Abstract
The centriole is the core structure of centrosome and cilium. Failure to restrict centriole duplication to once per cell cycle has serious consequences and is commonly observed in cancer. Despite its medical importance, the mechanism of centriole formation is poorly understood. Asl was previously reported to be a centrosomal protein essential for centrosome function. Here we identify mecD, a severe loss-of-function allele of the asl gene, and demonstrate that it is required for centriole and cilia formation. Similarly, Cep152, the Asl ortholog in vertebrates, is essential for cilia formation and its function can be partially rescued by the Drosophila Asl. The study of Asl localization suggests that it is closely associated with the centriole wall, but is not part of the centriole structure. By analyzing the biogenesis of centrosomes in cells depleted of Asl, we found that, while pericentriolar material (PCM) function is mildly affected, Asl is essential for daughter centriole formation. The clear absence of several centriolar markers in mecD mutants suggests that Asl is critical early in centriole duplication.
CENTRIOLES are microtubule-rich cylindrical structures surrounded by pericentriolar material (PCM), which nucleates astral microtubules during mitosis. In ciliated cells, the centriole migrates to the plasma membrane and becomes a basal body, the seed of the ciliary axoneme. Centrioles duplicate only once per cell cycle by forming a daughter centriole perpendicularly to the mother centriole. Although the discovery of the centriole dates back >100 years (van Beneden 1876; Boveri 1887), little is known about its molecular composition and the mechanism that controls its formation. Understanding centriole duplication is crucial as centriole overduplication is a commonly observed feature of cancer cells (Doxsey 1998; Brinkley 2001; Fukasawa 2007).
Multiple studies using a variety of approaches have identified components of the centriole and cilium (O'Connell et al. 1998; Gonczy et al. 2000; Andersen et al. 2003; Avidor-Reiss et al. 2004; Li et al. 2004; Keller et al. 2005; Goshima et al. 2007; Kilburn et al. 2007). Recent studies in Caenorhabditis elegans have established the chronology and hierarchy of a group of molecules involved in centriole assembly (Delattre et al. 2006; Pelletier et al. 2006). Spd2 and the kinase Zyg1 trigger centriole formation. Then two coiled-coil proteins, Sas5 and Sas6, are recruited and mediate the formation and elongation of the central tube. This is followed by the addition of Sas4, which is required for the formation of the microtubule wall. Additionally, it has been shown that PCM proteins such as Spd2, Spd5, and γ-tubulin have roles in centriole duplication (Dammermann et al. 2004; Pelletier et al. 2004; Shang et al. 2005; Delattre et al. 2006). However, if Spd2 is absolutely required for centriole duplication, Spd5 and γ-tubulin play only partial roles (Dammermann et al. 2004; Pelletier et al. 2004). In flies, two other centriolar proteins, Ana1 and Ana2, identified in an RNA interference screen in S2 cells, are involved in centriole duplication (Goshima et al. 2007).
Recently, the gene mutated in asterless (asl) has been cloned (Varmark et al. 2007). The asl gene CG2919 encodes a coiled-coil protein of 994 amino acids. Barbara Wakimoto originally isolated asterless mutants in a screen for male sterile mutants (Bonaccorsi et al. 1998). Three alleles of asterless (asl1, asl2, and asl3) have been studied and analysis of the allele asl1 found that Asl is not essential for centriole formation and that its major role is in PCM assembly/maintenance (Bonaccorsi et al. 1998, 2000; Varmark et al. 2007). Cep152, the human homolog of Asl, was identified in a proteomic screen for centrosomal proteins (Andersen et al. 2003) and nothing is known about its function.
In this article we identify a new allele of asl, which we call aslmecD. We demonstrate that Asl is required for the initiation of centriole duplication. As a result, aslmecD flies do not have centrioles and therefore lack basal bodies and cilia. We studied cep152, the vertebrate homolog of asl in zebrafish, and show that its function in ciliogenesis is conserved. We identified a new function for Asl in centriole duplication and our results show that it acts very early in the process, providing new insights toward understanding its human homolog function.
MATERIALS AND METHODS
Mutant flies and transgenic fly constructs:
Asl1, 2 and 3 flies were obtained from C. Gonzalez's laboratory and were studied as hemizygotes over the deficiency ED5177 from the Bloomington fly collection (8103). The mecD mutant was cleaned for a second-site mutation by recombination to the bw;st isogenic line from which it was produced (Koundakjian et al. 2004). The generation of Drosophila reporter constructs was performed by cloning 2-kb upstream elements immediately adjacent to the predicted initiator methionine up to the stop codon into the p{UAST} vector. The asl, ana1, and dsas6 genes were introduced between EcoRI and NotI. For dbld10, the coding sequence from cDNA (LD35990) was placed after its own promoter. GFP, TAP-tag, or tdTomato were fused at the C terminus between NotI and XbaI. P-element-mediated germline transformations were performed by BestGene (Chino Hills, CA).
Antibody:
Anti-peptide antibodies to Asl were produced by Immunology Consultants Laboratory (Newberg, OR). N-terminal (2891) Asl antibody was generated against the peptide LDRQEEEEALQDQKRREEEL-C. The antibody against Asl C-terminal (AP1193) was generated against the peptide C-LERRSREKHRDKENV and purified by affinity.
Immunofluorescence staining and imaging:
Embryos aged between 0 and 3 hr were collected on grape agar plates. They were dechorionated and fixed according to Rothwell and Sullivan (2000). Brains or testes were dissected in saline solution (0.7% NaCl) and fixed 5 min in formaldehyde (3.7% in PBS). After squashing, the coverslip was removed using liquid nitrogen and slides were placed in methanol for 2 min. After washing in PBS, the preparations were permeabilized with PBS 0.1% TritonX-100 for 10 min and saturated with PBS 1% BSA, 0.1% TritonX-100. Antibody staining was performed for 1 hr at room temperature followed by three washes with PBS. The following primary antibodies were used: mouse anti-γ-tubulin (1:200; Sigma), rabbit anti-cnn (1/200; Megraw and Kaufman 2000), mouse anti-FasIII (1:50; Developmental Studies Hybridoma Bank), rabbit anti-Asl (AP1193; 1/200, homemade), rabbit anti-Asl (2891) (1/500, homemade), rabbit anti-α-tubulin (1:200; Lab Vision), or rat anti-α-tubulin (1/200, Chemicon). All fluorescent secondary antibodies were from Jackson Immunoresearch and used at 1/200: rhodamine goat anti-mouse, Cy5 goat anti-mouse, Cy5 donkey anti-rabbit, Cy5 donkey anti-rat, FITC goat anti-rabbit, and rhodamine goat anti-rabbit. Anti-Asl (AP1193) (1/200) immunostaining in testis was performed according to Varmark et al. (2007). DAPI (at 1 μg/ml; Sigma) was used to stain DNA. The slides were then mounted in mounting media (Biomedia) and examined using a Leica TCS SP5 scanning confocal microscope. Images were processed using Adobe Photoshop. We quantified the total intensity (pixel sum) of a defined region around one centriole with the Leica LAS AF software and statistical analysis was done with GraphPad Prism5 or Excel. Imaging of the sensory organ was performed as described previously (Avidor-Reiss et al. 2004). Zebrafish nasal pit basal bodies were visualized by staining with the mouse monoclonal anti-γ-tubulin antibody (1:1000) as described previously (Tsujikawa and Malicki 2004; Omori and Malicki 2006)
Electron microscopy of isolated centrosomes:
Drosophila embryo extract was prepared as described previously (Moritz et al. 1995). The centrosomal fractions were fixed in 3.7% paraformaldehyde plus 0.1% glutaraldehyde in 80 mm K-PIPES, pH 6.8, 1 mm MgCl2, 1 mm Na3EGTA at 4° for 10 min. The fixed centrosomes were sedimented onto a previously discharged ACLAR coverslip (Ted Pella, Redding, CA). The centrosomes were post-fixed with 3% glutaraldehyde for 10 min followed by 1% osmium tetroxide and 0.5% potassium ferricyanide in 0.1 m cacodylate buffer, pH 7.4, for 15 min at 4°. The specimens were stained overnight with 1% aqueous uranyl acetate at 4° followed by dehydration through graded cold alcohol series and brought to room temperature with absolute alcohol. The samples were subsequently embedded onto epon/araldite using standard protocols and remounted for thin serial sections. Thin sections of ∼70 nm were cut using the Reichert ultracut microtome and the thin sections were collected on Formvar-coated copper grids. The specimens were post-stained with 1% uranyl acetate in 50% methanol followed by aqueous lead citrate and viewed in a Tecnai G Spirit BioTWIN transmission electron microscope (FEI, Hillsboro, OR) operated at 80 kV.
