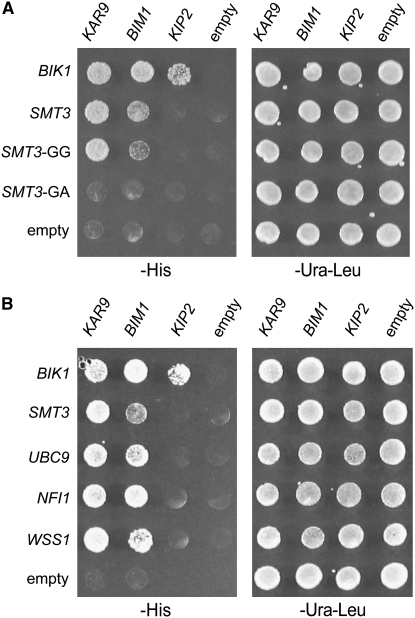
Figure 2.—
KAR9 and BIM1 interact with multiple proteins in the sumoylation pathway by two-hybrid analysis. Two-hybrid reporter strains (yRM1757/PJ69-4A) containing KAR9-BD (pRM1493), BIM1-BD (pRM3145), KIP2-BD (pRM3595), or empty BD (pRM1154) were mated to reporter strains (yRM1756/PJ69-4α) containing AD-BIK1 (pRM2627), AD-SMT3 (pRM4920), AD-SMT3-GG (pRM4382), AD-SMT3-GA (pRM4383), AD-UBC9 (pRM4495), AD-NFI1 (pRM4496), AD-WSS1 (pRM4597), or empty AD (pRM4380) plasmids. Diploids were selected on media lacking uracil and leucine (−ura −leu) and assayed for interactions on media lacking histidine (−his). These were scored after incubation at 30° for 2–3 days. (A) The yeast two-hybrid interaction is specific to forms of SUMO that are competent for conjugation. AD-SMT3 is the full-length construct. AD-SMT3-GG truncates the last three amino acids of SMT3, leaving glycine 98 as the last amino acid. In the AD-SMT3-GA construct, glycine 98 was mutated to an alanine residue, creating a conjugation-incompetent form of Smt3p. The kinesin Kip2p, which transports Kar9p to the plus end, is included as an additional negative control, indicating that not all proteins involved in spindle positioning interact with SUMO/Smt3p. Bik1p has been shown to interact with Kip2p previously (Carvalho et al. 2004) and therefore the BIK1-KIP2 interaction demonstrates that the KIP2-BD construct is functional. (B) KAR9 and BIM1 interact with other proteins in the sumoylation pathway. Ubc9p is an E2 enzyme and Nfi1p is an E3. Wss1p was identified as a weak suppressor of a temperature-sensitive allele of SMT3 (Biggins et al. 2001).