were retained as ibr5 suppressors.
Suppressor lines used for the initial IBA retests were analyzed as the progeny of the original isolates. Most mutant lines (MS34, MS72, MS115, MS182, MS252, MS339) used in subsequent phenotypic analyses were from the first backcross to the parental ibr5-1 line. Other mutants (MS5, MS109, MS371) were analyzed as the progeny of the original isolates.
Phenotypic assays:
All assays were conducted at least twice with similar results. For auxin-responsive root elongation assays, seedlings were grown for 8 days on the indicated auxin concentrations and primary root lengths were measured. For 1-aminocyclopropane-1-carboxylic acid (ACC)-responsive root elongation assays, seedlings were grown for 10 days on medium supplemented with either ethanol or 100 nm ACC under white light and primary root lengths were measured. For ABA-responsive root elongation assays, imbibed seeds were incubated at 4° for 4 days in the dark and then plated on unsupplemented medium. Plates were incubated in the light at 22° for an additional 4 days to allow efficient germination. Seedlings then were transferred to medium supplemented with either ethanol or 10 μm ABA and primary root lengths were measured after an additional 4 days of growth in the light.
For lateral root assays, 4-day-old seedlings grown on unsupplemented medium were transferred to medium supplemented with either ethanol or 10 μm IBA and grown for an additional 4 days. Lateral roots were counted under a dissecting microscope; primordia emerging from the primary root were counted as lateral roots.
Double-mutant isolation:
The ibr5-1 mutant (Monroe-Augustus et al. 2003) was crossed to sar3-3 (Parry et al. 2006), pdr9-1, and pdr9-2 (Ito and Gray 2006), all in the Col-0 accession. The tir1-1 mutant (Ruegger et al. 1998) was crossed to sar3-3 (Parry et al. 2006); tir1-1 pdr9-1 was a gift from William Gray (Ito and Gray 2006). The axr1-3 mutant (Lincoln et al. 1990) was crossed to sar3-3. Double mutants were identified by PCR analysis of DNA prepared from the F2 plants. Amplification of SAR3 with SAR3-1 (5′-AACATAACTCCTTGGCTTCC-3′) and SAR3-2 (5′-ACTTGGGCTGTGTTGTCATC-3′) yields a 400-bp product in wild type and no product in sar3-3. SAR3 amplification with SAR3-2 and LB1-SALK (5′-CAAACCAGCGTGGACCGCTTGCTGCAACTC-3′) yields a 333-bp product in sar3-3 and no product in wild type. PDR9 amplification with PDR9-13 (5′-GCTTTCCCCTCTGTTGCTTGGTTC-3′) and PDR9-16 (5′-ATCTCACCGTAACTCAAAGG-3′) yields a 390-bp product with two MspI restriction sites in wild type and one in pdr9-72. PDR9 amplification with the derived cleaved amplified polymorphic sequence (dCAPS; Michaels and Amasino 1998; Neff et al. 1998) primers PDR9-HinPI (5′-TGGATGAGCCAACGACGGGGCTAGGC-3′; underlined nucleotide indicates an introduced mutation for dCAPS) and PDR9-17 (5′-TGTAGATCATGCGACCACCTC-3′) yields a 270-bp product with one HinPI restriction site in wild type and none in pdr9-1. PDR9 amplification with PDR9-1 (5′-CAACGTTTTCTCTGATTACAC-3′) and PDR9-2 (5′-GCTACCAACGCCCTGACAACGAG-3′) yields a 1472-bp product in wild type and no product in pdr9-2. PDR9 amplification with PDR9-1 and LB1-SALK yields an ∼1-kbp product in pdr9-2 and no product in wild type. PCR-based identification of axr1-3 (Strader et al. 2008), ibr5-1 (Monroe-Augustus et al. 2003), and tir1-1 (Strader et al. 2008) alleles was as described previously.
Genetic analysis:
The ibr5-1 mutation, originally in the Col-0 background, was introgressed into the Wassilewskija (Ws-2) accession by crossing ibr5-1 to Ws-2 three times. Outcrossing was monitored using genetic markers (Konieczny and Ausubel 1993; Bell and Ecker 1994) polymorphic between Col-0 and Ws-2. Ws-2-introgressed ibr5-1 was homozygous for Ws-2 DNA at markers nga59, nga63, nga280, nga111, RGA1, nga168, nga172, nga112, SC5, nga249, GA3, and MBK-5.
Several ibr5-1 suppressors (in the Col-0 background) were outcrossed to Ws-introgressed ibr5-1 for mapping. F2 seedlings from the MS34 and MS115 outcrosses were screened on 10 μm IBA, and F2 seedlings from the MS72 and MS182 outcrosses were screened on 2 μm 2,4-dichlorophenoxybutyric acid (2,4-DB). DNA from sensitive individuals was isolated (Celenza et al. 1995) for mapping using published genetic markers (Konieczny and Ausubel 1993; Bell and Ecker 1994) and newly developed PCR-based markers (Table 1). New markers were identified by PCR amplifying and sequencing ∼1.6-kbp genomic DNA fragments from Ws-2 and identifying polymorphisms that altered restriction enzyme recognition sites. To ensure that those individuals in the mapping population that exhibited a short root on IBA or 2,4-DB had restored sensitivity, rather than merely delayed germination or general growth defects, progeny from mapping plants were retested on PNS with and without 10 μm IBA or 2 μm 2,4-DB.
TABLE 1.
New markers used in ibr5 suppressor mapping
| Size of products (bp)
|
||||||
|---|---|---|---|---|---|---|
| Marker | Nearest gene | Enzyme | Col-0 | Ler-0 | Ws-2 | Oligonucleotidesa |
| LCS104 | At1g53645 | EcoNI | 185 | 165, 20 | 165, 20 | CAAAGTAGGCCACCATCTCCTCTTG |
| AGGCTCACACTCAATCTGCAAACCAAAATAG | ||||||
| SNP3 | At1g60950 | HinfI | 190 | 160, 30 | 160, 30 | AGTCAACTTCTAATGGCCTTTCAGTACATG |
| ATCAACCGATGTAGATGGTCTCATACTCGACT | ||||||
| LCS301 | At3g52910 | AclI | 385 | 365, 20 | 365, 20 | AGTAGATTTGGTTAATTACAAAC |
| TGTGTTAATAAGAGGAAGTGGTTGC | ||||||
| LCS302 | At3g54050 | EciI | 462 | ND | 440 | ATCAGGCCCAACTCTTTATTATC |
| CTCGCCGCCGTTTTCGTCTC | ||||||
| LCS320 | At3g53400 | FokI | 190, 30 | 220 | 220 | GGTAGACAACAAAAAAATGGATCTTTGGAT |
| CAACACCTCAAAGCCCATAGTAG | ||||||
| LCS304 | At3g51530 | HinfI | 168, 130 | ND | 298 | GACGGCGATATGACTAGAGAAGAAC |
| TCCACGGTTGACTGAGAAGAG | ||||||
| GLL340 | At3g52510 | ApoI | 399, 295, 47 | 399, 342 | 399, 342 | AAAAGGAGAAAGAGGAAGAAGATACTACTG |
| CATTTTACTTTTAGGCGTTGAGGTGAC | ||||||
| T8M16 | At3g56770 | ApoI | 309, 96 | 405 | 405 | CCCGACAAAGTGATTATCAGCTTCAGAG |
| CATATTCTTCAGTACTCGTCTAAACATGC | ||||||
ND, not determined.
Underlined nucleotide is the introduced mutation for this dCAPS marker (Michaels and Amasino 1998; Neff et al. 1998).
