Abstract
Chemokine receptors, and in particular CXCR4 and CCR5 play a key role in the neuropathogenesis of Human Immunodeficiency Virus-1 (HIV)4 associated dementia (HAD). Thus, new insight into the expression of CXCR4 in the central nervous system may help develop therapeutic compounds against HAD. Brain-derived neurotrophic factor (BDNF) is neuroprotective in vitro against two strains of the HIV envelope protein gp120 that binds to CXCR4 or CCR5. Therefore, we examined whether BDNF modulates chemokine receptor expression in vivo. The content of CXCR4 mRNA and proteins was determined in the cerebral cortex and hippocampus of 6-month-old BDNF heterozygous mice and wild type littermates by using polymerase chain reaction and immunohistochemistry, respectively. BDNF heterozygous mice exhibited an increase in CXCR4 mRNA compared to wild type. Histological analyses revealed an up-regulation of CXCR4 immunoreactivity mainly in neurons. Most of these neurons were positive for TrkB, the BDNF receptor with a tyrosine kinase activity. Increases in CXCR4 mRNA levels were observed in 18-month-old BDNF heterozygous mice but not in 7 day-old mice, suggesting that the modulatory role of BDNF occurs only in mature animals. To determine whether BDNF affects also CXCR4 internalization, SH-SY5Y neuroblastoma cells were exposed to BDNF and cell surface CXCR4 levels were measured at various times. BDNF induced CXCR4 internalization within minutes. Lastly, BDNF heterozygous mice showed higher levels of CCR5 and CXCR3 mRNA than wild type in the cerebral cortex, hippocampus and striatum. Our data indicate that BDNF may modulate the availability of chemokine receptors implicated in HIV infection.
Keywords: BDNF, HIV dementia, CXCR4, CCR5, gp120, TrkB
1. Introduction
Chemokine and chemokine receptors were originally identified for their chemoattractant and pro-adhesive roles in inflammation, wound repair, and cell migration [rev in (Miller and Tran, 2005)]. During the development of the central nervous system (CNS), the chemokine receptor CXCR4 plays a crucial role in the migration of cerebrocortical interneurons (Stumm et al., 2003) as well as granule cells of the cerebellum (Ma et al., 1998) and the hippocampal dentate gyrus (Bagri et al., 2002; Lu et al., 2002). Indeed, the lack of CXCR4 results in an improper formation of neuronal layers in these areas. However, CXCR4 is also abundant in the adult CNS. For instance, pyramidal cells of the cortex, dopaminergic neurons of the substantia nigra and cholinergic neurons of the basal forebrain are especially enriched in CXCR4 in rats (Banisadr et al., 2002). Astrocytes and microglia cells of different brain regions also express CXCR4 (Banisadr et al., 2002; Lavi et al., 1997) even in the absence of inflammatory responses (Lazarini et al., 2003). Thus, the anatomical profile and abundance of CXCR4 and other chemokines in the adult CNS suggest a broader function than the originally proposed neuroinflammation and migration role.
CXCR4 has recently been recognized as crucial mediators of Human Immunodeficiency Virus-1 (HIV) entry into immunocompetent target cells [rev in (Moore et al., 2004)]. Sequential binding of the HIV envelope glycoprotein 120 (gp120) to CD-4 and chemokine receptors on the surface of immunocompetent cells facilitates HIV entry into cells (Berson et al., 1996; Feng et al., 1996; He et al., 1997). Different strains of gp120 exhibit affinity for specific chemokine receptors differentially expressed on certain cell types. For example, gp120 associated with M-tropic HIV binds specifically to CCR5, which is expressed on macrophages, whereas gp120 associated with T-tropic HIV binds to CXCR4, which is abundant in T-cells (Moore et al., 2004). Thus, availability of chemokine receptors is crucial for the infectious phase of the disease.
There is also growing evidence for the role of chemokines in the neurotoxicity of HIV in the CNS. Neurons are susceptible to massive apoptotic cell death in the presence of T-tropic gp120 or other viral proteins both in vitro and in vivo (Bansal et al., 2000; Kaul and Lipton, 1999; Kruman et al., 1998; Meucci et al., 1998; Nosheny et al., 2004). Moreover, mice overexpressing T-tropic gp120 (Toggas et al., 1994) exhibit several features that mirror the neuropathology of HAD. The neurotoxic effect of gp120 involves CXCR4-mediated activation of pro-apoptotic signal transduction cascades, including caspase-3 (Bachis et al., 2003; Biard-Piechaczyk et al., 2000; Hesselgesser et al., 1998; Zheng et al., 1999). These results imply a fundamental role for CXCR4 in HIV-Associated Dementia (HAD) pathogenesis, which includes neuronal apoptosis and atrophy (Gray et al., 2001; James et al., 1999; Masliah et al., 2000; McArthur, 2004), and suggest that targeted therapeutic interventions for HAD should include a down-regulation of chemokine receptor function.
Brain-derived neurotrophic factor (BDNF) is a member of the neurotrophin family of trophic factors that includes nerve growth factor, neurotrophin-3 and NT4/5 (Chao, 2003). There is evidence that BDNF reduces the neurotoxicity of gp120 in vitro (Bachis and Mocchetti, 2005) and in vivo (Nosheny et al., 2007). The neuroprotective effect correlates with its ability to reduce CXCR4 levels in vitro (Bachis et al., 2003). However, little is known about the modulation of CXCR4 in vivo. Moreover, little is known about the ability of BDNF to induce CXCR4 internalization. In the present study, we used BDNF heterozygous (+/−) mice as well as in vitro studies in an attempt to characterize a relationship between BDNF and modulation of chemokine receptor availability. Such details about BDNF activity may provide an avenue for better treatment or prevention of HAD.