For immunolabeling, the fixed centrosomes were initially blocked with a buffer of 2% BSA in Tris buffer saline containing 0.1% cold-water fish-skin gelatin. The samples were subsequently labeled using mouse monoclonal anti-GFP (Clontech) diluted to 1:100 in the same buffer and then with rabbit anti-mouse followed by protein A with 5 nm colloidal gold. To enhance the visualization of small gold particles, we have recorded the tilted image.
Tissue processing for electron microscopy:
EM analysis of macrochaetae and fly testis was done as previously described (Eberl et al. 2000; Avidor-Reiss et al. 2004). For EM analysis of stem sperm cells, testes of wild-type and aslmecD pupae were dissected and processed by fixing them with 2.5% glutaraldehyde, post-fixed with 1% OsO4, and embedded according to standard EM procedures. The osmium-fixed tissues were further incubated overnight with 1% aqueous uranyl acetate at 4° followed by dehydration with 50 and 100% cold alcohol. Ultrathin sections of 70 nm were cut using Leica UltraCut UCT ultramicrotome. The sections on slot grids were counterstained with 1% uranyl acetate and lead citrate and viewed with Tecnai G2 Spirit BioTWIN at 80 kV. Zebrafish electron microscopy was performed according to standard protocols (Tsujikawa and Malicki 2004).
Functional studies in zebrafish:
The structure of the cep152 gene in zebrafish is based on publicly available genomic data. In knockdown experiments, morpholinos were injected into wild-type AB embryos at the one- to two-cell stage as described previously (Tsujikawa and Malicki 2004). The following morpholinos were used: TTGACTGGCTGACCTGCAGGGC TTT targeted to the cep152 splice site and CCTCTTACCT CAGTTACAATTTATA as a control. The efficiency of morpholino knockdown was determined by RT–PCR using primers flanking the target intron as described previously (Tsujikawa and Malicki 2004). To prepare dcep152 mRNA for rescue experiments, the full-length fly asl sequence was inserted into the pXT7 vector and used as template for in vitro transcription performed with a message machine kit (Ambion). asl and GFP mRNA were injected into embryos along with morpholinos as described previously (Tsujikawa and Malicki 2004).
Western blot:
The equivalent of five brains were collected from third instar larvae, boiled in SDS sample buffer, and run on an 8% acrylamide gel. Blotted membranes were incubated with anti-Asl antibody (AP1193) (1/20,000) or anti-Asl (2891) (1/10,000) for 1 hr at room temperature. After three washes with PBS 0.2% Tween 20, secondary antibody HRP-anti-rabbit IgG (1/5000; Lab Vision) was applied for 1 hr. Blots were washed as before and treated with chemiluminescent substrate (Pierce) to reveal peroxidase activity. Chemiluminescent signal was detected with auto-radiographic film.
RESULTS AND DISCUSSION
Asl/cep152 is a conserved gene required for ciliogenesis:
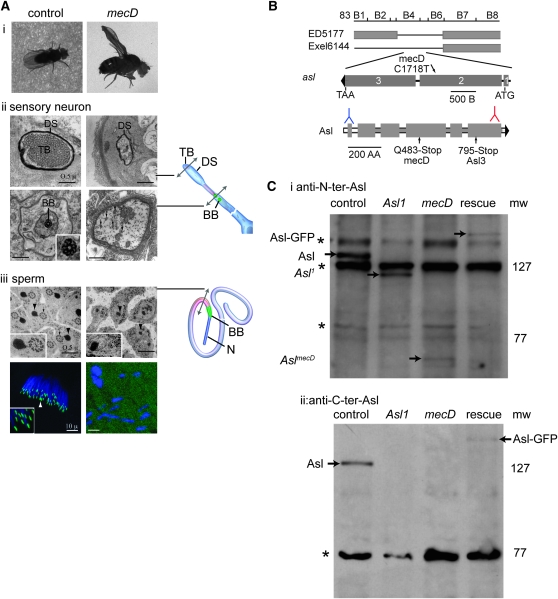
In addition to their role in cellular division, centrioles transform into basal bodies that give rise to cilia. In flies, cilia perform mechanosensory or chemosensory functions in neurons and a motile function as flagella. To identify genes that function in centriole and cilium formation, we screened a collection of ∼600 potential candidate mechanosensory mutants (Avidor-Reiss et al. 2004). The mechanosensory mutant D (mecD) exhibited all of the characteristics of a ciliary defect with a severe phenotype of uncoordination (Figure 1A, i) and nonmotile sperm tails (data not shown). We confirmed the absence of cilia by serial-section electron microscopy (EM) of the sensory neurons and the sperm tails (Figure 1A, ii and iii). In wild-type sensory neurons, the tip of the cilium had a tubular body and a pair of basal bodies at the base of the sensory cilium (see diagram in Figure 1A, ii). The basal bodies and sensory cilium were missing in mecD (Figure 1A, ii). Spermatid tails were still formed in mecD but contained abnormal mitochondrial derivatives and the axonemes were missing completely (Figure 1A, iii). Analysis using light microscopy showed that in control spermatids each nucleus is associated with a basal body [GFP–PACT (pericentrin/AKAP450 centrosomal targeting); Martinez-Campos et al. 2004] and that in mecD spermatids no basal bodies were found and the tissue is disorganized (Figure 1A, iii). These findings firmly implicate mecD in cilia and basal bodies biogenesis.