Identification of the pdr9-72 mutation:
A candidate gene (PDR9/ABCG37/At3g53480) within the MS72 mapping interval was examined for defects in the mutant. Genomic DNA extracted from MS72 mutant plants was amplified using six oligonucleotide pairs [PDR9-1 (5′-CAACGTTTTCTCTGATTACAC-3′) and PDR9-2 (5′-GCTACCAACGCCCTGACAACGAG-3′); PDR9-3 (5′-AAAGCCAGGAAGGTTAGTAGTTG-3′) and PDR9-4 (5′-CATAGGATTCTGGGGCGGGTTG-3′); PDR9-5 (5′-TCAACCCGCCCCAGAATCCTATG-3′) and PDR9-6 (5′-TGAAGAGCACAGTGAAACCCAACAAG-3′); PDR9-7 (5′-ACTGGGTATCATTATGTGCCTTGTTGG-3′) and PDR9-8 (5′-CTCTTGCGTCTAGCCCCGTCGTTG-3′); PDR9-9 (5′-CCGTCGATTATATTTATGGATGAGC-3′) and PDR9-10 (5′-ATGAAGTTTGGCGTGATGGAGAC-3′); PDR9-11 (5′-ATCGGTTTCTATCCTTCAGCCTAC-3′) and PDR9-12 (5′-AGTTAACTATTGCCCATTTTTCTTGATTTG-3′)]. The resulting overlapping fragments covered the gene from 282 bp upstream of the putative translation start site to 358 bp downstream of the stop codon. Amplification products were purified using a QIAquick PCR purification kit (QIAGEN) and sequenced directly (Lone Star Labs, Houston) with the primers used for amplification.
Auxin accumulation assays:
Primary root tips (5 mm) from 8-day-old light-grown Col-0, aux1-7, pdr9-1, pdr9-2, and pdr9-72 seedlings were excised and incubated in 40 μl uptake buffer (20 mm 2-[N-morpholino]ethanesulfonic acid, 10 mm sucrose, 0.5 mm CaSO4, pH 5.6) for 10 min at room temperature. An additional 40 μl uptake buffer containing radiolabeled auxins was added to a final concentration of 25 nm [3H]-indole-3-acetic acid (20 Ci/mmol; American Radiolabeled Chemicals, St. Louis) or 25 nm [3H]-indole-3-butyric acid (25 Ci/mmol; American Radiolabeled Chemicals) and incubated at room temperature for 1 hr. Root tips were briefly rinsed with three changes of uptake buffer and placed in a fourth change of uptake buffer. After 20 min, root tips were removed from the buffer, placed in 3 ml Cytoscint scintillation cocktail (Fisher Scientific), and analyzed by scintillation counting.
RESULTS
sar3 fails to suppress ibr5 phenotypes:
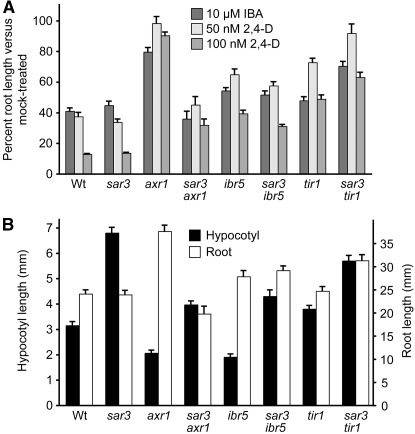
Both sar1 (Cernac et al. 1997) and sar3 (Parry et al. 2006) were isolated as axr1 suppressors and display pleiotropic phenotypes. An additional sar3 allele (mos3) was isolated in a screen for suppressors of a mutant that displays constitutive pathogenesis phenotypes (Zhang and Li 2005), and another sar1 allele (atnup160-1) was isolated in a screen for mutants with impaired cold responsiveness (Dong et al. 2006). SAR1 (At1g33410) and SAR3 (At1g80680) encode nucleoporins related to human NUP160 and NUP96, respectively, and may suppress axr1 phenotypes by excluding the Aux/IAA transcriptional repressors, which are stabilized in axr1 (Gray et al. 2001; Zenser et al. 2001), from the nucleus (Parry et al. 2006). Because ibr5 differs from axr1 in that it appears to affect auxin responses without stabilizing Aux/IAA proteins (Strader et al. 2008), we were interested in determining whether loss of SAR3 could suppress ibr5 phenotypes. We crossed ibr5-1 to the sar3-3 T-DNA disruption allele and isolated the double mutant. For controls, we crossed axr1-3 and tir1-1 to sar3-3 and isolated the corresponding double mutants. As previously reported for sar3-1 (Parry et al. 2006), we found that the sar3-3 allele restored axr1-3 2,4-D responsiveness (Figure 1A). Similarly, sar3-3 restored axr1-3 IBA responsiveness (Figure 1A). In contrast, sar3-3 did not fully rescue the reduced responses of ibr5 to the inhibitory effects of 2,4-D or IBA on root elongation (Figure 1A). Unexpectedly, sar3 appeared to enhance, rather than suppress, tir1 auxin-response defects (Figure 1A).
Figure 1.—
sar3 ibr5 auxin response. (A) Normalized primary root lengths of 8-day-old Col-0 (Wt), sar3-3, axr1-3, sar3-3 axr1-3, ibr5-1, sar3-3 ibr5-1, tir1-1, and sar3-3 tir1-1 seedlings grown under yellow-filtered light at 22° on medium supplemented with the indicated concentrations of IBA or 2,4-D. Data were normalized by comparing auxin-treated root lengths to the mean root length on mock-supplemented media (n ≥ 13). (B) Hypocotyl and root lengths of seedlings grown at 22° under continuous yellow-filtered light on unsupplemented medium (n ≥ 15). Error bars represent standard errors of the means.
Light-grown axr1 (Lincoln et al. 1990), ibr5 (Monroe-Augustus et al. 2003), and tir1 (Ruegger et al. 1998) have long primary roots in the absence of exogenous hormone. We found that sar3 suppressed the axr1 long primary root but did not decrease ibr5 or tir1 root lengths (Figure 1B). Light-grown sar3-3 exhibits a long hypocotyl (Parry et al. 2006), whereas axr1 (Lincoln et al. 1990) and ibr5 (Monroe-Augustus et al. 2003) have short hypocotyls in the light, and tir1 hypocotyls are similar in length to wild type when grown at 20° (Ruegger et al. 1998). We found that the sar3 axr1, sar3 ibr5, and sar3 tir1 double-mutant hypocotyl lengths were intermediate compared to their respective parents (Figure 1B). In contrast, sar3 early flowering was not suppressed by ibr5 or tir1 (data not shown).
Because sar3 did not restore ibr5 or tir1 auxin responsiveness, we concluded that the defects resulting from disruption of the SAR3 nucleoporin were likely to specifically affect AXR1 function rather than generally affect all mutants with decreased auxin responsiveness. The failure of the axr1 suppressor sar3-3 to suppress ibr5 auxin-response defects is consistent with IBR5 acting downstream of Aux/IAA repressor degradation (Strader et al. 2008) and suggested that a mutant screen for ibr5 suppressors might reveal novel factors involved in auxin responses in general and the IBR5 pathway in particular.
