Abstract
Infectious diseases during pregnancy can impact the development of fetal immunity, leading to reduced neonatal resistance to infection and decreased responses to pediatric vaccines. P. falciparum causes placental infection in low parity pregnant women and is among the pathogens that affect fetal immunity. Recognizing the relationship between malaria and γδ T lymphocytes in adults, we asked whether neonatal γδ T cells would be altered in malaria-endemic regions as a marker for changes in fetal immunity. Our initial studies compared cord blood γδ T cells from deliveries to HIV- mothers in Jos (Nigeria) where malaria is endemic, or in Rome (Italy). We noted substantial differences in the Vγ2 repertoire for cord blood collected in Jos or Rome; differences were consistent with a negative selection mechanism operating on the fetal Vγ2 chain repertoire in neonates from Jos. A specific disruption affected the fraction of γδ T cells that we expect will respond to Bacille Calmette-Guerin (BCG). Fetal γδ T cell depletion might be a mechanism for impaired neonatal immunity and lowered responses to pediatric vaccines.
Keywords: cord blood, gammadelta T lymphocytes, Vγ2 repertoire, Jγ1.2, Nigeria
Introduction
Microbial antigens can cross the placental barrier and prime fetal immunity, often creating immunological memory that persists into early childhood (Malhotra et al., 1999). However, in some cases prenatal priming tolerizes the neonatal immune system and lowers resistance to pathogen exposure in the infant. Placental malaria for example, generates fetal CD4 T-regulatory cells specific for plasmodial antigens and these cells, through IL10 production, suppress protective neonatal T cell responses against Plasmodium (Brustoski et al., 2006) and possibly B cell responses as well (Dent et al., 2006). Children born to mothers with placental malaria have increased susceptibility to malaria in early childhood (Le Hesran et al., 1997). Similarly, in utero sensitization to helminthes suppresses neonatal responses to helminthes (Hitch et al., 1997; Lammie et al., 1991; Steel et al., 1994) but it also decreases responses to mycobacterium BCG vaccination through a non-specific mechanism (Malhotra et al., 1999). Since we know that γδ T cells comprise an important component of T cell responses to BCG and also respond to plasmodia in adult individuals (Dieli et al., 2001), we decided to explore whether maternal infectious diseases impact neonatal γδ T cells. In utero sensitization to pathogens might affect fetal γδ T cell responses either directly, by altering the γδ T cell repertoire, or indirectly by altering neonatal accessory/antigen-presenting cells.
One subset of γδ T lymphocytes, Vγ2Vδ2+ cells are present at low frequency in thymus or cord blood (Casorati et al., 1989; Parker et al., 1990), where a dominant fraction expresses the Vδ1 chain paired mostly with Vγ1 (Morita et al., 1994). Cord blood also contains cells expressing gamma delta chain combinations that are infrequent in adult peripheral blood, such as Vγ2Vδ1 or Vγ1Vδ2 (Morita et al., 1994). Within a few years after birth, the number of Vγ2Vδ2 T cells increases because of strong selection for the Vγ2-Jγ1.2 rearrangement (Parker et al., 1990). It is believed that stimulation by self- or ubiquitous non-self antigens amplifies and maintains the Vγ2-Jγ1.2Vδ2 population, thus creating the biased adult repertoire (Davodeau et al., 1993b; Parker et al., 1990).
Vγ2Vδ2 T cells are stimulated by small non-peptidic compounds collectively called phosphoantigens (Tanaka et al., 1995; Tanaka et al., 1994). Many of these molecules are precursors of isoprenoid biosynthesis (Eberl et al., 2003) such as isopentenyl-pyrophosphate (IPP), and are ubiquitous in eukaryotes. Phosphoantigen recognition is TCR-dependent (Bukowski et al., 1998; Morita et al., 1995) but direct binding to the TCR has not been confirmed with biophysical studies and no antigen presenting molecules are known. Aminobisphosphonates also elicit robust Vγ2Vδ2 lymphocyte responses but act indirectly, by blocking isoprenoid biosynthesis and inducing IPP accumulation (Gober et al., 2003). Responses to phosphoantigens are MHC unrestricted (Morita et al., 1995; Schild et al., 1994) and depend on the Vγ2-Jγ1.2 rearrangement since clones bearing other J segments are less reactive (Davodeau et al., 1993a).
Cord blood (CB) Vγ2Vδ2 T cells are considered to be immature because they have naïve phenotypes and display poor proliferative (Montesano et al., 2001) or cytokine responses (Engelmann et al., 2006) to IPP stimulation. However, CB Vδ2 T cells proliferate robustly after treatment with pamidronate or live mycobacteria (BCG), thus they are not inherently unresponsive (Cairo et al., 2008). As a first step toward understanding the mechanisms controlling Vγ2Vδ2 T cells in CB, we compared the repertoire of Vγ2, Vδ2 and Vδ1 chains for CB samples collected in Jos, Nigeria and in Rome, Italy.
Malaria is endemic in Nigeria and prevalence among pregnant women in Jos was reported to be approximately 40% (Egwunyenga et al., 2001; Egwunyenga et al., 1997). In other areas of Nigeria, between 70 and 80% of pregnant women were positive for Plasmodium even in the absence of symptoms (Onyenekwe et al., 2002). Such a prevalence can be explained by a high incidence and/or long duration of infection. Treatment or previous exposures may decrease the duration of infection and re-infection in not easily distinguished from recrudescence in the absence of molecular typing for the parasites. In a setting that mimicked transmission to a naïve host in an endemic region, untreated P. falciparum infection lasted on average about 200 days (Macdonald, 1950; Sama et al., 2006) but some studies found evidence for longer (Sama et al., 2004) or shorter (Sama et al., 2006) parasite persistence in humans that were exposed repeatedly. If we consider a duration of approximately 200 days and a prevalence of 40%, we can appreciate that most women will suffer from Plasmodium infection at least once during pregnancy. Although we did not measure specific parasite burden in maternal blood, it is likely that our samples from Jos reflect this common situation.
Our goal in this study was to collect CB from a random sample of deliveries in a population at high risk for malaria, then look for similarities and differences between neonatal γδ T cells in high (central African) or low risk (European) populations. Our results are consistent with the idea that environmental factors alter fetal Vγ2Vδ2 T cells; likely mediators include malaria and other endemic tropical diseases.
Materials and Methods
Lymphocyte sample collection
Umbilical cord blood samples were obtained by venipuncture of the umbilical vein immediately after uncomplicated full-term vaginal deliveries. Samples were collected at San Pietro-Fatebenefratelli Hospital, in Rome, Italy, and Plateau State Hospital, in Jos, Nigeria. In both sites women donating CB were HIV-. Women enrolled in Rome did not have symptoms of infectious disease during pregnancy; infectious disease history was not known for women enrolled in Jos. HIV prevalence in Jos is approximately 10% among pregnant women (Sagay et al., 2006). HIV serostatus for Nigerian women was confirmed during labor using two different rapid tests, Determine (Abbott) and Stat-Pak (Chembio). In case of discordance, the Genie-II (Sanofi) test was used as a tie-breaker.
Informed consent was obtained from all women donating cord blood. Cord blood mononuclear cells (CBMC) were isolated by centrifugation over Ficoll-Hypaque (Pharmacia, Uppsala, Sweden) density gradient according to the manufacturer’s instructions.