Figure 1.—
Asl is required for cilia formation. (A) Ciliary defect in mecD mutants. (i) In contrast to control animals, mecD flies cannot stand on their legs, which are crossed, and their wings extend upward. (ii) EM cross sections of the cilia of mechanosensory neurons; in contrast to the control, in the mutants the dendrite tip marked by the dendritic sheath (DS) lacks the tubular body (TB) (top). At the base of sensory cilium (bottom), control but not mecD flies have basal bodies (BB) (inset). In mecD flies, only fragments of the tubular body (arrows) are found. (iii) EM cross sections of spermatid tails; the axoneme (arrows) is missing in the mutant and mitochondrial derivatives (arrowheads) are abnormal (top). Inset shows higher magnification of mitochondrial derivatives and axoneme. A Z-projection of a spermatid cyst where basal bodies labeled by GFP–PACT (arrowhead and inset) are sitting below the nuclei (DAPI, blue) in control and are absent in mutants (bottom). (B) Genetic map illustrating the chromosomal deficiencies (indicated by thin lines) that do not complement aslmecD. The aslmecD mutation introduces a C-to-T transformation near the end of the second exon. aslmecD mutation transforms Q 483 in the fourth coiled-coil domain (shaded squares) to an early stop codon. For comparison, the stop position in asl3 is indicated: in blue, the recognition site of the N-terminal antibody (2891), and in red, the recognition site of the C-terminal antibody (AP1193). (C) Endogenous and GFP fusion proteins are well recognized by both antibodies. The N-terminal antibody (i) detects a faint band of the short Asl protein in aslmecD and a modest level of a larger protein in asl1. Nothing is detected in mutants with the C-terminal antibody (ii). Asterisk is a nonspecific band.
We mapped mecD and found a nonsense mutation in the gene CG2919, which transforms CAG (Q483) into the stop codon TAG (Figure 1B) at position 1718 of the coding sequence. An independent study showed that CG2919 is the gene mutated in the three asterless mutants but mutation in the coding region was found only in asl3 (Varmark et al. 2007). We generated two antibodies, one against the N-terminal part (amino acids 21–40) and the other against the C terminus (aa 958–972). Both antibodies recognize in Western blot the endogenous protein and Asl-GFP fusion (Figure 1C). In accordance with the mutation location in mecD, the C terminus antibody did not detect any Asl protein in this allele. Using the N-terminal antibody, we detect a faint band ∼60 kDa in mecD mutants, indicating that the predicted truncated Asl protein is very unstable. Similar to previous study (Varmark et al. 2007) using the C-terminal antibody, we did not detect a specific Asl band in asl1 mutants. Surprisingly, in asl1 homozygotes, the N-terminal antibody identifies a truncated protein ∼3/4 of the full-length protein that is expressed at moderate level. This result demonstrates that, whatever the asl1 mutation is, an abnormal Asl protein is present while in mecD, Asl is almost completely missing.
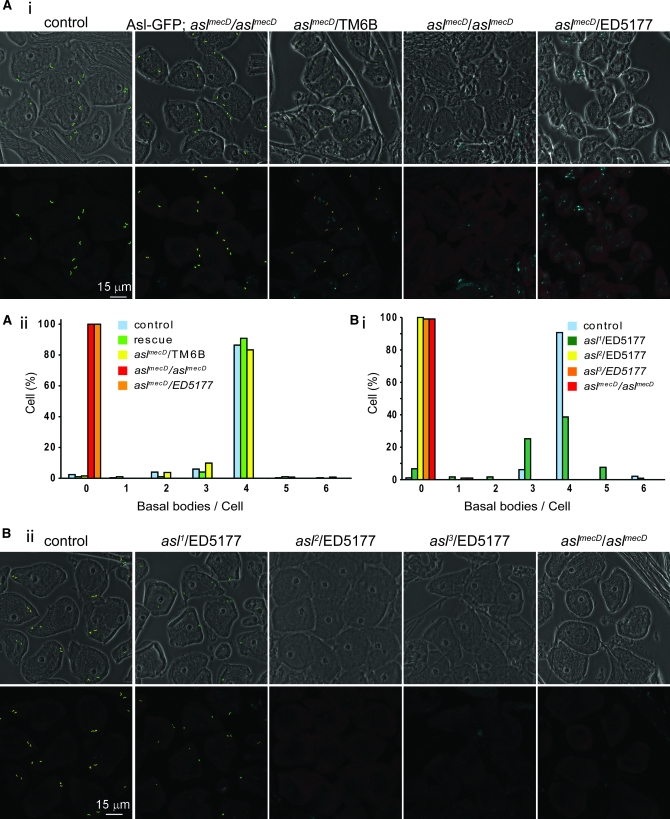
Consistent with the protein data, the phenotype of aslmecD over a deficiency is identical to the aslmecD homozygotes, suggesting that aslmecD is a genetically null or a severe loss-of-function allele (Figure 2A). We generated transgenic flies expressing Asl-GFP under the control of the asl/dcep152 promoter. This construct rescues the phenotype of aslmecD and generates a healthy stock, indicating that it is a functional reporter (Figure 2A). Heterozygote and homozygote flies expressing the wild-type copy of Asl tagged with GFP (Asl-GFP, rescue) have a similar phenotype to the control, ruling out the possibility that aslmecD is a gain-of-function allele (Figure 2A). When placed over a deficiency, the alleles asl2 and asl3 have been shown to have an uncoordinated phenotype (Varmark et al. 2007) similar to that of aslmecD, and asl3 has an early stop codon at position 795 (Varmark et al. 2007), suggesting that these two alleles are also severe loss-of-function alleles. To verify this, we expressed the centriolar protein Ana1 (Goshima et al. 2007) fused to GFP to look for the presence of basal bodies in spermatocytes of asterless mutants over the deficiency ED5177 (Figure 2B). As we expected, as with aslmecD, asl2 and asl3 show a total absence of basal bodies. In asl1 mutants, as previously reported, basal bodies are present (Varmark et al. 2007); however, >1/3 of the cells present a loss of basal bodies (Figure 2B, i).
Figure 2.—
Allelic series analysis of Asl mutations (A) (i) Phase contrast (top) and fluorescent (bottom) photos show that aslmecD homozygous mutants and hemizygotes are devoid of basal bodies whereas heterozygotes, rescue, and control flies have a similar number of centrioles. (ii) Quantification of the number of basal bodies per cell for one representative experiment; for each genotype, ∼100 cells have been counted and two to three flies have been checked. (B) (i) Ana1-GFP labeling shows that basal bodies are present in asl1 and control spermatocytes but are absent in asl2,3 and mecD. Graph shows quantification of basal bodies per cell; for each genotype, ∼100 cells have been counted (ii). Similar results were obtained using Ana1 antibody (data not shown).