Isolation of ibr5 suppressors with restored IBA responsiveness:
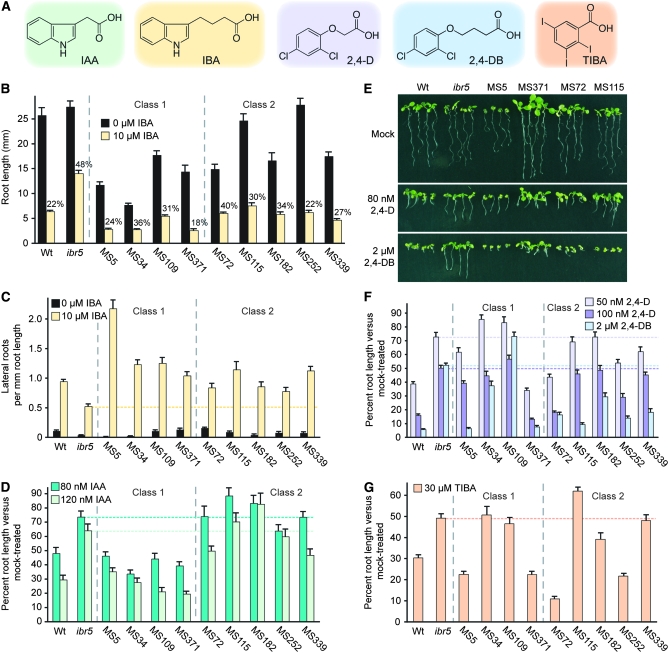
IBA inhibits primary root elongation in Arabidopsis (Zolman et al. 2000), and ibr5 mutants exhibit a long root on exogenous IBA (Zolman et al. 2000; Monroe-Augustus et al. 2003). To isolate suppressors of ibr5 IBA-resistant root growth, we generated 32 pools of EMS-mutagenized ibr5-1 seed and screened ∼48,000 of the resultant M2 progeny for seedlings with restored IBA responsiveness. We selected 371 putative suppressor mutants exhibiting a short root on IBA. Of these, 62 died, 32 were infertile, and 23 were wild-type contaminants. Progeny of the 254 remaining putative mutants were rescreened for restored sensitivity to IBA; 212 of those mutants had notably short roots with or without auxin and displayed a percentage of root elongation on IBA vs. unsupplemented medium similar to ibr5. These mutants were discarded, as mutations in these lines may have affected general seedling growth rather than auxin responsiveness. The 42 mutants that displayed a percentage of root elongation on IBA-supplemented vs. unsupplemented medium similar to wild type were retained as ibr5 suppressors. Some of these mutants displayed partial defects in root elongation even without auxin. Nine of the IBA-sensitive suppressor lines were characterized in detail (Figures 2 and 3). Because these mutants came from eight different M2 seed pools (Table 2), the mutants represent at least eight independent mutagenic events. All 42 suppressors retained the original ibr5-1 lesion and were thus expected to be extragenic, as the ibr5-1 parent contains an early stop codon (Monroe-Augustus et al. 2003).
Figure 2.—
Auxin responses of ibr5 suppressors. (A) Compounds used to monitor auxin responses. IAA is a naturally occurring auxin, IBA is a naturally occurring IAA precursor, 2,4-D is a synthetic auxin, 2,4-DB is a 2,4-D precursor, and TIBA is a synthetic auxin transport inhibitor. (B) Primary root lengths of 8-day-old Col-0 (Wt), ibr5-1, and various ibr5 suppressor (MS lines) seedlings grown under yellow-filtered light at 22° on medium supplemented with ethanol (0 μm IBA) or 10 μm IBA (n ≥ 17). Numbers above bars represent the percentage of root length on IBA compared to the control. (C) Number of lateral roots per millimeter of root length 4 days after transfer of 4-day-old seedlings to medium supplemented with either ethanol (0 μm IBA) or 10 μm IBA (n ≥ 13). (D) Primary root lengths of 8-day-old Col-0 (Wt), ibr5-1, and various ibr5 suppressor (MS lines) seedlings grown under yellow-filtered light at 22° on medium supplemented with 80 or 100 nm IAA shown normalized to the mean root length of each genotype on medium lacking IAA (n ≥ 11). (E) Photograph of 8-day-old Col-0 (Wt), ibr5-1, and various ibr5 suppressor (MS lines) seedlings grown under white light at 22° on medium supplemented with ethanol (mock), 80 nm 2,4-D, or 2 μm 2,4-DB. (F) Normalized primary root lengths of 8-day-old Col-0 (Wt), ibr5-1, and various ibr5 suppressor (MS lines) seedlings grown under yellow-filtered light at 22° on medium supplemented with 50 nm 2,4-D, 100 nm 2,4-D, or 2 μm 2,4-DB (n ≥ 16). (G) Normalized primary root lengths of 8-day-old Col-0 (Wt), ibr5-1, and various ibr5 suppressor (MS lines) seedlings grown under yellow-filtered light at 22° on medium supplemented with 30 μm TIBA (n ≥ 17). Error bars represent standard errors of the means.
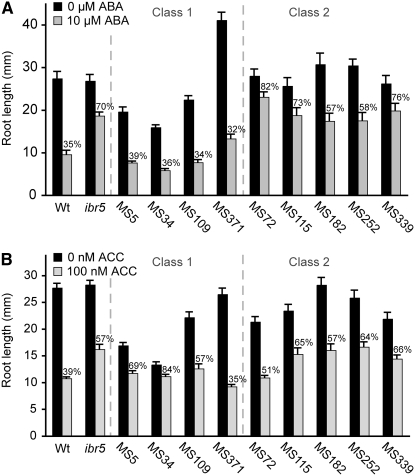
Figure 3.—
ABA and ACC response of ibr5 suppressors. (A) Primary root lengths of Col-0 (Wt), ibr5-1, and various ibr5 suppressor (MS lines) seedlings 4 days after transfer of 4-day-old seedlings to medium supplemented with either ethanol (0 μm ABA) or 10 μm ABA (n ≥ 13). The percentage of root length of seedlings transferred to ABA compared to control seedlings is indicated above the bars. (B) Primary root lengths of 10-day-old Col-0 (Wt), ibr5-1, and various ibr5 suppressor (MS lines) seedlings grown under white light at 22° on medium supplemented with either ethanol (0 nm ACC) or 100 nm ACC (n ≥ 16). Error bars represent standard errors of the means.
TABLE 2.
Classification of ibr5 suppressors
| Hormone response in root elongationb
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Classa | Isolate | M2 pool | IBA | IAA | 2,4-D | 2,4-DB | TIBA | ABA | ACC |
| — | Wild type | — | S | S | S | S | S | S | S |
| — | ibr5-1 | — | R | R | R | R | R | R | R |
| 1 | MS5 | 3 | S | S | R | S | S | S | R |
| 1 | MS34 | 4 | S | S | R | R | R | S | R |
| 1 | MS109 | 6 | S | S | R | R | R | S | R |
| 1 | MS371 | 19 | S | S | S | S | S | S | S |
| 2 | MS72 | 5 | I | R | S | S | S | R | R |
| 2 | MS115 | 6 | S | R | R | S | R | R | R |
| 2 | MS182 | 12 | S | R | R | I | I | R | R |
| 2 | MS252 | 16 | S | R | S | S | S | R | R |
| 2 | MS339 | 18 | S | R | R | S | R | R | R |
Class 1 suppressors restore IBA-, IAA-, and ABA-responsive root elongation inhibition to ibr5; class 2 suppressors restore IBA responses but remain IAA and ABA resistant.
Additional auxin phenotypes of ibr5 suppressors:
The basis of the suppressor screen was restored sensitivity to the inhibitory effect of IBA on root elongation (Figure 2B). In addition to inhibiting primary root growth, IBA promotes lateral root production in Arabidopsis (Zolman et al. 2000), and ibr5 produces fewer lateral roots than wild type with and without auxin treatment (Monroe-Augustus et al. 2003). All nine suppressors restored IBA-responsive lateral root production to ibr5, and one suppressor (MS5) appeared to be more sensitive than wild type to IBA-promoted lateral root production (Figure 2C).
In Arabidopsis, genetic evidence suggests that the auxin activity of IBA requires carboxyl sidechain shortening to IAA in peroxisomes (Zolman et al. 2000, 2007). ibr5 is resistant to inhibition of root elongation caused by either IBA or IAA, reflecting general auxin resistance (Monroe-Augustus et al. 2003). We examined the ibr5 suppressors on IAA and found that MS5, MS34, MS109, and MS371 exhibited restored IAA-responsive root elongation inhibition, while the remaining five mutants remained IAA resistant (Figure 2D) despite displaying restored IBA responsiveness (Figure 2, B and C). We designated the mutants exhibiting restored response to both IBA and IAA as class 1 mutants and those exhibiting restored response to IBA but not to IAA as class 2 mutants (Figure 2, B–D; Table 2).