RNA extraction, RT- PCR, PCR
Total RNA was extracted from 1–10×106 cells using the RNeasy mini Kit (Qiagen, Valencia, CA), as described by the manufacturer. One µg of total RNA was then converted into cDNA using the reverse transcription system kit (Promega, Madison, WI), as described previously (Cairo et al., 2007). Polymerase chain reactions were performed as described (Cairo et al., 2007) using the following primers: for the Vγ2 (Vγ9 according to the IMGT nomenclature) chain, oligo Vγ2 (5’ ATC AAC GCT GGC AGT CC 3’) and oligo Cγ-1 (5’ GTT GCT CTT CTT TTC TTG CC 3’); for the Vδ2 chain, oligo Vδ2 (5’ GCC ATT GAG TTG GTG CCT GAA C 3’) and oligo Cδ-1 (5’ TGG CAG TCA AGA GAA AAT TG 3’); for the Vδ1 chain oligo Vδ1 (5’ TTA ACC ATT TCA GCC TTA CAG C 3’) and oligo Cδ-1 (5’ TGG CAG TCA AGA GAA AAT TG 3’). PCR products were separated on 1% agarose/Tris-acetate-EDTA buffer (TAE) gels containing 0.5µg/ml ethidium bromide.
Run-off reaction
Primer extension reactions were performed as described (Evans et al., 2001). Each reaction contained 2µl of PCR product, 3 mM MgCl2, 0,2 mM dNTP, 0.1 µM of 6-carboxyfluorescein (6-FAM) labeled primer (Cγ-6: 5’-6-FAM–AAT AGT GGG CTT GGG GGA AAC-3’; Jγ1.2: 5’-6-FAM–; Cδ: 5’-6-FAM–ACG GAT GGT TTG GTA TGA GG-3’), approximately 0.2 units of Taq DNA polymerase (Promega, Madison, WI), 10 mM Tris pH8.8, and 50 mM KCl. 4µl of run-offs products were diluted with 6µl deionized formamide (Applied Biosystems, Foster City, CA), and 0.7µl of GeneScan-500 ROX size standard were added to each sample. After a denaturation step (5 min at 95°C followed by immediate quenching on ice) products were loaded on a 3130 genetic analyzer (Applied Biosystems, Foster City, CA) and run on a performance-optimized polymer (POP-7). Molecular size and relative frequency of extension products were determined using GeneMappaer software (Applied Biosystems, Foster City, CA). In order to standardize the data irrespective of the run-off primer position, CDR3 length variation was expressed in terms of the total Vγ2, Vδ2 and Vδ1 coding region lengths. Run-off product lengths were corrected by adding the length of the known mRNA coding region outside the run-off primer-binding site. For Vδ2 and Vδ1 chains, the sequences used as reference for calculations are those listed in the IMGT website (accession number: M22198 for Vδ1; X15207 for Vδ2; M22148, M22149 and M22150 for Cδ), plus the sequence for the human alpha/delta locus (accession NG 001332). For the Vγ2 chain, according to our convention (Evans et al., 2001) the peak at 993 nucleotides corresponds to a product 444 nucleotide long.
Cloning and sequencing of Vγ2 chains
PCR products for the Vγ2 chain were purified by gel extraction, using QIAquick gel extraction kits (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Purified products were denatured (1 minute at 94°C), then incubated for 30 minutes at 72°C with 2mM MgCl2, 0.2mM dATPs, and 2.5 units of Amplitaq gold, then ligated into a pCR2.1 vector. Ligated vector was transfected into TOP 10F’ competent cells (TA cloning kit, Invitrogen, Carlsbad, CA), and bacterial colonies representing a library of Vγ2 chain sequences were grown overnight on agar plates containing 50µg/ml ampicillin, 500 µM IPTG and 80µg/ml X-Gal. Colonies containing recombinant plasmids were cultured overnight in LB media and plasmid DNA were