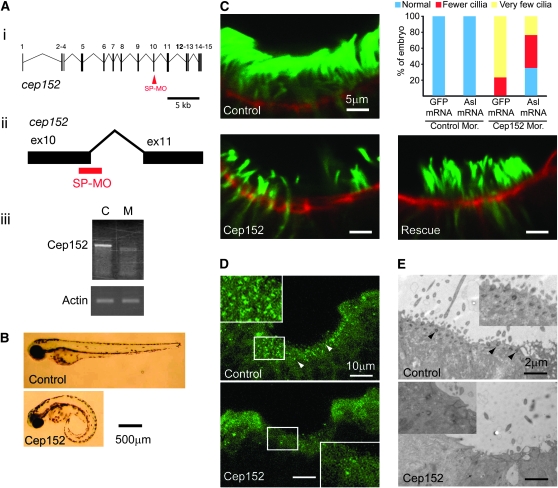
To investigate whether asl could serve as a model for the vertebrate cep152 function, we downregulated cep152 activity in zebrafish and examined cilia formation. An anti-splice site morpholino that targets the zebrafish embryonic cep152 RNA eliminates a 92-bp exon and produces a frameshift in the open reading frame (Figure 3A). Approximately 50% (46/86) of embryos treated with this compound feature a curly tail (Figure 3B), a phenotype characteristic of ciliary defects in zebrafish (Tsujikawa and Malicki 2004; Zhao and Malicki 2007). In addition, the number of cilia in the nasal epithelium is reduced in morphant animals by >50% and the remaining cilia are reduced in length by ∼45% (Figure 3C; data not shown). In another experiment, we analyzed the number of basal bodies in the nasal pits of cep152 morphant animals by staining with anti-γ-tubulin antibody (Figure 3, D and E). In the wild type, the number of basal bodies per arbitrary unit of surface area is 0.58 (n = 399, 11 nasal pits), while in morphant siblings this number is reduced by ∼70% to 0.19 (n = 126, 11 nasal pits) (Figure 3D). We confirmed by electron microscopy that the number of basal bodies and cilia were reduced in the morphant nasal epithelium (Figure 3E). While cross sections through nasal pits of control animals feature ∼1.1 basal bodies per an arbitrary unit of length, ∼0.5 basal bodies are found in cep152 morphants (data based on electron microscopy of tissue from two morphant and two control animals). In addition, we were able to partially rescue cilia length and number by expressing Drosophila asl in cep152 morphant animals (Figure 3C; data not shown). These findings indicate that Asl/Dcep152 function is conserved between insects and vertebrates.
Figure 3.—
Asl function is conserved in zebrafish. (A) Anti-Cep152 morpholino treatment produces a shorter product that lacks 92 bp and contains a frameshift. (i and ii) Location of morpholino target site (red) in the zebrafish Cep152 gene. Gene structure is predicted largely on the basis of publicly available genomic data. (iii) RT–PCR amplification of the Cep152 or Actin transcript at 24 hr post-fertilization in embryos treated with a control (C) or anti-Cep152 morpholino (M). (B) Curly tail, a phenotype characteristic of ciliary defects, is observed in Cep152 morphant larvae. (C) Quantification of cilia number in olfactory pits of morphant embryos. GFP or Asl mRNA are injected along with a control (control mor.) or anti-Cep152 splice site morpholino (Cep152 mor.). Anti-acetylated tubulin antibody (green) was used to stain cilia, and phaloidin (red) marks the apical surface of the tissue. Colored bars indicate the relative quantities of embryos in three phenotypic categories: blue, 50–100% of the wild-type cilia number; red, 25–50%; and yellow, <25%. For each group, n ≥ 10. (D) Staining of basal bodies (arrowheads) in the nasal pits of control and Cep152 morphant embryo with antibodies to γ-tubulin. Insets show higher magnification of areas in white boxes. (E) The apical surface of nasal pits in control and Cep152 morphant embryos visualized by electron microscopy. Basal bodies are indicated with arrowheads. Insets show higher magnification of this tissue.
Asl is closely associated with the centriole wall but is not part of its structure:
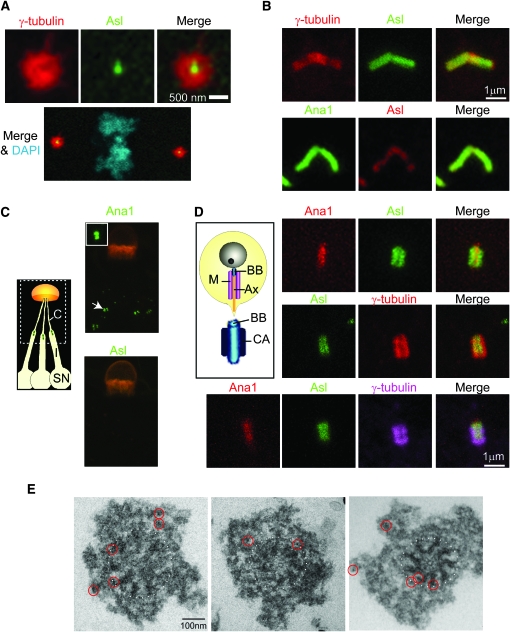
To gain insight into Asl function, we studied its localization using Asl-GFP or the antibody. Like its vertebrate ortholog (Andersen et al. 2003) and consistent with Varmark et al. (2007), we found that Asl is a component of the centrosomes (Figure 4A) and spermatocyte basal bodies (Figure 4B). However, unlike the centriolar/basal body protein Ana1 (Goshima et al. 2007), in sensory neurons Asl is absent from basal bodies (Figure 4C), showing that Asl is not a permanent component of centrioles/basal bodies. We also checked Asl localization in basal bodies in spermatids and we found that Asl surrounds the basal body and does not overlap with Ana1 (Figure 4D). At this stage, EM studies describe the formation of a PCM structure called “centriolar adjunct” (diagram in Figure 4D) that surrounds the basal body and contains γ-tubulin (Tates 1971; Wilson et al. 1997). Staining with anti-γ-tubulin antibody shows that Asl is part of the centriolar adjunct but is localized more centrally than γ-tubulin (Figure 4D). This result indicates that Asl localizes at the boundary between the centriole and the PCM.
Figure 4.—
Asl is a pericentriolar protein closely associated with the centriole wall. (A) In the early embryo, Asl-GFP localizes to the center of the centrosome. The mitotic PCM is marked by γ-tubulin. DAPI stains chromosomes. (B) In spermatocytes, Asl-GFP (top) and endogenous Asl labeled with antibody (bottom) mark the basal body along its length. (C) Unlike the centriolar protein Ana1, Asl is not a component of the basal body of sensory neurons (arrow). The cuticular structure is highlighted by auto-fluorescence (orange). Inset: zoom on basal body. Diagram depicting the location of the basal body (green) and cilia (C) in the sensory organ (SN). (D) Diagram depicting in early spermatids the PCM/centriolar adjunct (CA) surrounding the basal body (BB). M, mitochondria, Ax, axoneme. Asl-GFP (green) is localized to the centriolar adjunct surrounding the basal body marked by Ana1-tdTomato (red) (top). γ-Tubulin staining labels the centriolar adjunct but is even more peripheral than Asl (middle and bottom). (E) Immuno-EM of centrosomes purified from fly embryos expressing Asl-GFP; the 5-nm gold particles are circled in red. Dashed lines indicate the border of the centriole.
In centrioles, the 100- to 200-nm-thick PCM that closely associates with the centriole wall is hard to distinguish from the centriole using light microscopy. Therefore, to directly examine the localization of Asl at higher resolution, we performed immunoelectron microscopy using an anti-GFP antibody on biochemically purified centrosomes (Figure 4E). We observed gold particles in close proximity to the centriole wall but also some gold particles located farther away in the PCM cloud. No gold particles were observed in wild-type centrosomes that did not contain GFP. In agreement with Varmark et al. (2007), we predominantly detected Asl at the periphery of the centriole but, in addition, our preparation reveals a localization of Asl farther in the PCM.