We also examined the suppressor responses to the auxinic compounds 2,4-D and 2,4-DB (Figure 2A). As with IBA conversion to IAA, 2,4-DB requires chain shortening to 2,4-D for auxin activity (Hayashi et al. 1998). We found that MS371, MS72, and MS252 displayed nearly wild-type 2,4-D responsiveness, whereas the remaining suppressors displayed 2,4-D resistance similar to ibr5 (Figure 2, E and F). In contrast, most suppressors restored ibr5 2,4-DB responsiveness (Figure 2, E and F); only MS34 and MS109 displayed 2,4-DB resistance similar to ibr5, and MS182 showed intermediate 2,4-DB responsiveness (Figure 2F).
In addition to resistance to the effects of auxin and auxinic compounds, ibr5 is resistant to the effects of the auxin transport inhibitor 2,3,5-triiodobenzoic acid (TIBA; Monroe-Augustus et al. 2003). We tested the ibr5 suppressors on TIBA and found that MS5, MS371, MS72, and MS252 had restored responsiveness to 30 μm TIBA, whereas MS34, MS109, MS115, and MS339 remained resistant, and MS182 displayed an intermediate phenotype (Figure 2G).
ABA responsiveness of ibr5 suppressors:
Mutations in IBR5 confer resistance to the inhibitory effects of the phytohormone ABA on root elongation (Monroe-Augustus et al. 2003). We examined the nine IBA-sensitive ibr5 suppressors to determine if they also restored ABA responses and found that all four class 1 mutants (MS5, MS34, MS109, and MS371) exhibited restored ABA-induced root elongation inhibition whereas all five class 2 mutants remained resistant to the inhibitory effects of ABA on root elongation, although comparison of the percentage of elongation on ABA vs. on unsupplemented medium revealed that some mutants in the latter class were no longer as dramatically ABA resistant as the ibr5 parent (Figure 3A).
Ethylene responsiveness of ibr5 suppressors:
Like several other auxin-resistant mutants (Stepanova et al. 2007), ibr5 is weakly resistant to the inhibitory effects of the ethylene precursor ACC on root elongation (Figure 3B). We examined the effects of ACC on the nine ibr5 suppressors and found that only one line, MS371, restored wild-type ACC responsiveness to ibr5 (Figure 3B). The other eight suppressors remained resistant to the inhibitory effects of ACC on root elongation (Figure 3B).
A mutation in the gene encoding the PDR9/ABCG37 2,4-D transporter suppresses a subset of ibr5 phenotypes:
MS72 was isolated as an ibr5 suppressor on IBA (Figure 2B), but subsequent testing revealed that the suppression of ibr5 auxin-resistant root elongation was more apparent on 2,4-DB and 2,4-D than on IBA or IAA (Figure 2). We used restored 2,4-DB responsiveness to map the recessive ibr5-suppressing lesion in MS72 to a 215-kbp region on the lower arm of chromosome 3 between LCS320 and LCS302 (Figures 4 and 5A). This region contains PLEIOTROPIC DRUG RESISTANCE9 (PDR9/ABCG37), a dominant mutation of which has been identified as eta4 in a tir1 enhancer screen (Ito and Gray 2006). In contrast to the gain-of-function eta4/pdr9-1 mutation, which enhances tir1 2,4-D resistance, loss of PDR9 function in the pdr9-2 T-DNA insertion allele results in 2,4-D hypersensitivity (Ito and Gray 2006), making pdr9 a reasonable candidate for an ibr5 suppressor. We PCR amplified and sequenced PDR9 from MS72 genomic DNA and identified a G-to-A base change at position 3072 (where the A of the ATG is at position 1) that causes a Gly-to-Asp missense mutation in a conserved amino acid (Figure 5, B and D). Because the identified nucleotide change destroys an MspI site, we confirmed the mutation by amplifying and digesting this region of PDR9 from wild-type and mutant genomic DNA. We named the identified mutation in MS72 pdr9-72.
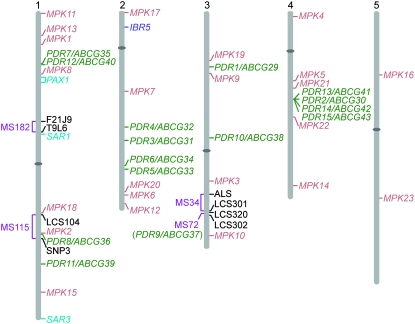
Figure 4.—
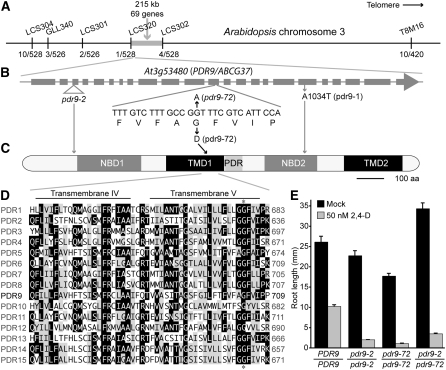
Map positions of IBA-sensitive ibr5 suppressors. Approximate map positions of molecular markers (in black type), IBR5 (in dark purple type; Monroe-Augustus et al. 2003), the 23 MPK genes (in tan type; Tena et al. 2001), and the 15 PDR/ABCG genes (in green type; Verrier et al. 2008) are shown to the right of each chromosome. Map positions of the previously isolated suppressors SAR1 (Parry et al. 2006), SAR3 (Parry et al. 2006), and the pax1 mapping interval (Tanimoto et al. 2007) are in aqua type. The interval to which each ibr5-suppressing mutation maps is shown to the left of the chromosomes in light purple type. The ibr5-suppressing mutation in MS182 maps to chromosome 1 south of F21J9 (Leclere et al. 2004) and north of T9L6 (Magidin 2002) with 5/120 and 1/120 recombinants, respectively. The recessive ibr5-suppressing mutation in MS115 maps to chromosome 1 south of LCS104 and north of SNP3 with 16/482 and 5/270 recombinants, respectively. The recessive ibr5-suppressing mutation in MS34 maps to chromosome 3 south of ALS (http://www.arabidopsis.org) and north of LCS301 with 8/76 and 5/76 recombinants, respectively. And the recessive ibr5-suppressing mutation in MS72 (pdr9-72) maps south of LCS320 and north of LCS302 (Figure 5).
Figure 5.—
Positional cloning of PDR9/ABCG37. (A) Recombination mapping with PCR-based markers T8M16, LCS304, GLL340, LCS301, LCS320, and LCS302 (Table 1) localized the ibr5-suppressing mutation in MS72 between LCS320 and LCS302 with 1/526 north and 4/528 south recombinants. (B) Examination of the PDR9 (At3g53480) gene in this region revealed a G-to-A mutation at position 3072 in MS72 DNA that destroys an MspI site and results in a Gly704-to-Asp substitution. pdr9-2 carries a T-DNA insert in the third exon of PDR9 (Ito and Gray 2006). pdr9-1 results in an Ala1034-to-Thr substitution (Ito and Gray 2006). (C) PDR9 schematic based on output from the domain-predicting program SMART (Schultz et al. 1998). PDR9 contains two NBDs, two TMDs each containing six transmembrane spans, and a PDR signature motif. (D) The pdr9-72 mutation disrupts a conserved glycine in the fifth predicted transmembrane span of TMD1. The alignment shows the fourth and fifth predicted transmembrane spans of the 15 Arabidopsis PDR family members. Sequences were aligned using the MegAlign program (DNAStar, Madison, WI). Amino acid residues identical in at least eight sequences are against a solid background; chemically similar residues in at least eight sequences are shaded. The position of the pdr9-72 mutation is indicated with an asterisk. (E) Complementation test showing primary root lengths of 8-day-old Col-0 wild-type (PRD9/PDR9), pdr9-2/pdr9-2, pdr9-72/pdr72-2, and pdr9-2/pdr9-72 seedlings grown under yellow-filtered light at 22° on medium supplemented with ethanol (mock) or 50 nm 2,4-D. Error bars represent standard errors of the means (n ≥ 13).