purified using the REAL Minipreps DNA purification kit (Qiagen, Valencia, CA). Sequencing reactions were done with a Big Dye v3.1 fluorescent sequencing kit (Applied Biosystems, Foster City, CA), with both M13F and M13R oligonucleotide primers for each sample. Sequences were loaded on an automated sequencer ABI3700 and analyzed using Sequencher and MacClade softwares. We were unable to unambiguously identify C1 and C2 chains based on the C sequence that we determined, hence the Jγ1.3 and Jγ2.3 segments (J1 and J2, respectively, according to the IMGT nomenclature) were not distinguished and were pooled together as Jγ1.3/2.3
Statistical analysis
Differences among groups were evaluated using two-tailed Student’s t test. p< 0.05 was considered significant.
Results
γδ T cells for CB collected in Rome or Jos differed in Vγ2 chain length distributions
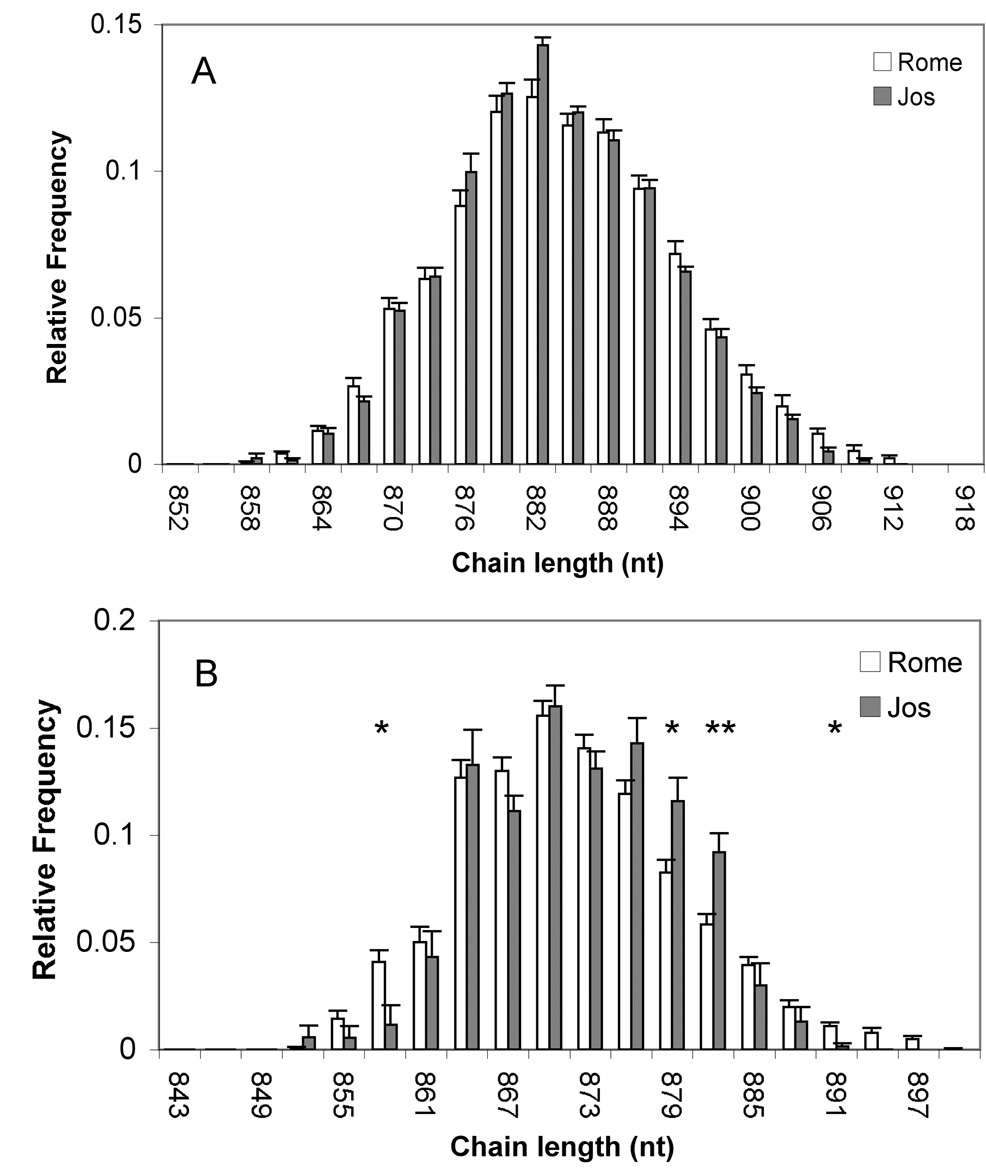
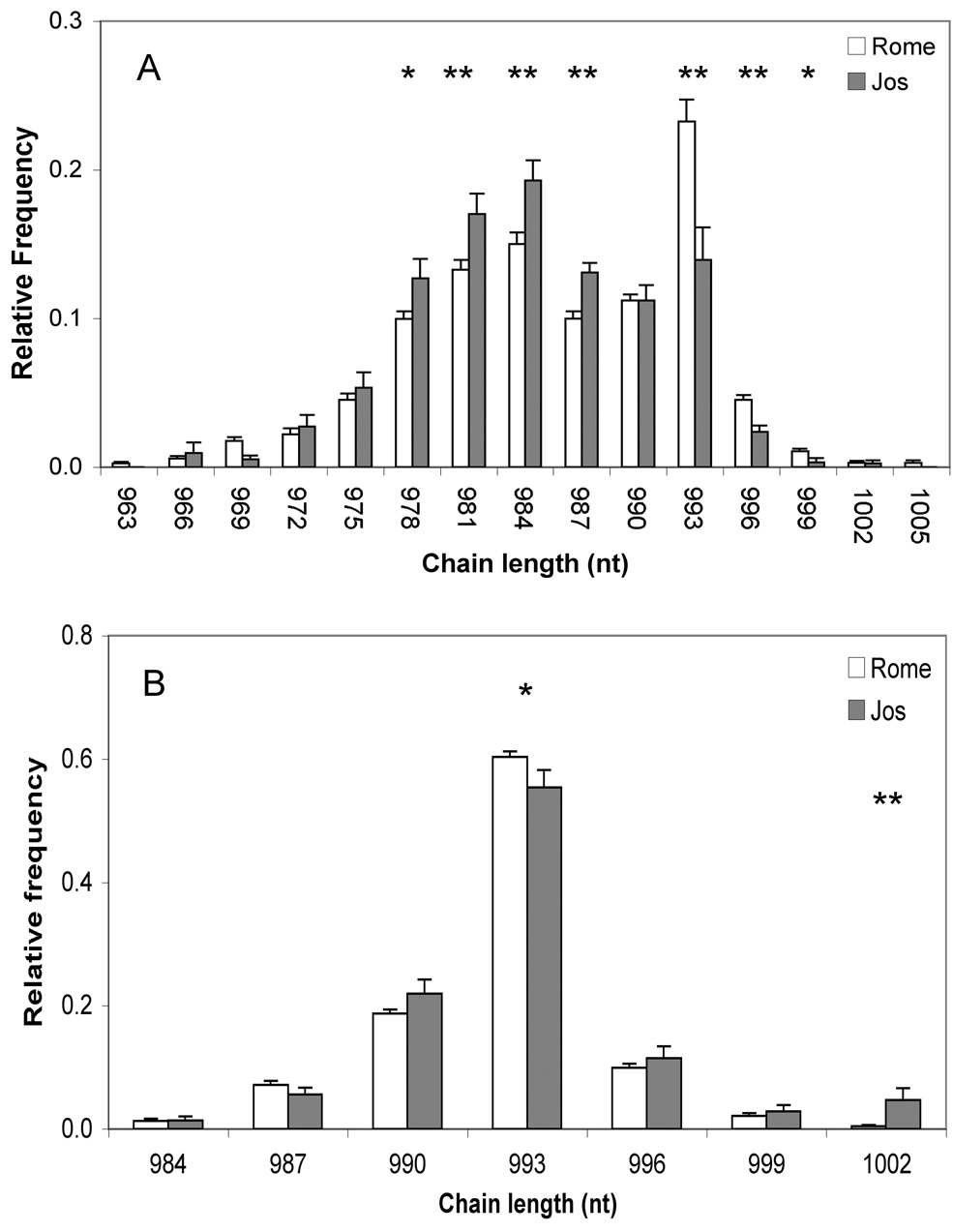
As a test for differences in the neonatal γδ TCR repertoire between CB samples collected in Rome (CB-R) or Jos (CB-J), we compared spectratype data for 35 Italian and 17 Nigerian CB specimens. The Vδ1 chain length distributions were similar for both groups, with mode at 882 nucleotides (nt) (Fig 1A). The Vδ2 chain lengths were also similar (modal value = 870 nt) although some longer chains were more frequent in CB-J (Fig 1B). The Vγ2 (Vγ9 according to the IMGT nomenclature) chain length distributions for both CB groups were bimodal, the first peak being 984 and the second 993 nt (Fig 2A). CB-R had a higher frequency of chains between 990 and 996 nt compared to CB-J (p=0.01); this was of interest because most Vγ2 chains in the 990–996 length range from adults include the Jγ1.2 (JP according to the IMGT nomenclature) segment (Evans et al., 2001). About 90% of the Jγ1.2+ chains fell in the 990–996 length range for both CB groups (Fig 2B).
Figure 1. Vδ1 and Vδ2 chain length distributions are similar for CB-R and CB-J.
Vδ1 (A) and Vδ2 (B) chain length distributions were analyzed by spectratyping for 34 CB collected in Rome and 17 CB collected in Jos. Panels show, for each chain length, mean frequency + SE. *=p<0.01; **=p<0.001
Figure 2. The frequency of Vγ2 chains in the 900–996 nt range is higher for CB-R.
Vγ2 chain length distribution (A) was analyzed by spectratyping for 34 CB collected in Rome and 17 CB collected in Jos. The distribution of Jγ1.2+ chains (B) was also examined by using a Jγ1.2 specific primer. Panels show, for each chain length, mean frequency + SE. *=p<0.05; **=p<0.01
The Vγ2 repertoire for CB samples collected in Rome had higher Jγ1.2 frequency and lower population complexity
Based on spectratyping results, we chose 6 representative CB-R and 5 CB-J specimens, then created Vγ2 cDNA libraries from mRNA. For each sample, we sequenced between 200 and 300 Vγ2 cDNA clones in order to have sequences for at least 100 productively rearranged chains from each specimen. For all CB samples, Vγ2 chain length distributions determined by cDNA sequencing agreed with distributions measured by spectratyping (data not shown). In particular, 92.2%±3.1% (for CB-R) and 84.2%±12.5% (for CB-J) of the Vγ2-Jγ1.2 chains fell into the 990–996 length range. Among all Vγ2 chains between 990 and 996 nt long, 79.3%±9.8% were Jγ1.2+ for CB-R, but only 45.6%±11.8% were Jγ1.2+ for CB-J (p<0.001).