To further test if Asl is firmly attached to the centriole, we examined Asl biochemically. In a sucrose gradient with 100 mm KCl, Asl is found as a component of the centrosome (high density) and in the cytosol (low density) (supplemental Figure 1A). Under these conditions, the PCM proteins Cnn and γ-tubulin are also cofractionated with the centrosome. A high concentration of KCl eliminates the centrosomal fraction of γ-tubulin, Cnn, and Asl (supplemental Figure 1A). In contrast, Ana1 is only partially stripped from the centrosomes under these conditions. This result shows that Asl is loosely associated with the centriole wall or is part of the PCM.
Finally, fluorescent study shows that Asl does not reorganize during cellular division like Cnn or γ-tubulin (mitosis or meiosis, supplemental Figure 1B). However, it was previously reported that the PCM is not homogeneous and that some centrosomal proteins are closely associated with the centriole to form a distinct PCM compartment called the “PCM tube” while others are more peripheral (Bornens 2002; Ou et al. 2003). Taken together, we have a similar conclusion to that of Varmark et al. (2007)—that Asl is localized at the periphery of the centriole—but we showed that it is not a structural component of the centriole. In addition, some Asl is found farther in the PCM compartment and it is possible that Asl is a PCM tube protein.
Asl is required for the initiation of centriole duplication:
The observation that Asl is a centrosomal protein and that aslmecD lacks basal bodies prompted us to test if Asl was essential for centriole formation, maintenance, or function. We analyzed wild-type and mutant larval brain cells during mitosis and confirmed the absence of Asl in aslmecD (supplemental Figure 2A). Unlike in control cells, the centriolar marker GFP–PACT and PCM markers γ-tubulin and Cnn do not label any centrosomal structure in aslmecD (Figure 5A). We repeated this analysis using Ana-1-GFP, Dsas6-GFP, and Dbld10/Cep135-GFP, which were previously shown to be centriolar (Goshima et al. 2007; Hiraki et al. 2007; Kleylein-Sohn et al. 2007; Rodrigues-Martins et al. 2007a). This analysis confirmed our original observations (supplemental Figure 2, B–D) and demonstrated that aslmecD brain cells are missing centrioles and centrosomes. This analysis also confirmed previous publications showing that the absence of centrosomes does not impair the cell's ability to undergo normal mitosis (Heald et al. 1997; Bonaccorsi et al. 2000; La Terra et al. 2005; Basto et al. 2006).
Figure 5.—
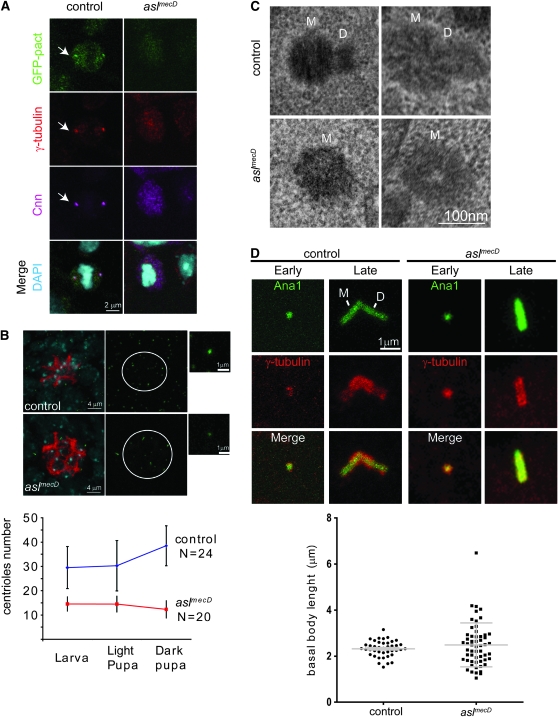
Asl is essential for daughter centriole formation. (A) In control metaphase cells, the centriolar marker GFP–PACT colocalizes with γ-tubulin and Cnn at the spindle pole (12 of the 12 cells analyzed). In aslmecD, chromosomes (DAPI) are normal but centrosomes are absent (8 of the 8 cells analyzed). (B) Asl is not essential for centriole maintenance. In wild-type testes, cells are filled with centrioles (Ana1-GFP, green) while in aslmecD a population of MC centrioles (green) is found at the tip of the testes (white circle) marked with anti-FasIII (red). DAPI-stained DNA is blue; insets provide magnification of centrioles. Graph below shows quantification of the number of centrioles in the tip of the testes at different stages of development. (C) EM sections of the tip of the testis show a pair of mother (M) and daughter (D) centrioles as control and a single centriole (M) in aslmecD testis. (D) Spermatocytes expressing Ana1-GFP (green) and stained for γ-tubulin (red) show mother and daughter centrioles in control as a 𝒱-shape. In aslmecD, γ-tubulin staining is normal but only one centriole can be seen. The basal body length is similar in control and aslmecD (graph below).
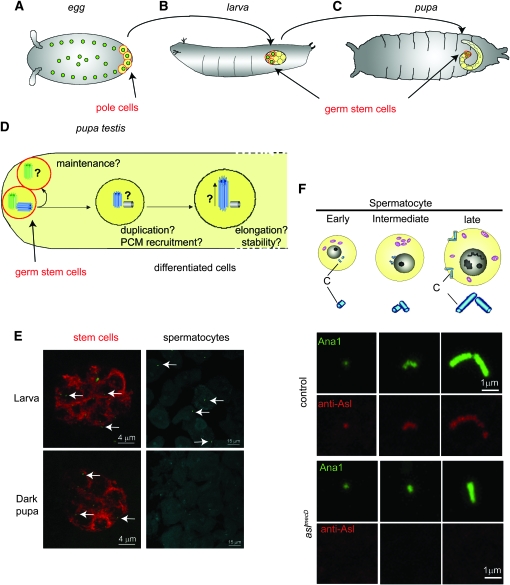
To determine the origin of the acentriolar phenotype in aslmecD, we developed a method that allowed us to study wild-type centrioles in a mutant context (Figure 6). In flies, the egg contains a stock of centrosomal proteins (Rodrigues-Martins et al. 2007b) from the mother (maternal contribution) that is used for centriole duplication until transcription starts. Consequently, in aslmecD, the wild-type Asl protein provided by the mother allows for the formation of normal centrioles. We call these “maternally contributed” centrioles (MC centrioles). Over the course of development the wild-type Asl protein will be lost, placing these MC centrioles in a mutant context where we can observe the centriolar functions that require the Asl protein. We followed the fate of the MC centrioles in the testis, which has several advantages: (1) stem cells in the tip of the testis always conserve the mother centriole, which depends on the PCM to do so (Yamashita et al. 2007); (2) the cellular differentiation of germ cell is linear along the testis, so we can follow the MC centrioles through distinct stages of differentiation; and (3) centrioles in testis elongate to form basal bodies that become very easy to see in light microscopy.