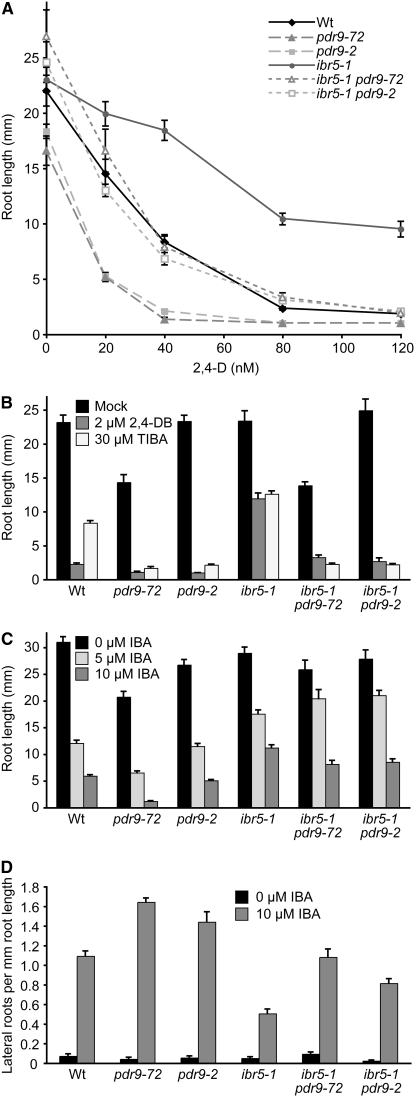
To test whether the observed MS72 phenotypes were caused by the pdr9-72 lesion, we crossed MS72 to wild type and isolated the homozygous pdr9-72/pdr9-72 mutant in a wild-type IBR5/IBR5 background. We then crossed pdr9-72 to the previously described loss-of-function pdr9-2 allele (Ito and Gray 2006) and tested 2,4-D responsiveness in the pdr9-2/pdr9-72 F1 progeny. We found that pdr9-72 failed to complement the pdr9-2 hypersensitivity to root growth inhibition by 2,4-D (Figure 5E). Because both pdr9-2 (Ito and Gray 2006) and pdr9-72 (data not shown) are recessive, this lack of complementation indicates that the lesion that we identified in pdr9-72 confers a PDR9 loss of function.
To verify that loss of PDR9 could suppress ibr5 auxin resistance in MS72, we crossed the pdr9-2 loss-of-function mutant (Ito and Gray 2006) to ibr5-1 and compared ibr5 pdr9-2 auxin responses to those of ibr5 pdr9-72 and the single mutants. We found that the pdr9-72 and pdr9-2 single mutants were similarly hypersensitive to the inhibitory effects of 2,4-D, 2,4-DB, and TIBA on primary root elongation and that pdr9-2 and pdr9-72 restored 2,4-D, 2,4-DB, and TIBA responsiveness to ibr5 to a similar extent (Figure 6, A and B). As previously reported (Ito and Gray 2006), pdr9-2 responded similarly to wild type to the inhibitory effects of IBA on root elongation (Figure 6C). Moreover, we found that pdr9-2 failed to restore ibr5 root elongation inhibition in response to IBA (Figure 6C). However, both pdr9-72 and pdr9-2 appeared to be more sensitive than wild type to IBA-promoted lateral root production and at least partially restored ibr5 IBA-induced lateral root induction (Figure 6D). Because pdr9-72 responded similarly to the pdr9-2 likely null allele (Ito and Gray 2006) in these assays and because both alleles similarly restored ibr5 responsiveness to 2,4-D, 2,4-DB, and TIBA (Figure 6, A and B), we concluded that the pdr9-72 lesion reduced PDR9 function and was responsible for suppression of a subset of ibr5 phenotypes in MS72.
Figure 6.—
ibr5 pdr9-72 and tir1 pdr9-2 auxin response. (A) Primary root lengths of 8-day-old Col-0 (Wt), pdr9-72, pdr9-2, ibr5-1, ibr5-1 pdr9-72 (MS72), and ibr5-1 pdr9-2 seedlings grown under yellow-filtered light at 22° on medium supplemented with ethanol (mock) or various concentrations of 2,4-D (n ≥ 9). (B) Primary root lengths of 8-day-old seedlings grown under yellow-filtered light at 22° on medium supplemented with ethanol (mock), 2 μm 2,4-DB, or 30 μm TIBA (n = 15). (C) Primary root lengths of 8-day-old seedlings grown on medium supplemented with ethanol (0 μm IBA) or IBA (n = 15). (D) Lateral roots were counted 4 days after transfer of 4-day-old seedlings to medium supplemented with either 0 (ethanol control) or 10 μm IBA (n = 12). Error bars represent standard errors of the means.
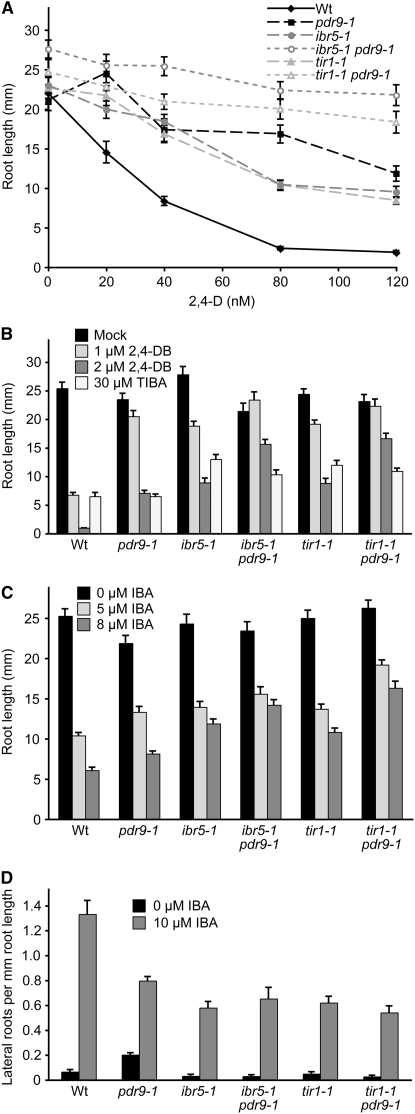
To determine whether the gain-of-function pdr9-1 allele (Ito and Gray 2006) could enhance ibr5 phenotypes, we crossed pdr9-1 to ibr5-1 and compared the resultant ibr5 pdr9-1 double mutant to the single mutants and the previously described (Ito and Gray 2006) tir1 pdr9-1 mutant. As previously reported (Ito and Gray 2006), the pdr9-1 single mutant was resistant to the inhibitory effects of 2,4-D on primary root elongation (Figure 7A). In addition, we found that pdr9-1 was resistant to 2,4-DB (Figure 7B) and slightly resistant to the auxin precursor IBA in both root elongation inhibition and lateral root promotion (Figure 7, C and D). In contrast to the heightened TIBA sensitivity of the pdr9 loss-of-function alleles (Figure 6B), pdr9-1 resembled wild type in sensitivity to the auxin transport inhibitor TIBA (Figure 7B). In the double mutants, we found that pdr9-1 enhanced ibr5-1 resistance to root elongation inhibition by 2,4-D (Figure 7A), conferring similar 2,4-D resistance as the tir1 pdr9-1 double mutant. In addition to enhancing 2,4-D resistance, we found that pdr9-1 enhanced tir1 and ibr5 resistance to 2,4-DB (Figure 7B) and IBA (Figure 7C) in root elongation inhibition. However, pdr9-1 failed to enhance tir1 or ibr5 resistance to IBA in lateral root initiation (Figure 7D) or to TIBA in root elongation inhibition (Figure 7B) in the conditions tested.