Table 1 shows an overview of sequencing results for productively rearranged Vγ2 chains from both CB groups. The frequency of Vγ2-Jγ1.2 nucleotide sequences (or nucleotypes) was significantly higher for CB collected in Rome (41.5%±15.3% vs. 13.9%±8.5%, p=0.01). The Vγ2 repertoire complexity (defined as: number of different Vγ2 nucleotypes ÷ number of total Vγ2 sequence entries) was higher for CB-J. Because of the way complexity is defined, a repertoire dominated by nucleotypes that are present multiple times (repeated) will have lower complexity, while a repertoire dominated by unique nucleotypes (that occur only once in the pool) will have higher complexity. The two CB groups showed similar complexities for the subset of Vγ2 chains that include Jγ1.3 or 2.3 segments (Vγ2-Jγ1.3/2.3 complexity = number of different Jγ1.3/2.3 nucleotypes ÷ number of total Jγ1.3/2.3 sequence entries). When we examined the subset with Vγ2-Jγ1.2 chains, complexity was significantly lower for CB-R (0.55±0.1 for CB-R vs 0.8±0.17 for CB-J, p=0.015). Lower complexity often indicates a repertoire bias caused either by preferential recombination or positive selection mechanisms that allow individual V-J rearrangement to accumulate.
Table 1.
CB samples collected in Nigeria have a lower Jγ1.2 frequency and higher population complexity
| CCB22 | CCB23 | CCB29 | CCB35 | CCB38 | CCB42 | Average | ||
|---|---|---|---|---|---|---|---|---|
| Jγ1.2 | 51 (36.7%) | 33 (29.2%) | 93 (66.4%) | 67 (45.3%) | 17 (18.9%) | 76 (52.4%) | 41.5% | |
| Jγ1.3/2.3 | 81 (58.3%) | 72 (63.7%) | 43 (30.7%) | 78 (52.7%) | 63 (70%) | 57 (39.3%) | 52.5% | |
| Jγ1.1 | 6 (4.3%) | 8 (7.1%) | 4 (2.9%) | 3 (2%) | 5 (5.5%) | 8 (5.5%) | 4.6% | |
| Jγ2.1 | 1 (0.7%) | nd | nd | nd | 5 (5.5%) | 4 (2.8%) | 1.5% | |
| Total # | 139 | 113 | 140 | 148 | 90 | 145 | 129 | |
| Vγ2 Complexity | 0.75 | 0.72 | 0.68 | 0.79 | 0.73 | 0.71 | 0.73 | |
| Jγ1.2 Complexity | 0.65 | 0.36 | 0.56 | 0.63 | 0.53 | 0.55 | 0.55 | |
| Jγ1.3/2.3 Complexity | 0.79 | 0.88 | 0.91 | 0.92 | 0.81 | 0.88 | 0.87 |
| NCB4 | NCB5 | NCB7 | NCB18 | NCB19 | Average | |||
|---|---|---|---|---|---|---|---|---|
| Jγ1.2 | 12 (9.5%) | 8 (7%) | 11 (8%) | 25 (16.1%) | 43 (28.9%) | 13.9% | ||
| Jγ1.3/2.3 | 108 (85.7%) | 104 (90.4%) | 120 (87%) | 119 (76.8%) | 91 (61%) | 80.2% | ||
| Jγ1.1 | 3 (2.4%) | 2 (1.7%) | 2 (1.5%) | 6 (3.9%) | 7 (4.7%) | 2.8% | ||
| Jγ2.1 | 3 (2.4%) | 1 (0.9%) | 4 (2.9%) | 4 (2.6%) | 8 (5.4%) | 2.8% | ||
| Total # | 126 | 115 | 138 | 155 | 149 | 137 | ||
| Vγ2 Complexity | 0.92 | 0.8 | 0.92 | 0.75 | 0.79 | 0.84 | ||
| Jγ1.2 Complexity | 0.92 | 1 | 0.82 | 0.64 | 0.61 | 0.80 | ||
| Jγ1.3/2.3 Complexity | 0.92 | 0.78 | 0.93 | 0.78 | 0.88 | 0.86 |
The table reports, for every CB specimen, the number (percentage) of chains bearing each J segment, the total number of productively rearranged Vγ2 chains and population complexity for the Vγ2 pool as well as for the two major subsets, the Jγ1.2 and the Jγ1.3/2.3. Complexity is calculated as: number of different nucleotyes in a sample set ÷ total number of nucleotypes in the sample set
Particular Jγ1.2 nucleotypes were abundant (Supplementary Material S1 and S2); the most common was a canonical sequence (where canonical refers to any sequence that lacks N or P nucleotides and is entirely encoded by the germ-line) that was detected previously in fetal liver (McVay and Carding, 1996) and in CB (Delfau et al., 1992). This nucleotype was the only one present in all CB samples and its frequency within the Vγ2-Jγ1.2 pool ranged from 7.8% to 45.5% (Table 2). The proportion of this canonical nucleotype within the entire Vγ2 pool was significantly lower for CB-J samples, although its proportion within the Jγ1.2 subset was similar in both groups (Table 2). The high frequency of a canonical nucleotype in CB-R specimens is consistent with preferential production and/or positive selection of these cell clones in CB-R specimens. Assuming that the same recombination/selection mechanisms would also occur in the CB-J samples, differences for the Jγ1.2 subsets are likely to result from negative selection in the CB-J specimens.
Table 2.
The canonical nucleotype (GGGAGGTG CAAGAG) frequency in the Vγ2 pool is lower for CB-J, but within the Jγ1.2 subset it is similar for CB-R and CB-J
| % within Jγ1.2 | % within Vγ2 | ||
|---|---|---|---|
| CCB22 | 7.8 | 3.6 | |
| CCB23 | 45.5 | 13.3 | |
| CCB29 | 33.3 | 22.1 | |
| CCB35 | 29.9 | 13.5 | |
| CCB38 | 41.2 | 7.7 | |
| CCB42 | 32.8 | 17.2 | |
| Average | 31.8 | 12.9 | |
| NCB4 | 8.3 | 0.8 | |
| NCB5 | 12.5 | 0.9 | |
| NCB7 | 27.3 | 2.2 | |
| NCB18 | 36 | 5.8 | |
| NCB19 | 39.5 | 11.4 | |
| Average | 24.7 | 4.2 | |
The number and proportion of public Vγ2-Jγ1.2 clonotypes is higher for CB collected in Rome
For each specimen, we analyzed the deduced aminoacid sequences (clonotypes). In the Jγ1.2 and Jγ1.3/2.3 subsets we observed clonotypes that were present in more than one CB specimen (public clonotypes). The number of public Jγ1.2 clonotypes was higher for CB-R than for CB-J (23 versus 5) and the fraction of Jγ1.2 chains encoding public clonotypes was also higher (on average 68.5% for CB-R and 38.3% for CB-J, p<0.001) (Table 3). CB-R had a similar number of public Jγ1.3/2.3 clonotypes compared to CB-J (21 versus 26), and the fraction of Jγ1.3/2.3 chains encoding public clonotypes was the same for both (Table 4). Several public Jγ1.2 clonotypes were abundant, while public Jγ1.3/2.3 clonotypes were found only 3 times or less in any sample.
Table 3.