Figure 6.—
Maternally contributed centrioles: a method for studying centriolar protein function. (A) In the embryos of flies, a stock of wild-type proteins is provided by the heterozygous mother until transcription starts. It allows for the formation of a certain number of wild-type centrioles: MC centrioles (in green). Polar cells (red) localized in the posterior part of the embryos contain MC centrioles and give rise to germ stem cells. (B and C) Polar cells become the germline stem cells of the larva and the pupa. In pupae, they are localized at the tip of the testis and can be stained with anti-fasIII antibody (E and Figure 5B). (D) In stem cells, the mother centriole (green) stays attached to the cell while the daughter centriole (blue) is inherited by differentiating cells. In mutants, we can ask if MC centrioles are maintained in the stem cells. We can study the fate of the daughter centrioles (blue) during differentiation and ask if they are capable of duplicating or elongating. Are they stable and conserve the same size during time? Are they able to recruit PCM? While new centrioles are not formed after Asl depletion, the fate of the few daughter centrioles formed during the time of maternal contribution can be analyzed by dissecting animals at larval, early, and late pupal stages. (E) In aslmecD, MC centrioles (green) stay in the tip of the testis marked by fasIII antibody (red) from larval to dark pupal stages. In contrast, in spermatocytes some MC centrioles are present in larvae, but at the pupal stage they have disappeared as the spermatocytes that had a centriole in larvae have differentiated into spermatids. (F) During spermatocyte growth, centrioles (C) duplicate and elongate (diagram). Spermatocytes at different stages stained by anti-Cter-Asl (AP1193) show absence of Asl on MC centrioles in aslmecD.
Using Ana1-GFP to visualize centrioles in aslmecD, we observed MC centrioles in a pool that was always present at the tip of the testis (Figure 5B and Figure 6E). Using specific anti-fasIII antibodies, we confirmed that the cells containing MC centrioles were the stem cells (Yamashita et al. 2007). Additionally, the MC centrioles remained in the stem cells until late pupal stages (Figure 6E), indicating that, in aslmecD, MC centrioles are stable over time and conserve the ability to stay attached to the stem cells. In aslmecD, unlike in the control, the differentiating sperm cells did not show any Ana1-GFP staining. This indicates that, in the absence of Asl, either the MC centrioles lost the capacity to duplicate or the newly formed daughter centrioles failed to incorporate Ana1.
To discriminate between these two possibilities, we performed serial-section EM of the tip of the testis in control and aslmecD (Figure 5C). We found that in wild-type testes the mother centriole was frequently (n = 9/13) associated with a daughter centriole. In aslmecD, no daughter centriole was found near the MC centrioles (n = 0/9). These findings demonstrate that Asl is required for the initiation of centriole duplication. Interestingly, no centriole intermediates were observed in EM of aslmecD, which correlates with the total absence of any centriolar markers tested in neuroblasts (supplemental Figure 2). The combination of these results firmly indicates that Asl acts very early in centriole duplication.
The MC centrioles in aslmecD are morphologically normal in EM and stay in the stem cells, suggesting that they are structurally normal and functional. To confirm this impression, we tested their ability to elongate and recruit PCM. In testis, centrioles duplicate in early spermatocytes and then elongate to reach 2 μm before meiosis (diagram in Figure 6F), forming a 𝒱-shape-like pair where it is possible to distinguish the mother centriole from the daughter (Figure 5D, left). In aslmecD larvae, some MC centrioles are present in spermatocytes (Figure 6E). Later, in aslmecD pupae, MC centrioles become basal bodies of the mature sperm and the new spermatocytes are totally devoid of centrioles (Figure 6E). We performed an immunostaining on these MC centrioles, using anti-Asl antibody to show that there is no residual Asl (Figure 6F, bottom). This correlates with the fact that Asl shows a rapid turnover in centrioles after photo bleaching (Varmark et al. 2007). In aslmecD, only one elongated centriole could be seen in the late spermatocyte stage (Figure 5D, right), confirming that it has failed to duplicate. We measured their size and found that they were similar in aslmecD (2.49 ± 0.95 μm, n = 56) compared to the control (2.32 ± 0.35 μm, n = 41) (Figure 5D). However, the length variability is higher in aslmecD possibly because MC centrioles on average are much older than the normal population of centrioles. We confirmed that Asl is needed for duplication but it is dispensable for the elongation of the centriole.
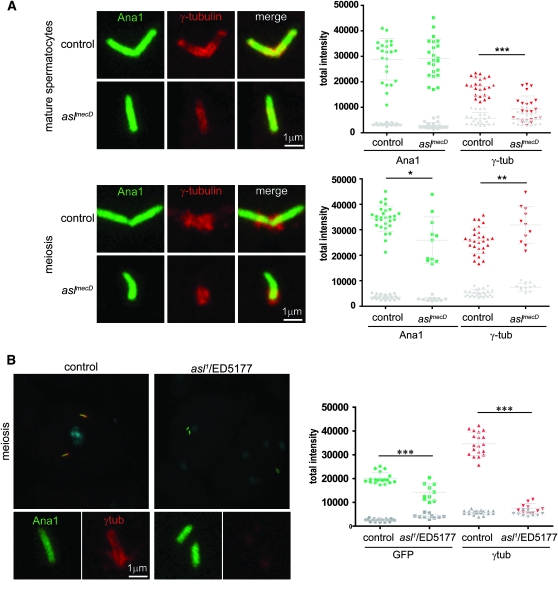
To test if Asl plays a role in PCM recruitment, we immunolabeled the MC centrosomes for the PCM proteins γ-tubulin and Cnn (Figure 7A; data not shown). We found that in both control and aslmecD the PCM decorates the centriole before and after elongation. Quantification of the intensity of γ-tubulin staining found that a substantial amount of PCM accumulates in meiosis and that there is a reduction in aslmecD mutants in mature spermatocytes. Additionally, in aslmecD meiosis, the DNA associates most often with the MC centriole, suggesting that MC centrioles lacking Asl are able to function as microtubule organization center (supplemental Figure 3A). This indicates that Asl is not required for PCM recruitment, which correlates with the fact that MC centrioles are maintained in the stem cells, which in turn is dependent on a functional PCM (Yamashita et al. 2007). However, in aslmecD the shape of the PCM recruited in meiosis is slightly different from that of the control (supplemental Figure 3B). In the control, γ-tubulin spreads largely around the centriole, while in aslmecD cells, it is more concentrated near the centriole wall. Considering the localization of Asl near the centriole wall, it is possible that Asl has a role in organizing the proteins in the PCM periphery.
Figure 7.—
PCM recruitment is not impaired in aslmecD. (A) In aslmecD, quantification of the total intensity of the γ-tubulin (red) signal per centriole shows a significant reduction in mature spermatocytes but normal amounts in meiosis. In the graphs (right), green indicates the intensity of Ana1-GFP used as a control and gray the intensity of the background. (B) In contrast and as previously reported, asl1 shows a dramatic reduction of the total intensity of γ-tubulin signal per centriole in meiosis.