Figure 7.—
ibr5 pdr9-1 and tir1 pdr9-1 auxin response. (A) Primary root lengths of 8-day-old Col-0 (Wt), pdr9-1, ibr5-1, ibr5-1 pdr9-1, tir1-1, and tir1-1 pdr9-1 seedlings grown under yellow-filtered light at 22° on medium supplemented with ethanol (mock) or various concentrations of 2,4-D (n = 12). (B) Primary root lengths of 8-day-old seedlings grown under yellow-filtered light at 22° on medium supplemented with ethanol (mock) or various concentrations of 2,4-DB or 30 μm TIBA (n ≥ 12). (C) Primary root lengths of 8-day-old seedlings grown on medium supplemented with ethanol (0 μm IBA) or various concentrations of IBA (n ≥ 12). (D) Lateral roots were counted 4 days after transfer of 4-day-old seedlings to medium supplemented with either 0 (ethanol control) or 10 μm IBA (n = 12). Error bars represent standard errors of the means.
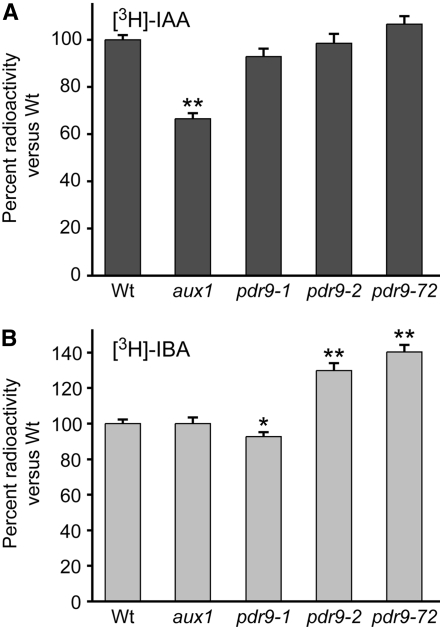
Using an excised root-tip auxin transport assay, Ito and Gray (2006) demonstrated that pdr9-1 root tips accumulate less [14C]-2,4-D than wild type, whereas pdr9-2 root tips accumulate more [14C]-2,4-D than wild type, consistent with a role for PDR9 in 2,4-D efflux that is supported by the root elongation phenotypes of the pdr9 alleles on 2,4-D-supplemented media. Because we found that pdr9 alleles also display altered IBA responsiveness (Figure 6D and Figure 7, C and D), we sought to determine whether PDR9 also might play a role in IBA efflux. We assessed [3H]-IAA and [3H]-IBA accumulation in excised root tips from 8-day-old seedlings. As previously reported (Ito and Gray 2006), we found that pdr9 mutants displayed wild-type [3H]-IAA accumulation in this assay (Figure 8A). We included the aux1 IAA influx mutant as a control and found reduced [3H]-IAA accumulation in aux1 root tips (Figure 8A), as expected. In addition, we found that aux1 mutant root tips displayed wild-type [3H]-IBA accumulation (Figure 8B), consistent with the normal [3H]-IBA transport reported in aux1 roots (Rashotte et al. 2003) and the inability of excess IBA to compete with [3H]-IAA uptake by AUX1 expressed in Xenopus oocytes (Yang et al. 2006). In contrast to aux1, we found that root tips of both pdr9-2 and pdr9-72 clearly hyperaccumulated [3H]-IBA (Figure 8B). Moreover, we observed a small but statistically significant reduction in [3H]-IBA accumulation in pdr9-1 root tips (Figure 8B). These results are consistent with the possibility that PDR9 facilitates IBA efflux from root cells.
Figure 8.—
[3H]-IAA and [3H]-IBA accumulation in pdr9 mutants. Root tips of 8-day-old Col-0 (Wt), aux1-7, pdr9-1, pdr9-2, and pdr9-72 seedlings were incubated for 1 hr in buffer containing 25 nm [3H]-IAA (A) or 25 nm [3H]-IBA (B), rinsed three times, and incubated for an additional 20 min in buffer. Root tips were then removed and analyzed by scintillation counting. Data were averaged from two (A) or four (B) independent experiments, each with eight replicates of five root tips of each genotype. Data were normalized by comparison to the mean radioactivity of wild-type samples, which ranged from 13,083 to 15,406 cpm for the [3H]-IAA experiments (A) and from 17,144 to 22,167 cpm for the [3H]-IBA experiments (B). Error bars represent standard errors of the means, and asterisks indicate significant differences from wild type in two-tailed t-tests assuming equal variance (*P ≤ 0.01; **P ≤ 0.001).
Mapping second-site mutations in additional ibr5 suppressors:
In addition to MS72, we used recombination mapping with PCR-based markers to localize three additional recessive ibr5-suppressing mutations (MS34, MS115, and MS182) to three distinct chromosomal regions. None of the mapped suppressors appeared to be allelic, as none mapped to the same interval (Figure 4). In addition, none of the mapping intervals include the previously isolated auxin-resistance-suppressing mutations pax1 (Tanimoto et al. 2007), sar1, or sar3 (Parry et al. 2006), suggesting that additional novel ibr5-suppressing pathways remain to be identified. Map-based cloning of the defective genes in these ibr5-suppressing mutants is ongoing.
DISCUSSION
PDR9 role in auxin response:
PDR subfamily members of ABC transporters are found only in fungi and plants and, like other full-sized ABC transporters, contain two apparent nucleotide-binding domains (NBD) and two transmembrane domains (TMD) consisting of six membrane-spanning sequences each (reviewed in Crouzet et al. 2006; Verrier et al. 2008). Fifteen PDR/ABCG genes have been identified in Arabidopsis (Sanchez-Fernandez et al. 2001; Martinoia et al. 2002; van den Brule and Smart 2002; Verrier et al. 2008), but only a few have been functionally characterized in genetic studies. PDR9/ABCG37 is a 2,4-D efflux facilitator localized in the plasma membrane (Ito and Gray 2006); the gain-of-function pdr9-1 mutant is 2,4-D resistant and hypoaccumulates 2,4-D, whereas the loss-of-function pdr9-2 mutant displays increased 2,4-D sensitivity and hyperaccumulates 2,4-D (Ito and Gray 2006).
We isolated the pdr9-72 mutant as a class 2 ibr5 suppressor (Figure 2; Table 2). To our knowledge, this is the first example of a mutation in an auxin transporter suppressing the phenotype of an auxin-resistant mutant. The identical 2,4-D, 2,4-DB, and TIBA hypersensitivity of the pdr9-2 likely null allele (Ito and Gray 2006) and the pdr9-72 allele that we isolated as an ibr5 suppressor (Figure 6, A and B) suggests that the pdr9-72 Gly704-to-Asp change abolishes PDR9 function.
Although ibr5 is 2,4-D resistant, it is not completely 2,4-D insensitive, as it responds to high 2,4-D concentrations (Monroe-Augustus et al. 2003; Strader et al. 2008). pdr9 may counteract ibr5 2,4-D resistance by allowing 2,4-D to accumulate to higher levels within cells. We envision that when PDR9 is disrupted, applied 2,4-D is less efficiently removed from cells (Ito and Gray 2006), and the consequent 2,4-D hyperaccumulation allows the ibr5 pdr9 double mutant to respond to 2,4-D similarly to wild type (Figure 6A).
We found that pdr9 TIBA responses resembled previously reported responses of pdr9 alleles to the auxin transport inhibitor 1-N-naphthylphthalamic acid (NPA). The loss-of-function pdr9-2 is hypersensitive to NPA (Ito and Gray 2006) and TIBA (Figure 6B), whereas the gain-of-function pdr9-1 responds similarly to wild type to NPA (Ito and Gray 2006) and TIBA (Figure 7B). Because pdr9-2 and pdr9-72 mutants are TIBA hypersensitive and completely abolish the TIBA resistance of ibr5 (Figure 6B), it is possible that the PDR9 transporter may efflux TIBA in addition to 2,4-D. Why the gain-of-function pdr9-1 allele is resistant to some (2,4-D and 2,4-DB) but not all compounds to which the loss-of-function pdr9-2 allele is hypersensitive (2,4-D, 2,4-DB, TIBA, and NPA) remains unexplained.