The fraction of public Jγ1.2 clonotypes is lower for CB-J than for CB-R
| A | ||||||||
|---|---|---|---|---|---|---|---|---|
| Vγ2 | N | Jγ1.2 | CCB22 | CCB23 | CCB29 | CCB35 | CCB38 | CCB42 |
| CALWE----AL--QELGKKIKVFGPGTKLIIT | 1 | 1 | ||||||
| CALWE----D---QELGKKIKVFGPGTKLIIT | 1 | 1 | ||||||
| CALWE----E----ELGKKIKVFGPGTKLIIT | 1 | 1 | ||||||
| CALWE---------ELGKKIKVFGPGTKLIIT | 3 | 1 | 2 | |||||
| CALWE----G---QELGKKIKVFGPGTKLIIT | 1 | 1 | ||||||
| CALWE----I---QELGKKIKVFGPGTKLIIT | 1 | 1 | 2 | |||||
| CALWE----K----ELGKKIKVFGPGTKLIIT | 1 | 1 | ||||||
| CALWE----P---QELGKKIKVFGPGTKLIIT | 2 | 1 | 3 | |||||
| CALWE--------QELGKKIKVFGPGTKLIIT | 4 | 5 | 3 | 1 | ||||
| CALWE----R---QELGKKIKVFGPGTKLIIT | 2 | 2 | ||||||
| CALWE----S---QELGKKIKVFGPGTKLIIT | 1 | 1 | ||||||
| CALWEV---E----ELGKKIKVFGPGTKLIIT | 1 | 2 | 1 | |||||
| CALWEV--------ELGKKIKVFGPGTKLIIT | 3 | 2 | 3 | 3 | ||||
| CALWEV---G----ELGKKIKVFGPGTKLIIT | 3 | 1 | ||||||
| CALWEV---H----ELGKKIKVFGPGTKLIIT | 2 | 1 | 1 | |||||
| CALWEV---K----ELGKKIKVFGPGTKLIIT | 3 | 1 | 1 | |||||
| CALWEV---L----ELGKKIKVFGPGTKLIIT | 1 | 2 | 1 | |||||
| CALWEV-------QELGKKIKVFGPGTKLIIT | 7 | 15 | 33 | 22 | 10 | 27 | ||
| CALWEV---Q-----LGKKIKVFGPGTKLIIT | 3 | 1 | 1 | |||||
| CALWEV---R----ELGKKIKVFGPGTKLIIT | 5 | 4 | 1 | 7 | ||||
| CALWEV---RG---ELGKKIKVFGPGTKLIIT | 1 | 1 | ||||||
| CALWEV---R---QELGKKIKVFGPGTKLIIT | 1 | 1 | ||||||
| CALWEV---RS---ELGKKIKVFGPGTKLIIT | 1 | 2 | ||||||
| Total # of public Jγ1.2 | 28 | 25 | 60 | 37 | 15 | 55 | ||
| Total # of Jγ1.2 | 51 | 33 | 93 | 67 | 17 | 76 | ||
| Fraction of public Jγ1.2 (%) | 54.9 | 75.8 | 64.5 | 55.2 | 88.2 | 72.4 | ||
| B | |||||||
|---|---|---|---|---|---|---|---|
| Vγ2 | N | Jγ1.2 | NCB4 | NCB5 | NCB7 | NCB18 | NCB19 |
| CALWE----A---QELGKKIKVFGPGTKLIIT | 1 | 1 | |||||
| CALWE----D---QELGKKIKVFGPGTKLIIT | 1 | 1 | |||||
| CALWE----------LGKKIKVFGPGTKLIIT | 1 | 1 | |||||
| CALWE--------QELGKKIKVFGPGTKLIIT | 1 | 3 | |||||
| CALWEV-------QELGKKIKVFGPGTKLIIT | 1 | 1 | 3 | 10 | 17 | ||
| Total # of public Jγ1.2 | 3 | 3 | 4 | 10 | 22 | ||
| Total # of Jγ1.2 | 12 | 8 | 11 | 25 | 43 | ||
| Fraction of public Jγ1.2 (%) | 25.0 | 37.5 | 36.4 | 40.0 | 51.2 | ||
All public Vγ2-Jγ1.2 clonotypes are listed for both CB-R (A) and CB-J (B). The number of occurrences is reported for every clonotype and the fraction of Jγ1.2 sequences coding public clontypes is calculated as: total number of Jγ1.2 sequences ÷ total number of Jγ1.2 sequences coding public clonotypes.
Table 4.
The fraction of public Jγ1.3/2.3 clonotypes is similar for CB-J than for CB-R.
| A | ||||||||
|---|---|---|---|---|---|---|---|---|
| Vγ2 | N | Jγ1.3/2.3 | CCB22 | CCB23 | CCB29 | CCB35 | CCB38 | CCB42 |
| CALWE---AP---NYYKKLFGSGTTLVVT | 1 | 1 | ||||||
| CALWE---D-------KKLFGSGTTLVVT | 1 | 1 | ||||||
| CALWE---D-----YYKKLFGSGTTLVVT | 1 | 2 | 1 | |||||
| CALWE---G-------KKLFGSGTTLVVT | 1 | 1 | ||||||
| CALWE---LS---NYYKKLFGSGTTLVVT | 1 | 1 | 1 | |||||
| CALWE---M----NYYKKLFGSGTTLVVT | 1 | 1 | ||||||
| CALWEV--E--------KLFGSGTTLVVT | 1 | 2 | ||||||
| CALWEV--F-----YYKKLFGSGTTLVVT | 1 | 1 | ||||||
| CALWEV--GG----YYKKLFGSGTTLVVT | 1 | 3 | ||||||
| CALWEV--G------YKKLFGSGTTLVVT | 1 | 2 | 1 | |||||
| CALWEV--H------YKKLFGRGTTLVVT | 2 | 1 | ||||||
| CALWEV-----------KKLFGSGTTLVVT | 1 | 3 | ||||||
| CALWEV--L-----YYKKLFGSGTTLVVT | 2 | 1 | 3 | |||||
| CALWEV-------NYYKKLFGSGTTLVVT | 1 | 1 | 1 | 2 | 2 | |||
| CALWEV--QG-------KLFGSGTTLVVT | 1 | 1 | ||||||
| CALWEV--RG-----YKKLFGSGTTLVVT | 1 | 1 | ||||||
| CALWEV--R-----YYKKLFGSGTTLVVT | 1 | 2 | 1 | |||||
| CALWEV--T-----YYKKLFGSGTTLVVT | 1 | 1 | ||||||
| CALWEV--V------YKKLFGSGTTLVVT | 1 | 1 | ||||||
| CALWEV--------YYKKLFGSGTTLVVT | 2 | 1 | 1 | 1 | ||||
| CALWE---------YYKKLFGSGTTLVVT | 1 | 2 | ||||||
| Total # of public Jγ1.3/2.3 | 12 | 11 | 8 | 12 | 10 | 10 | ||
| Total # of Jγ1.3/2.3 | 81 | 72 | 43 | 78 | 57 | 57 | ||
| Fraction of public Jγ1.3/2.3 (%) | 14.8 | 15.3 | 18.6 | 15.4 | 23.8 | 17.5 | ||
| B | |||||||
|---|---|---|---|---|---|---|---|
| Vγ2 | N | Jγ1.3/2.3 | NCB4 | NCB5 | NCB7 | NCB18 | NCB19 |
| CALWE---A----NYYKKLFGSGTTLVVT | 1 | 1 | 1 | ||||
| CALWE---A-----YYKKLFGSGTTLVVT | 1 | 2 | |||||
| CALWE---D-----YYKKLFGSGTTLVVT | 1 | 2 | 1 | ||||
| CALWE---G----NYYKKLFGSGTTLVVT | 1 | 1 | |||||
| CALWE---G------YKKLFGSGTTLVVT | 1 | 1 | |||||
| CALWE---G-----YYKKLFGSGTTLVVT | 1 | 1 | |||||
| CALWE---LD----YYKKLFGSGTTLVVT | 1 | 1 | |||||
| CALWE---L----NYYKKLFGSGTTLVVT | 1 | 1 | |||||
| CALWE---LP----YYKKLFGSGTTLVVT | 1 | 1 | |||||
| CALWE--------NYYKKLFGSGTTLVVT | 2 | 1 | 1 | ||||
| CALWE---PR---NYYKKLFGSGTTLVVT | 1 | 1 | |||||
| CALWE---S----NYYKKLFGSGTTLVVT | 1 | 1 | |||||
| CALWE---S-----YYKKLFGSGTTLVVT | 1 | 1 | 1 | ||||
| CALWEV--G------YKKLFGSGTTLVVT | 1 | 1 | |||||
| CALWEV--H-----YYKKLFGSGTTLVVT | 1 | 1 | 1 | ||||
| CALWEV--IN------KKLFGSGTTLVVT | 1 | 1 | |||||
| CALWEV----------KKLFGSGTTLVVT | 2 | 1 | |||||
| CALWEV--KY------KKLFGSGTTLVVT | 1 | 1 | |||||
| CALWEV--L------YKKLFGSGTTLVVT | 1 | 1 | |||||
| CALWEV--L-----YYKKLFGSGTTLVVT | 3 | 1 | |||||
| CALWEV-------NYYKKLFGSGTTLVVT | 2 | 1 | |||||
| CALWEV--P-----YYKKLFGSGTTLVVT | 3 | 3 | 1 | ||||
| CALWEV--QG----YYKKLFGSGTTLVIT | 1 | 1 | |||||
| CALWEV--R-------KKLFGSGTTLVVT | 2 | 1 | 2 | ||||
| CALWEV--R------YKKLFGSGTTLVVT | 2 | 2 | 2 | 1 | |||
| CALWEV--------YYKKLFGSGTTLVVT | 2 | 1 | |||||
| Total # of public Jγ1.3/2.3 | 20 | 15 | 23 | 10 | 10 | ||
| Total # of Jγ1.3/2.3 | 108 | 104 | 120 | 119 | 91 | ||
| Fraction of public Jγ1.3/2.3 (%) | 18.5 | 14.4 | 19.2 | 8.4 | 11.0 | ||
All public Vγ2-Jγ1.3/2.3 clonotypes are listed for both CB-R (A) and CB-J (B). The number of occurrences is reported for every clonotype and the fraction of Jγ1.3/2.3 sequences coding public clontypes is calculated as: total number of Jγ1.3/2.3 sequences ÷ total number of Jγ1.2/2.3 sequences coding public clonotypes.