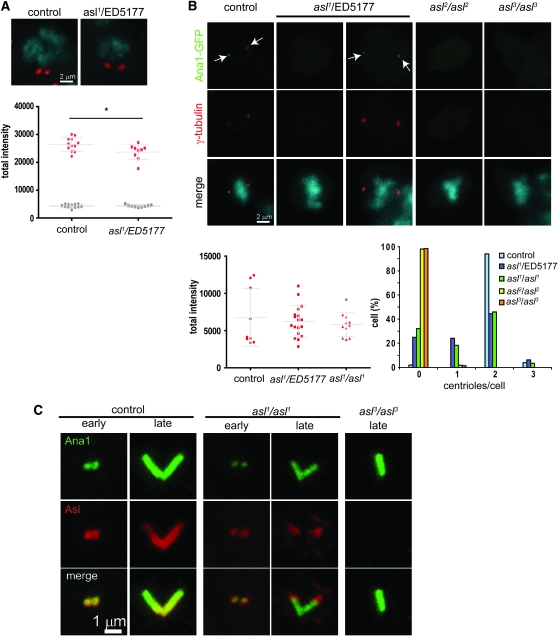
It was reported previously that in asl1 PCM assembly/stability was strongly impaired (Bonaccorsi et al. 1998; Varmark et al. 2007), a phenotype that is missing in aslmecD. To rule out the possibility that the difference between these two results is due to a technical reason, we checked the presence of PCM in asl1 over a deficiency. Similar to Varmark et al. (2007), we found a large reduction of PCM recrutment in asl1 spermatocytes in interphase (data not shown) as well as in meiosis (Figure 7B). We also observe a reduction of the GFP signal, which can be explained by the fact that basal bodies are smaller in asl1: 1.34 ± 0.12 μm (n = 16) vs. 2.15 ± 0.10 μm (n = 19) in the control. We also noted that, in spermatogonia, the PCM are able to assemble in mitosis and we measured just a slight reduction in the amount (Figure 8A). We hypothesize that the PCM phenotype observed in asl1 is restricted to differentiated germinal cells. To confirm this, we immunostained neuroblasts of asl1 over a deficiency with an anti-γ-tubulin antibody (Figure 8B). Like the control, mitotic cells in asl1 mutants accumulate normal amounts of PCM. We observed similar results for asl1 homozygous mutants (Figure 8B). Like aslmecD, asl2 and asl3 homozygous mutants are totally devoid of centrioles (Figure 8B). As we observed in testis, we noted that half of the mitotic cells in the asl1 mutant exhibit fewer than two centrosomes (Figure 8B). This result suggests that centriole duplication is also impaired in asl1 although in a less severe way compared to the other mutants. But since PCM assembly is not affected in the neuroblast, it indicates that the role of Asl in centriole duplication is not a consequence of the absence of PCM.
Figure 8.—
asl1 accumulates a normal amount of PCM in mitosis. (A) Spermatogonia in mitosis of asl1 hemizygotes are able to assemble PCM. Quantification of the total intensity of γ-tubulin signal shows a slight reduction in asl1compared with control. (B) Mitotic cells in larval brains of asl1 homozygotes accumulate normal amounts of PCM. However, half of the cells contain fewer than two centrosomes. Like aslmecD mutants (Figure 5A), asl2 and asl3 are devoid of centrosomes. (C) In asl1 mutant testis expressing Ana1-GFP, the anti-N-terminal Asl antibody detects the Asl1 protein on centrioles in young spermatocytes, but shows abnormal localization of Asl1 to the distal part of the basal body in mature spermatocytes. MC centrioles present in asl3 larval testis are not stained by anti-Asl.
Since Asl1 protein is detected in Western blot by the N-terminal antibody, we decided to check the presence of Asl in asl1 mutant testis. Similar to the control, we observed in spermatogonia and young spermatocytes that the Asl1 protein colocalizes on centrioles with Ana1-GFP (Figure 8C). In the control, when the centrioles start to elongate to form basal bodies, Asl localizes all along the length of the basal body. In asl1 mutants during this same period, the protein is restricted to the distal part of the basal body. This result suggests that the asl1 mutation affects the regulation of the protein. In asl3 mutants, we did not detect any labeling on the MC centrioles present in larval testis (Figure 8C). This shows, as previously, that the maternal Asl protein has been depleted and that, if any truncated protein is synthesized, it is unable to localize to the centriole.
Our analysis of the different alleles show that asl1 expresses an abnormal Asl protein and presents a less severe phenotype than the other three alleles, aslmecD, asl2, and asl3, which are severe loss-of-function alleles. To date, most studies that describe Asl function in detail have been done on the asl1 allele (Bonaccorsi et al. 1998; Varmark et al. 2007), which explains why Asl was not implicated in centriole/cilia formation. Considering the entirety of our work, we showed that Asl is required for centriole duplication. The PCM stability phenotype in asl1 may be the consequence of an abnormal regulation of Asl protein during spermatocyte differentiation. Alternatively, it is formally possible that absence of some aspect of Asl function early in centriole duplication can lead later to a defect in PCM stability/accumulation and that this phenotype is not observed in aslmecD mutant since MC centrioles are formed at a time when Asl was present.
Conclusion:
In this article we have identified a new allele of the asl gene. We demonstrated that Asl is involved in the initiation of centriole duplication and that it is dispensable for PCM recruitment in meiosis in MC centrioles. We show that Asl is essential for centriole formation in brain cells and spermatocytes. Without centrioles, cells do not have basal bodies and cannot grow cilia, which is the cause of the adult lethality of these animals. Similar results are observed in zebrafish, indicating that Asl function in centriole duplication is conserved in animals. The absence of any intermediate centriolar structure in EM, plus the lack of centriolar markers such as Sas6, which is known to be involved in the early steps of centriole duplication, strongly suggests that Asl acts very early in the process. We excluded the possibility that the failure in centriole duplication is due to growth abnormalities of the mother centriole since it is surrounded by functional PCM and is able to elongate. Asl is closely associated with the centriole wall and could be part of the PCM tube. EM studies show that procentriole formation occurs at 30 nm from the mother centriole wall (Anderson and Brenner 1971), which correlates with the localization of Asl. Therefore it will be interesting in the future to see if the role of Asl and the PCM tube is to anchor and stabilize the daughter centriole nucleation site. This would be a simple mechanism to ensure that centriole duplication occurs in the vicinity of the mother centriole.
Acknowledgments
We thank the Harvard Medical School Nikon Imaging Center, EM facility, Peg Coughlin, and Susumu Ito for valuable help with fluorescent and electron microscopy; scientific illustration by Jeff Dixon; Developmental Studies Hybridoma Bank at the University of Iowa (supported by the National Institute of Child Health and Human Development); and C. Goodman for anti-FasIII antibody, J. W. Raff for the GFP–PACT flies, T. C. Kaufman for Cnn antibodies, and C. Gonzalez for asl flies. We also thank Charles S. Zuker, Tom Rapoport, Michael Friedlander, and John Flanagan for suggestions and comments. This work was supported partially by a grant from the Giovanni Armenise-Harvard Foundation. S. B. was supported by a Fondation pour la Recherche Medicale grant. A.P. was supported by Russian Foundation of Basic Research (RFBR) grant 07-04-01127.