Loss of PDR9 does not restore all ibr5 defects. Although pdr9-72 restores ibr5 responsiveness to 2,4-D, 2,4-DB, and TIBA, responses to IAA, ABA, and ACC appear largely unaffected (Figures 2 and 3). The IBA response of pdr9 is more complex. We isolated MS72 (ibr5-1 pdr9-72) in a screen for ibr5 suppressors displaying a short root when grown on IBA, but subsequent analyses revealed that our initial MS72 line displayed a short root even on unsupplemented medium, which undoubtedly contributed to MS72 isolation. In contrast, the pdr9-2 root elongates normally and is not hypersensitive to IBA-induced root elongation inhibition (Figure 6; Ito and Gray 2006). Moreover, pdr9-2/pdr9-72 seedlings were 2,4-D hypersensitive but did not display root elongation defects on unsupplemented medium (Figure 5E), indicating that the short root of the initial MS72 line was likely caused by extraneous recessive mutations. Interestingly, however, both pdr9-2 and pdr9-72 mutants displayed heightened sensitivity to IBA in lateral root induction and partially restored ibr5 lateral rooting defects (Figure 6D), and the pdr9-1 mutant displayed slight resistance to IBA in root elongation inhibition under our conditions (Figure 7C), consistent with the possibility that PDR9 effluxes substrates in addition to 2,4-D. Indeed, we found that root tips of both pdr9-2 and pdr9-72 hyperaccumulated [3H]-IBA and the gain-of-function pdr9-1 allele displayed slightly reduced [3H]-IBA accumulation in an auxin transport assay, suggesting that PDR9 may promote IBA efflux.
ibr5 suppressors restore distinct subsets of ibr5 phenotypes:
We identified and characterized ibr5 suppressors with the anticipation that analysis of the genes defective in these suppressors will help elucidate the role of IBR5 in auxin, ABA, and ethylene responses. IBR5 is a putative MAP kinase phosphatase (Monroe-Augustus et al. 2003), and IBR5 phosphatase activity appears to be required for full auxin and ABA responsiveness (Strader et al. 2008). Although we expect that mutation of a substrate MAP kinase might suppress some ibr5 defects, MPK2 and MPK18 are the only MPK genes in or near our current ibr5 suppressor mapping intervals (Figure 4), and these genes are not mutated in MS115 (data not shown), demonstrating that there are means to restore ibr5 hormone responsiveness that do not involve MPK mutations. Although additional backcrossing will be needed to ensure that all of the phenotypes observed in this initial analysis result from disruptions in single loci, the diversity of ibr5 suppressor phenotypes (Table 2) suggests that several mechanisms can restore IBA responsiveness to ibr5.
We found that all of the suppressor mutants that restored ibr5 root elongation inhibition in response to IBA (Figure 2B) also restored ibr5 defects in IBA-responsive lateral root production (Figure 2C). We classified the suppressors on the basis of the response to the natural auxin IAA (Table 2). The class 1 mutants (MS5, MS34, MS109, MS371) restored IAA-responsive root elongation inhibition to ibr5, whereas the class 2 mutants (MS72, MS115, MS182, MS252, MS339) remained IAA resistant.
Although the ibr5 suppressors can be divided into two broad classes, mutants within each class have varied phenotypes, suggesting that they restore auxin responses differently from one another. All of the class 1 mutants regained the ability to respond to ABA, but only MS371 exhibited restored response to all hormones tested. Moreover, MS371 was the only suppressor that fully restored ibr5 responses to the ethylene precursor ACC (Figure 3B). These data suggest that the gene disrupted in MS371 might act closely with IBR5.
MS109 and MS34 displayed restored response to naturally occurring auxins (IAA and IBA) but remained resistant to the synthetic compounds 2,4-D, 2,4-DB, and TIBA. This dichotomy suggests that these suppressors might impact a process that can differentiate between natural and synthetic auxins, such as transport or metabolism. For example, a mutant that reduces IAA efflux or inactivation might render plants more sensitive to IAA (and IBA, which can be converted to IAA) without affecting responses to synthetic auxins.
The class 2 suppressors restored ibr5 responses to IBA but not to IAA; none of these mutants restored ABA responses (Table 2). The class 2 mutants also displayed diverse phenotypes in auxin response assays. Like MS72 (ibr5 pdr9-72), MS252 regained sensitivity to 2,4-D and 2,4-DB, but not to IAA or ABA. However, MS252 can be distinguished from MS72: although MS252 restored ibr5 TIBA sensitivity, MS252 does not appear to be more TIBA sensitive than wild type, as are both ibr5 pdr9-72 and ibr5 pdr9-2 (Figures 2G and 6B).
The MS115 and MS339 class 2 suppressors specifically increase sensitivity to IBA and 2,4-DB, which are four-carbon side-chain auxins (Figure 2A) that require peroxisomal chain shortening for auxin activity (Hayashi et al. 1998; Zolman et al. 2000). MS115 and MS339 might increase the efficiency of IBA-to-IAA and 2,4-DB-to-2,4-D conversion and thereby restore IBA and 2,4-DB responses to ibr5 to near wild-type levels without restoring IAA, 2,4-D, TIBA, or ABA responses. One mechanism to increase the efficiency of IBA-to-IAA conversion might be to block IBA efflux, and it is interesting that a PDR/ABCG gene is found in the MS115 mapping interval (Figure 4).
Intriguingly, the examined suppressors exhibiting restored IAA response (class 1) also displayed restored ABA response, whereas the suppressors that remained IAA resistant (class 2) also remained ABA resistant (Figure 2A). Indeed, all previously examined IAA-resistant mutants also exhibit ABA resistance (Wilson et al. 1990; Tiryaki and Staswick 2002; Monroe-Augustus et al. 2003; Strader et al. 2008). Because IAA is an active form of auxin in the plant, this correlation suggests that response to endogenous IAA is necessary for root elongation inhibition in response to exogenous ABA.
Disruption of many genes can restore ibr5 auxin responsiveness:
Previous genetic screens for suppressors of auxin-resistant mutants have yielded the axr3 suppressor pax1 (Tanimoto et al. 2007) and the axr1 suppressors sar1 and sar3 (Cernac et al. 1997; Parry et al. 2006). Although PAX1 has not been cloned, both SAR1 and SAR3 encode nucleoporins (Parry et al. 2006). We found that sar3 fails to suppress the auxin resistance of ibr5 or tir1 (Figure 1A), suggesting that the means of restoring auxin responsiveness may not be the same for every mutant.
Our screen for ibr5 suppressors with restored response to IBA has identified 42 confirmed mutants, and the 9 mutants that we describe here comprise at least four distinct loci (Figure 4). The disparate phenotypes and distinct mapping positions of the characterized mutants suggest that we have not identified many alleles of any particular gene. Our data are consistent with the possibility that lesions in various genes can restore distinct subsets of ibr5 defects. Strikingly, all but one of the suppressors restored ibr5 responses to only some hormones (Table 2). In particular, many of the suppressors remained resistant to 2,4-D, a commonly used synthetic auxin, and thus would not have been identified had we used 2,4-D in our primary screen. It is possible that similarly screening for restored responsiveness to other auxins, auxin precursors, ABA, or ACC might yield additional novel ibr5 suppression pathways. Moreover, characterizing the ability of ibr5 suppressors to restore auxin responsiveness to other mutants, such as tir1 or axr1, may illuminate different auxin-signaling mechanisms. Future cloning and characterization of the genes defective in the ibr5 suppressors identified here will contribute to our understanding of auxin metabolism, transport, and interactions with other hormones and also may allow identification of IBR5 substrates that contribute to the pleiotropic phenotypes of ibr5.