We also noticed that some clonotypes (mostly public) were encoded by more than one nucleotype; these are examples of sequence convergence. For CB-R, convergence occurred mainly among Jγ1.2 chains; for CB-J it occurred mostly among Jγ1.3/2.3 clonotypes (Supplementary Material, S3); this may be due to an overall lower proportion of Jγ1.2 clonotypes in CB-J. These data suggest that positive selection was operating mainly on the Vγ2-Jγ1.2 subset for CB-R specimens. Assuming that the same positive selection was operating in CB-J, the differences between Vγ2Jγ1.2 subsets in CB-R and CB-J are consistent with negative selection that limited or reversed the impact of an underlying positive selection on Vγ2-Jγ1.2 chains in CB-J.
Discussion
Using Vγ2Vδ2 T lymphocytes as a model population, we sought to understand how the fetal immune system is affected by maternal infectious diseases. In this work, we used detailed molecular characterization of CB γδ T cells to measure similarities and differences between specimens collected in Jos, Nigeria, or Rome, Italy. This is our initial step toward understanding environmental factors that affect fetal immunity.
In both CB groups, the Jγ1.3/2.3 subset showed characteristics of an unselected, naïve repertoire: high complexity, absence of prominent clones, and a low fraction of nucleotypes encoding public clonotypes. Importantly, the Jγ1.3/2.3 subset was very similar between the two groups. These results argue strongly against inherent differences in the Vγ2-Jγ1.3/2.3 cell populations and support the view that other differences are likely the result of selection mechanisms. We cannot exclude the impact of genetic factors on the observed repertoire differences. At present, we are studying the cord blood repertoire from African Americans to search for possible genetic confounders. Comparative studies to date of adult Vγ2 repertoire in European, North American and Chinese populations has not revealed any relationship between ethnicity and γδ T cells.
CB-R specimens (cord blood from Rome) had higher Vγ2-Jγ1.2+ chain frequencies and lower complexity compared to CB-J. These differences were due mostly to the low frequency of the dominant Jγ1.2 canonical nucleotype in CB-J specimens. CB-R specimens were characterized by higher proportions of Vγ2-Jγ1.2+ nucleotypes that encode public clonotypes, including the dominant canonical sequence. Though CB γδ T cell populations are still immature compared to the adult repertoire, CB-R specimens showed a pattern consistent with positive selection operating on the Vγ2 chain repertoire and mainly selecting for Vγ2-Jγ1.2+ cells.
The dominant canonical Vγ2-Jγ1.2 sequence, first produced during early ontogeny and detectable in prethymic fetal liver (McVay and Carding, 1996), is common to many human cord or adult blood samples (Davodeau et al., 1993b; Delfau et al., 1992; McVay and Carding, 1999) and its homolog is also common in macaques (Cairo et al., 2007; MacDougall et al., 2001; Rakasz et al., 2000). This sequence, like other canonical nucleotypes, could be generated by homology-mediated recombination (Asarnow et al., 1993; Itohara et al., 1993; Zhang et al., 1995). Such a mechanism may explain why this sequence is common even across species. Few of our Jγ1.2 and 1.3/2.3 canonical nucleotypes are compatible with a directed recombination mechanism. When present, sequences that might result from directed recombination were at frequencies lower than the dominant Jγ1.2 canonical nucleotype. Recombination mechanisms alone do not dictate the frequency of canonical sequences. It is likely that preferential survival allows expansion of clones expressing canonical Vγ2 chains and thus, shapes the fetal Vγ2 repertoire.
Whether the dominant canonical sequence arises by preferential recombination, positive selection or both, we expect processes shaping the Vγ2 repertoire to be similar for fetuses in Jos and Rome. However, a surfeit of stimulation occurring in the presence of high antigen concentration could result in depletion of the fetal Vγ2Jγ1.2+ subset. This mechanism would give the appearance of induced tolerance, because reactive Vγ2+ clones would be lost due to activation-induced cell death. Thus, environmental factors in Jos might be driving specific depletion of Vγ2Jγ1.2+ cells during fetal development and the differences between Jos and Rome might be explained by the type and intensity of signaling among fetal lymphocytes. The absence of effects on Vγ2-Jγ1.3/2.3 cells, which do not react to phosphoantigens, is consistent with an antigen-specific tolerance mechanism.
We focused on malaria as a possible source of Vg2Vd2 T cell stimulation, for three main reasons. First, malaria prevalence is high in Jos (Egwunyenga et al., 2001; Egwunyenga et al., 1997). Second, malaria has documented effects on adult γδ T cells, including expansion of peripheral blood subsets for endemic infections (Hviid et al., 2001; Worku et al., 1997) and specific depletion of Vγ2Jγ1.2 cells in naïve individuals (traveler’s malaria) (Martini et al., 2003); fetuses are immunologically naïve and plasmodia exposure might have an impact similar to infection of naïve adults. Third, P. falciparum produces small stimulatory phosphoantigens (Behr et al., 1996) (approximately 200 dalton) that might easily cross the placental barrier and trigger fetal responses.
Tolerance is the functional manifestation of negative selection that occurs when antigen-stimulated T cell clones are deleted. It is likely that fetal exposure to pathogens and their phosphoantigens will activate Vγ2-Jγ1.2+ cells including those with canonical or public clonotypes, and this potentially prolonged stimulation may promote tolerance, that we observe here as repertoire disruption. The importance of these mechanisms may be most apparent during the first year of life. A Vγ2-Jγ1.2 repertoire depletion might remove a T cell subset critical for the response to pediatric vaccination against tuberculosis. Documented poor performance of the tuberculosis vaccine (BCG) in equatorial vs. non-equatorial regions (Colditz et al., 1994) suggests some effects of environment on host response; tolerance in the γδ T cell population may be one mechanism for affecting newborn immunity.