References
- Andersen, J. S., C. J. Wilkinson, T. Mayor, P. Mortensen, E. A. Nigg et al., 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426 570–574. [DOI] [PubMed] [Google Scholar]
- Anderson, R. G., and R. M. Brenner, 1971. The formation of basal bodies (centrioles) in the Rhesus monkey oviduct. J. Cell Biol. 50 10–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor-Reiss, T., A. M. Maer, E. Koundakjian, A. Polyanovsky, T. Keil et al., 2004. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell 117 527–539. [DOI] [PubMed] [Google Scholar]
- Basto, R., J. Lau, T. Vinogradova, A. Gardiol, C. G. Woods et al., 2006. Flies without centrioles. Cell 125 1375–1386. [DOI] [PubMed] [Google Scholar]
- Bonaccorsi, S., M. G. Giansanti and M. Gatti, 1998. Spindle self-organization and cytokinesis during male meiosis in asterless mutants of Drosophila melanogaster. J. Cell Biol. 142 751–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi, S., M. G. Giansanti and M. Gatti, 2000. Spindle assembly in Drosophila neuroblasts and ganglion mother cells. Nat. Cell Biol. 2 54–56. [DOI] [PubMed] [Google Scholar]
- Bornens, M., 2002. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 14 25–34. [DOI] [PubMed] [Google Scholar]
- Boveri, T., 1887. Ueber die Befruchtung. der Eier von Ascaris megalocephala. Sitz.-Ber. Ges. Morph. Phys. Mçnchen. 3 394–443. [Google Scholar]
- Brinkley, B. R., 2001. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol. 11 18–21. [DOI] [PubMed] [Google Scholar]
- Dammermann, A., T. Muller-Reichert, L. Pelletier, B. Habermann, A. Desai et al., 2004. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell 7 815–829. [DOI] [PubMed] [Google Scholar]
- Delattre, M., C. Canard and P. Gonczy, 2006. Sequential protein recruitment in C. elegans centriole formation. Curr. Biol. 16 1844–1849. [DOI] [PubMed] [Google Scholar]
- Doxsey, S., 1998. The centrosome: a tiny organelle with big potential. Nat. Genet. 20 104–106. [DOI] [PubMed] [Google Scholar]
- Eberl, D. F., R. W. Hardy and M. J. Kernan, 2000. Genetically similar transduction mechanisms for touch and hearing in Drosophila. J. Neurosci. 20 5981–5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa, K., 2007. Oncogenes and tumour suppressors take on centrosomes. Nat. Rev. Cancer 7 911–924. [DOI] [PubMed] [Google Scholar]
- Gonczy, P., C. Echeverri, K. Oegema, A. Coulson, S. J. Jones et al., 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408 331–336. [DOI] [PubMed] [Google Scholar]
- Goshima, G., R. Wollman, S. S. Goodwin, N. Zhang, J. M. Scholey et al., 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald, R., R. Tournebize, A. Habermann, E. Karsenti and A. Hyman, 1997. Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 138 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraki, M., Y. Nakazawa, R. Kamiya and M. Hirono, 2007. Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr Biol. 17 1778–1783. [DOI] [PubMed] [Google Scholar]
- Keller, L. C., E. P. Romijn, I. Zamora, J. R. Yates, III and W. F. Marshall, 2005. Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 15 1090–1098. [DOI] [PubMed] [Google Scholar]
- Kilburn, C. L., C. G. Pearson, E. P. Romijn, J. B. Meehl, T. H. Giddings, Jr. et al., 2007. New Tetrahymena basal body protein components identify basal body domain structure. J. Cell Biol. 178 905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn, J., J. Westendorf, M. Le Clech, R. Habedanck, Y. D. Stierhof et al., 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell 13 190–202. [DOI] [PubMed] [Google Scholar]
- Koundakjian, E. J., D. M. Cowan, R. W. Hardy and A. H. Becker, 2004. The Zuker collection: a resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics 167 203–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Terra, S., C. N. English, P. Hergert, B. F. McEwen, G. Sluder et al., 2005. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J. Cell Biol. 168 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. B., J. M. Gerdes, C. J. Haycraft, Y. Fan, T. M. Teslovich et al., 2004. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117 541–552. [DOI] [PubMed] [Google Scholar]
- Martinez-Campos, M., R. Basto, J. Baker, M. Kernan and J. W. Raff, 2004. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 165 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw, T. L., and T. C. Kaufman, 2000. The centrosome in Drosophila oocyte development. Curr. Top. Dev. Biol. 49 385–407. [DOI] [PubMed] [Google Scholar]
- Moritz, M., M. B. Braunfeld, J. C. Fung, J. W. Sedat, B. M. Alberts et al., 1995. Three-dimensional structural characterization of centrosomes from early Drosophila embryos. J. Cell Biol. 130 1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, K. F., C. M. Leys and J. G. White, 1998. A genetic screen for temperature-sensitive cell-division mutants of Caenorhabditis elegans. Genetics 149 1303–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori, Y., and J. Malicki, 2006. oko meduzy and related crumbs genes are determinants of apical cell features in the vertebrate embryo. Curr. Biol. 16 945–957. [DOI] [PubMed] [Google Scholar]
- Ou, Y. Y., M. Zhang, S. Chi, J. R. Matyas and J. B. Rattner, 2003. Higher order structure of the PCM adjacent to the centriole. Cell Motil. Cytoskeleton 55 125–133. [DOI] [PubMed] [Google Scholar]
- Pelletier, L., N. Ozlu, E. Hannak, C. Cowan, B. Habermann et al., 2004. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr.. Biol. 14 863–873. [DOI] [PubMed] [Google Scholar]
- Pelletier, L., E. O'Toole, A. Schwager, A. A. Hyman and T. Muller-Reichert, 2006. Centriole assembly in Caenorhabditis elegans. Nature 444 619–623. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins, A., M. Bettencourt-Dias, M. Riparbelli, C. Ferreira, I. Ferreira et al., 2007. a DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr. Biol. 17 1465–1472. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins, A., M. Riparbelli, G. Callaini, D. M. Glover and M. Bettencourt-Dias, 2007. b Revisiting the role of the mother centriole in centriole biogenesis. Science 316 1046–1050. [DOI] [PubMed] [Google Scholar]
- Rothwell, W. F., and W. Sullivan, 2000. Fluorescent analysis of Drosophila embryos, pp. 141–157 in Drosophila Protocols, edited by W. Sullivan, M. Ashburner and R. S. Hawley. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shang, Y., C. C. Tsao and M. A. Gorovsky, 2005. Mutational analyses reveal a novel function of the nucleotide-binding domain of gamma-tubulin in the regulation of basal body biogenesis. J. Cell Biol. 171 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tates, A. D., 1971. Cytodifferentiation During Spermatogenesis in Drosophila melanogaster: An Electron Microscope Study. Rijksuniversiteit de Leiden, Leiden, The Netherlands.
- Tsujikawa, M., and J. Malicki, 2004. Intraflagellar transport genes are essential for differentiation and survival of vertebrate sensory neurons. Neuron 42 703–716. [DOI] [PubMed] [Google Scholar]
- van Beneden, E., 1876. Recherches sur les Dicyemides, sur-vivants actuels d'un embranchement des mesozoaires. Bull. Acad. R. Belg. A Sci. (Ser. 2) 41 1160–1205. [Google Scholar]
- Varmark, H., S. Llamazares, E. Rebollo, B. Lange, J. Reina et al., 2007. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr. Biol. 17 1735–1745. [DOI] [PubMed] [Google Scholar]
- Wilson, P. G., Y. Zheng, C. E. Oakley, B. R. Oakley, G. G. Borisy et al., 1997. Differential expression of two gamma-tubulin isoforms during gametogenesis and development in Drosophila. Dev. Biol. 184 207–221. [DOI] [PubMed] [Google Scholar]
- Yamashita, Y. M., A. P. Mahowald, J. R. Perlin and M. T. Fuller, 2007. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science 315 518–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C., and J. Malicki, 2007. Genetic defects of pronephric cilia in zebrafish. Mech. Dev. 124 605–616. [DOI] [PubMed] [Google Scholar]