Acknowledgments
We are grateful to William Gray for pdr9-1, pdr9-2, and pdr9-1 tir1-1 seeds; Mark Estelle for axr1-3; the Arabidopsis Biological Resource Center at Ohio State University for sar3-3 (SALK_109959) and tir1-1; Bhavika Kaul for technical assistance; A. Raquel Adham for developing the T8M16 marker; Arthur Millius for developing the SNP3 marker; and Matthew Lingard, Naxhiely Martinez, Dereth Phillips, Sarah Ratzel, and Andrew Woodward for critical comments on the manuscript. This research was supported by the National Science Foundation (NSF; IBN-0315596 and MCB-0745122 to B.B.), the Robert A. Welch Foundation (C-1309 to B.B.), and the National Institutes of Health (NIH; F32-GM075689 to L.C.S.). M.M.-A. was supported in part by NIH Training grant T32-GM08362, K.C.R. was supported in part by the Rice-Houston Alliance for Graduate Education and the Professoriate Program (NSF HRD-0450363), and G.L.L. was supported in part by the Beckman Scholars Program funded by the Arnold and Mabel Beckman Foundation.
References
- Bell, C. J., and J. R. Ecker, 1994. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. [DOI] [PubMed] [Google Scholar]
- Bennett, M. J., A. Marchant, H. G. Green, S. T. May, S. P. Ward et al., 1996. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273 948–950. [DOI] [PubMed] [Google Scholar]
- Celenza, J. L., P. L. Grisafi and G. R. Fink, 1995. A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 9 2131–2142. [DOI] [PubMed] [Google Scholar]
- Cernac, A., C. Lincoln, D. Lammer and M. Estelle, 1997. The SAR1 gene of Arabidopsis acts downstream of the AXR1 gene in auxin response. Development 124 1583–1591. [DOI] [PubMed] [Google Scholar]
- Crouzet, J., T. Trombik, A. S. Fraysse and M. Boutry, 2006. Organization and function of the plant pleiotropic drug resistance ABC transporter family. FEBS Lett. 580 1123–1130. [DOI] [PubMed] [Google Scholar]
- Davies, P. J., 2004. Introduction, pp. 1–35 in Plant Hormones: Biosynthesis, Signal Transduction, Action!, edited by P. J. Davies. Kluwer Academic, Dordrecht, The Netherlands.
- Dong, C. H., X. Hu, W. Tang, X. Zheng, Y. S. Kim et al., 2006. A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol. Cell. Biol. 26 9533–9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W. M., S. Kepinski, D. Rouse, O. Leyser and M. Estelle, 2001. Auxin regulates SCFTIR1-dependent degradation of Aux/IAA proteins. Nature 414 271–276. [DOI] [PubMed] [Google Scholar]
- Haughn, G. W., and C. Somerville, 1986. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 204 430–434. [Google Scholar]
- Hayashi, M., K. Toriyama, M. Kondo and M. Nishimura, 1998. 2,4-dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell 10 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, H., and W. M. Gray, 2006. A gain-of-function mutation in the Arabidopsis pleiotropic drug resistance transporter PDR9 confers resistance to auxinic herbicides. Plant Physiol. 142 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and F. M. Ausubel, 1993. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Last, R. L., and G. R. Fink, 1988. Tryptophan-requiring mutants of the plant Arabidopsis thaliana. Science 240 305–310. [DOI] [PubMed] [Google Scholar]
- LeClere, S., R. A. Rampey and B. Bartel, 2004. IAR4, a gene required for auxin conjugate sensitivity in Arabidopsis, encodes a pyruvate dehydrogenase E1α homolog. Plant Physiol. 135 989–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, C., J. H. Britton and M. Estelle, 1990. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidin, M., 2002. Genetic analysis of auxin homeostasis: conjugate sensitivity and auxin supersensitivity. Ph.D. Thesis, Rice University, Houston.
- Martinoia, E., M. Klein, M. Geisler, L. Bovet, C. Forestier et al., 2002. Multifunctionality of plant ABC transporters—more than just detoxifiers. Planta 214 345–355. [DOI] [PubMed] [Google Scholar]
- Michaels, S. D., and R. M. Amasino, 1998. A robust method for detecting single-nucleotide changes as polymorphic markers by PCR. Plant J. 14 381–385. [DOI] [PubMed] [Google Scholar]
- Monroe-Augustus, M., B. K. Zolman and B. Bartel, 2003. IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell 15 2979–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M. M., J. D. Neff, J. Chory and A. E. Pepper, 1998. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 14 387–392. [DOI] [PubMed] [Google Scholar]
- Normanly, J., P. Grisafi, G. R. Fink and B. Bartel, 1997. Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 9 1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry, G., and M. Estelle, 2006. Auxin receptors: a new role for F-box proteins. Curr. Opin. Cell Biol. 18 152–156. [DOI] [PubMed] [Google Scholar]
- Parry, G., S. Ward, A. Cernac, S. Dharmasiri and M. Estelle, 2006. The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 18 1590–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte, A. M., J. Poupart, C. S. Waddell and G. K. Muday, 2003. Transport of the two natural auxins, indole-3-butyric acid and indole-3-acetic acid, in Arabidopsis. Plant Physiol. 133 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J. W., 2001. Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6 420–425. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., E. Dewey, W. M. Gray, L. Hobbie, J. Turner et al., 1998. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 12 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Fernandez, R., T. G. Davies, J. O. Coleman and P. A. Rea, 2001. The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J. Biol. Chem. 276 30231–30244. [DOI] [PubMed] [Google Scholar]
- Schultz, J., F. Milpetz, P. Bork and C. P. Ponting, 1998. SMART, a simple modular architecture research tool: identification of signalling domains. Proc. Natl. Acad. Sci. USA 95 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasinopoulos, T. C., and R. P. Hangarter, 1990. Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 93 1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova, A. N., J. Yun, A. V. Likhacheva and J. M. Alonso, 2007. Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19 2169–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader, L. C., M. Monroe-Augustus and B. Bartel, 2008. The IBR5 phosphatase promotes Arabidopsis auxin responses through a novel mechanism distinct from TIR1-mediated repressor degradation. BMC Plant Biol. 8 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto, M., J. Jowett, P. Stirnberg, D. Rouse and O. Leyser, 2007. pax1-1 partially suppresses gain-of-function mutations in Arabidopsis AXR3/IAA17. BMC Plant Biol. 7 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena, G., T. Asai, W. L. Chiu and J. Sheen, 2001. Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 4 392–400. [DOI] [PubMed] [Google Scholar]
- Tiryaki, I., and P. E. Staswick, 2002. An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol. 130 887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brule, S., and C. C. Smart, 2002. The plant PDR family of ABC transporters. Planta 216 95–106. [DOI] [PubMed] [Google Scholar]
- Verrier, P. J., D. Bird, B. Burla, E. Dassa, C. Forestier et al., 2008. Plant ABC proteins: a unified nomenclature and updated inventory. Trends Plant Sci. 13 151–159. [DOI] [PubMed] [Google Scholar]
- Wilson, A. K., F. B. Pickett, J. C. Turner and M. Estelle, 1990. A dominant mutation in Arabidopsis confers resistance to auxin, ethylene, and abscisic acid. Mol. Gen. Genet. 222 377–383. [DOI] [PubMed] [Google Scholar]
- Woodward, A. W., and B. Bartel, 2005. Auxin: regulation, action, and interaction. Ann. Bot. 95 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., U. Z. Hammes, C. G. Taylor, D. P. Schachtman and E. Nielsen, 2006. High-affinity auxin transport by the AUX1 influx carrier protein. Curr. Biol. 16 1123–1127. [DOI] [PubMed] [Google Scholar]
- Zenser, N., A. Ellsmore, C. Leasure and J. Callis, 2001. Auxin modulates the degradation rate of Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 98 11795–11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., and X. Li, 2005. A putative nucleoporin 96 is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1. Plant Cell 17 1306–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman, B. K., A. Yoder and B. Bartel, 2000. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156 1323–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman, B. K., M. Nyberg and B. Bartel, 2007. IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol. Biol. 64 59–72. [DOI] [PubMed] [Google Scholar]