Supplementary Material
Supplementary Material S1. Vγ2-Jγ1.2 repeated nucleotypes. All repeated Vγ2-Jγ1.2 nucleotypes are reported for both CB-R and CB-J, with the number of repeats listed on a side for each of them.
Supplementary Material S2. Vγ2-Jγ1.3/2.3 repeated nucleotypes. All repeated Vγ2-Jγ1.3/2.3 nucleotypes are reported for both CB-R (A) and CB-J (B), with the number of repeats listed on a side for each of them.
Supplementary Material S3. Some Vγ2 clonotypes are coded by multiple nucleotypes (convergence). Examples of convergence are reported for both Jγ1.2 (A) and Jγ1.3/2.3 (B) clonotypes.
Acknowledgements
We thank Laura De Leo and the “San Pietro” Hospital Transfusional Center for their invaluable assistance with cord blood collection.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asarnow DM, Cado D, Raulet DH. Selection is not required to produce invariant T-cell receptor gamma-gene junctional sequences. Nature. 1993;362:158–160. doi: 10.1038/362158a0. [DOI] [PubMed] [Google Scholar]
- Behr C, Poupot R, Peyrat MA, Poquet Y, Constant P, Dubois P, Bonneville M, Fournie JJ. Plasmodium falciparum stimuli for human gammadelta T cells are related to phosphorylated antigens of mycobacteria. Infect Immun. 1996;64:2892–2896. doi: 10.1128/iai.64.8.2892-2896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukowski JF, Morita CT, Band H, Brenner MB. Crucial role of TCR gamma chain junctional region in prenyl pyrophosphate antigen recognition by gamma delta T cells. J Immunol. 1998;161:286–293. [PubMed] [Google Scholar]
- Cairo C, Hebbeler AM, Propp N, Bryant JL, Colizzi V, Pauza CD. Innate-like gammadelta T cell responses to mycobacterium Bacille Calmette-Guerin using the public Vgamma2 repertoire in Macaca fascicularis. Tuberculosis (Edinb) 2007;87:373–383. doi: 10.1016/j.tube.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairo C, Mancino G, Cappelli G, Pauza CD, Galli E, Brunetti E, Colizzi V. Vd2 T lymphocyte responses in cord blood samples from Italy and Cote D'Ivoire. Immunology. 2008 doi: 10.1111/j.1365-2567.2007.02784.x. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casorati G, De Libero G, Lanzavecchia A, Migone N. Molecular analysis of human gamma/delta+ clones from thymus and peripheral blood. J Exp Med. 1989;170:1521–1535. doi: 10.1084/jem.170.5.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colditz GA, Brewer TF, Berkey CS, Wilson ME, Burdick E, Fineberg HV, Mosteller F. Efficacy of BCG vaccine in the prevention of tuberculosis. Metaanalysis of the published literature. Jama. 1994;271:698–702. [PubMed] [Google Scholar]
- Davodeau F, Peyrat MA, Hallet MM, Gaschet J, Houde I, Vivien R, Vie H, Bonneville M. Close correlation between Daudi and mycobacterial antigen recognition by human gamma delta T cells and expression of V9JPC1 gamma/V2DJC delta-encoded T cell receptors. J Immunol. 1993a;151:1214–1223. [PubMed] [Google Scholar]
- Davodeau F, Peyrat MA, Hallet MM, Houde I, Vie H, Bonneville M. Peripheral selection of antigen receptor junctional features in a major human gamma delta subset. Eur J Immunol. 1993b;23:804–808. doi: 10.1002/eji.1830230405. [DOI] [PubMed] [Google Scholar]
- Delfau MH, Hance AJ, Lecossier D, Vilmer E, Grandchamp B. Restricted diversity of V gamma 9-JP rearrangements in unstimulated human gamma/delta T lymphocytes. Eur J Immunol. 1992;22:2437–2443. doi: 10.1002/eji.1830220937. [DOI] [PubMed] [Google Scholar]
- Dent A, Malhotra I, Mungai P, Muchiri E, Crabb BS, Kazura JW, King CL. Prenatal malaria immune experience affects acquisition of Plasmodium falciparum merozoite surface protein-1 invasion inhibitory antibodies during infancy. J Immunol. 2006;177:7139–7145. doi: 10.4049/jimmunol.177.10.7139. [DOI] [PubMed] [Google Scholar]
- Dieli F, Troye-Blomberg M, Farouk SE, Sirecil G, Salerno A. Biology of gammadelta T cells in tuberculosis and malaria. Curr Mol Med. 2001;1:437–446. doi: 10.2174/1566524013363627. [DOI] [PubMed] [Google Scholar]
- Eberl M, Hintz M, Reichenberg A, Kollas AK, Wiesner J, Jomaa H. Microbial isoprenoid biosynthesis and human gammadelta T cell activation. FEBS Lett. 2003;544:4–10. doi: 10.1016/s0014-5793(03)00483-6. [DOI] [PubMed] [Google Scholar]
- Egwunyenga AO, Ajayi JA, Nmorsi OP, Duhlinska-Popova DD. Plasmodium/intestinal helminth co-infections among pregnant Nigerian women. Mem Inst Oswaldo Cruz. 2001;96:1055–1059. doi: 10.1590/s0074-02762001000800005. [DOI] [PubMed] [Google Scholar]
- Egwunyenga OA, Ajayi JA, Duhlinska-Popova DD. Transplacental passage of Plasmodium falciparum and seroevaluation of newborns in northern Nigeria. Southeast Asian J Trop Med Public Health. 1997;28:741–745. [PubMed] [Google Scholar]
- Engelmann I, Moeller U, Santamaria A, Kremsner PG, Luty AJ. Differing activation status and immune effector molecule expression profiles of neonatal and maternal lymphocytes in an African population. Immunology. 2006;119:515–521. doi: 10.1111/j.1365-2567.2006.02466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PS, Enders PJ, Yin C, Ruckwardt TJ, Malkovsky M, Pauza CD. In vitro stimulation with a non-peptidic alkylphosphate expands cells expressing Vgamma2-Jgamma1.2/Vdelta2 T-cell receptors. Immunology. 2001;104:19–27. doi: 10.1046/j.0019-2805.2001.01282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gober HJ, Kistowska M, Angman L, Jeno P, Mori L, De Libero G. Human T cell receptor gammadelta cells recognize endogenous mevalonate metabolites in tumor cells. J Exp Med. 2003;197:163–168. doi: 10.1084/jem.20021500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitch WL, Eberhard ML, Lammie PJ. Investigation of the influence of maternal infection with Wuchereria bancrofti on the humoral and cellular responses of neonates to filarial antigens. Ann Trop Med Parasitol. 1997;91:461–469. doi: 10.1080/00034989760824. [DOI] [PubMed] [Google Scholar]
- Hviid L, Kurtzhals JA, Adabayeri V, Loizon S, Kemp K, Goka BQ, Lim A, Mercereau-Puijalon O, Akanmori BD, Behr C. Perturbation and proinflammatory type activation of V delta 1(+) gamma delta T cells in African children with Plasmodium falciparum malaria. Infect Immun. 2001;69:3190–3196. doi: 10.1128/IAI.69.5.3190-3196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itohara S, Mombaerts P, Lafaille J, Iacomini J, Nelson A, Clarke AR, Hooper ML, Farr A, Tonegawa S. T cell receptor delta gene mutant mice: independent generation of alpha beta T cells and programmed rearrangements of gamma delta TCR genes. Cell. 1993;72:337–348. doi: 10.1016/0092-8674(93)90112-4. [DOI] [PubMed] [Google Scholar]
- Lammie PJ, Hitch WL, Walker Allen EM, Hightower W, Eberhard ML. Maternal filarial infection as risk factor for infection in children. Lancet. 1991;337:1005–1006. doi: 10.1016/0140-6736(91)92661-k. [DOI] [PubMed] [Google Scholar]
- Le Hesran JY, Cot M, Personne P, Fievet N, Dubois B, Beyeme M, Boudin C, Deloron P. Maternal placental infection with Plasmodium falciparum and malaria morbidity during the first 2 years of life. Am J Epidemiol. 1997;146:826–831. doi: 10.1093/oxfordjournals.aje.a009200. [DOI] [PubMed] [Google Scholar]
- Macdonald G. The analysis of malaria parasite rates in infants. Trop Dis Bull. 1950;47:915–938. [PubMed] [Google Scholar]
- MacDougall A, Enders P, Hatfield G, Pauza D, Rakasz E. V gamma 2 TCR repertoire overlap in different anatomical compartments of healthy, unrelated rhesus macaques. J Immunol. 2001;166:2296–2302. doi: 10.4049/jimmunol.166.4.2296. [DOI] [PubMed] [Google Scholar]
- Malhotra I, Mungai P, Wamachi A, Kioko J, Ouma JH, Kazura JW, King CL. Helminth- and Bacillus Calmette-Guerin-induced immunity in children sensitized in utero to filariasis and schistosomiasis. J Immunol. 1999;162:6843–6848. [PubMed] [Google Scholar]
- Martini F, Paglia MG, Montesano C, Enders PJ, Gentile M, Pauza CD, Gioia C, Colizzi V, Narciso P, Pucillo LP, Poccia F. V gamma 9V delta 2 T-cell anergy and complementarity-determining region 3-specific depletion during paroxysm of nonendemic malaria infection. Infect Immun. 2003;71:2945–2949. doi: 10.1128/IAI.71.5.2945-2949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVay LD, Carding SR. Extrathymic origin of human gamma delta T cells during fetal development. J Immunol. 1996;157:2873–2882. [PubMed] [Google Scholar]
- McVay LD, Carding SR. Generation of human gammadelta T-cell repertoires. Crit Rev Immunol. 1999;19:431–460. [PubMed] [Google Scholar]
- Montesano C, Gioia C, Martini F, Agrati C, Cairo C, Pucillo LP, Colizzi V, Poccia F. Antiviral activity and anergy of gammadeltaT lymphocytes in cord blood and immuno-compromised host. J Biol Regul Homeost Agents. 2001;15:257–264. [PubMed] [Google Scholar]
- Morita CT, Beckman EM, Bukowski JF, Tanaka Y, Band H, Bloom BR, Golan DE, Brenner MB. Direct presentation of nonpeptide prenyl pyrophosphate antigens to human gamma delta T cells. Immunity. 1995;3:495–507. doi: 10.1016/1074-7613(95)90178-7. [DOI] [PubMed] [Google Scholar]
- Morita CT, Parker CM, Brenner MB, Band H. TCR usage and functional capabilities of human gamma delta T cells at birth. J Immunol. 1994;153:3979–3988. [PubMed] [Google Scholar]
- Onyenekwe CC, Meludu SC, Dioka CE, Salimonu LS. Prevalence of asymptomatic malaria parasitaemia amongst pregnant women. Indian J Malariol. 2002;39:60–65. [PubMed] [Google Scholar]
- Parker CM, Groh V, Band H, Porcelli SA, Morita C, Fabbi M, Glass D, Strominger JL, Brenner MB. Evidence for extrathymic changes in the T cell receptor gamma/delta repertoire. J Exp Med. 1990;171:1597–1612. doi: 10.1084/jem.171.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakasz E, MacDougall AV, Zayas MT, Helgelund JL, Ruckward TJ, Hatfield G, Dykhuizen M, Mitchen JL, Evans PS, Pauza CD. Gammadelta T cell receptor repertoire in blood and colonic mucosa of rhesus macaques. J Med Primatol. 2000;29:387–396. doi: 10.1111/j.1600-0684.2000.290602.x. [DOI] [PubMed] [Google Scholar]
- Sagay AS, Musa J, Adewole AS, Imade GE, Ekwempu CC, Kapiga S, Sankale JL, Idoko J, Kanki P. Rapid HIV testing and counselling in labour in a northern Nigerian setting. Afr J Reprod Health. 2006;10:76–80. [PubMed] [Google Scholar]
- Sama W, Dietz K, Smith T. Distribution of survival times of deliberate Plasmodium falciparum infections in tertiary syphilis patients. Trans R Soc Trop Med Hyg. 2006;100:811–816. doi: 10.1016/j.trstmh.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Sama W, Killeen G, Smith T. Estimating the duration of Plasmodium falciparum infection from trials of indoor residual spraying. Am J Trop Med Hyg. 2004;70:625–634. [PubMed] [Google Scholar]
- Schild H, Mavaddat N, Litzenberger C, Ehrich EW, Davis MM, Bluestone JA, Matis L, Draper RK, Chien YH. The nature of major histocompatibility complex recognition by gamma delta T cells. Cell. 1994;76:29–37. doi: 10.1016/0092-8674(94)90170-8. [DOI] [PubMed] [Google Scholar]
- Steel C, Guinea A, McCarthy JS, Ottesen EA. Long-term effect of prenatal exposure to maternal microfilaraemia on immune responsiveness to filarial parasite antigens. Lancet. 1994;343:890–893. doi: 10.1016/s0140-6736(94)90009-4. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Morita CT, Nieves E, Brenner MB, Bloom BR. Natural and Synthetic Nonpeptide Antigens Recognized by Human Gamma-Delta T-Cells. Nature. 1995;375:155–158. doi: 10.1038/375155a0. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Sano S, Nieves E, De Libero G, Rosa D, Modlin RL, Brenner MB, Bloom BR, Morita CT. Nonpeptide ligands for human gamma delta T cells. Proc Natl Acad Sci U S A. 1994;91:8175–8179. doi: 10.1073/pnas.91.17.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worku S, Bjorkman A, Troye-Blomberg M, Jemaneh L, Farnert A, Christensson B. Lymphocyte activation and subset redistribution in the peripheral blood in acute malaria illness: distinct gammadelta+ T cell patterns in Plasmodium falciparum and P. vivax infections. Clin Exp Immunol. 1997;108:34–41. doi: 10.1046/j.1365-2249.1997.d01-981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cado D, Asarnow DM, Komori T, Alt FW, Raulet DH, Allison JP. The role of short homology repeats and TdT in generation of the invariant gamma delta antigen receptor repertoire in the fetal thymus. Immunity. 1995;3:439–447. doi: 10.1016/1074-7613(95)90173-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material S1. Vγ2-Jγ1.2 repeated nucleotypes. All repeated Vγ2-Jγ1.2 nucleotypes are reported for both CB-R and CB-J, with the number of repeats listed on a side for each of them.
Supplementary Material S2. Vγ2-Jγ1.3/2.3 repeated nucleotypes. All repeated Vγ2-Jγ1.3/2.3 nucleotypes are reported for both CB-R (A) and CB-J (B), with the number of repeats listed on a side for each of them.
Supplementary Material S3. Some Vγ2 clonotypes are coded by multiple nucleotypes (convergence). Examples of convergence are reported for both Jγ1.2 (A) and Jγ1.3/2.3 (B) clonotypes.