2. Results
BDNF heterozygous mice exhibit altered levels of CXCR4 mRNA
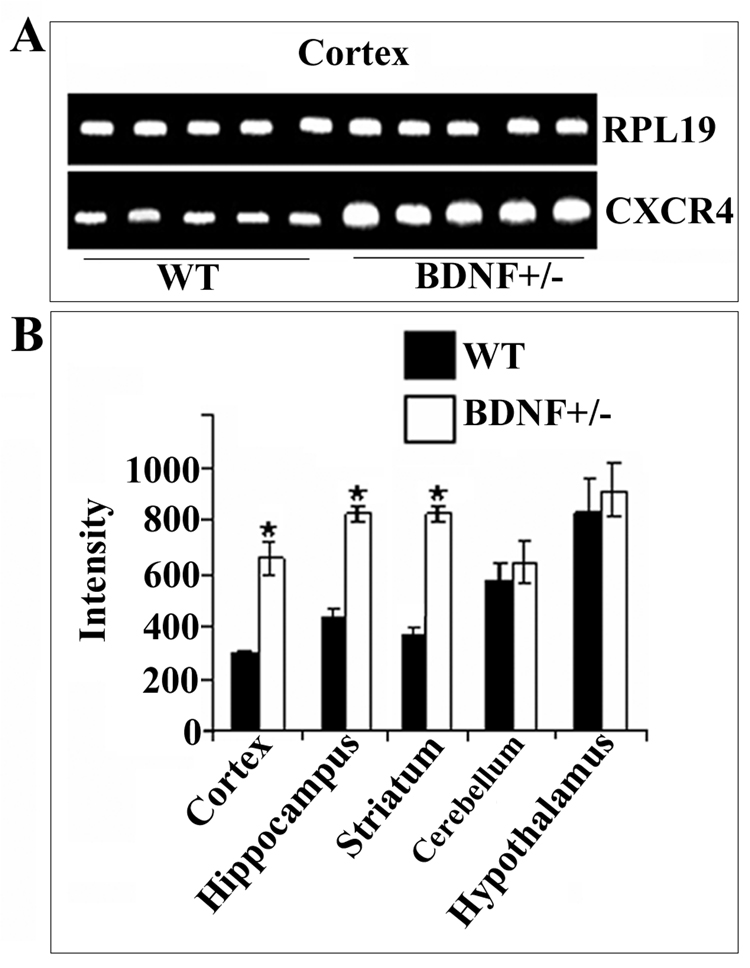
To examine the role of BDNF in regulating CXCR4 expression in vivo we utilized 6-month-old BDNF+/− mice and age-matched wild type (WT) littermates. One may predict that low levels of BDNF, as in those exhibited by BDNF+/− mice (Lyons et al., 1999), will cause CXCR4 up-regulation. CXCR4 mRNA was measured using a semi-quantitative reverse transcriptase polymerase chain reaction (RT-PCR) and specific primers for mouse CXCR4 (Table 1). Primers for ribosomial protein 19 (RPL19) were used as an internal control to determine relative levels of total mRNA and standardize CXCR4 mRNA levels. We found that BDNF+/− mice showed higher CXCR4 mRNA levels than WT in the cerebral cortex (Fig. 1A), hippocampus and striatum (Fig. 1B), while the levels of RPL19 mRNA remained unchanged. No differences in CXCR4 mRNA were found between WT and heterozygous mice in the hypothalamus and cerebellum (Fig. 1B), the two areas with highest levels of CXCR4 mRNA.
Table 1.
Sense and antisense primers used for RT-PCR.
| Chemokine | Species | gene bank number | Sequence |
|---|---|---|---|
| CXCR4 | Human | AK129916 | AGCGAGGTGGACATTCATCTGTT (1087–1109) |
| CGTGATTCACTACACGCTCTGG (1418–1439) | |||
| Cyclophilin | Human | NM_021130 | TCCTGCTTTCACAGAATTATTCC (234–256) |
| ATTCGAGTTGTCCACAGTCAGC (558–579) | |||
| RPL 19 | Mouse | BC010710 | GGTACTGCCAATGCTCGGAT (264~273) |
| TCCTTGGACAGAGTCTTGATGA (558~579) | |||
| CXCR4 | Mouse | U59760 | GGTCTGGAGACTATGACTCC (85~104) |
| CACAGATGTACCTGTCATCC (589~609) | |||
| CXCR3 | Mouse | AF045146 | GAGGTTAGTGAACGTCAAGTG (9–30) |
| GGGGTCCCTGCGGTAGATCTG (468–489) | |||
| CCR5 | mouse | AF019772 | TCAGTTCCGACCTATATCTATG (16–37) |
| GTGGAAAATGAGGACTGCATGT (535–556) |
Figure 1. BDNF heterozygous mice exhibit higher levels of CXCR4 mRNA than WT.
RNA was extracted from the indicated brain areas of 6-month-old BDNF+/− mice or WT littermates. RT-PCR was performed using primers designed to amplify mouse CXCR4 (see Experimental Procedures). PCR reaction products were analyzed by agarose gel-electrophoresis (see Experimental Procedures). A. Representative gel showing CXCR4 and RPL19 cDNAs from the cerebral cortex. B. Semi-quantitative analysis of CXCR4 cDNA was carried out by Quantity One 1-D Analysis as described in Experimental Procedures. RPL19 was used as an internal control to normalize gel loading. Data, expressed as intensity of the cDNA band, are the mean ± SEM of five independent samples. *p<0.05 vs control.
Changes in CXCR4 immunoreactivity in heterozygous mice occur in neurons
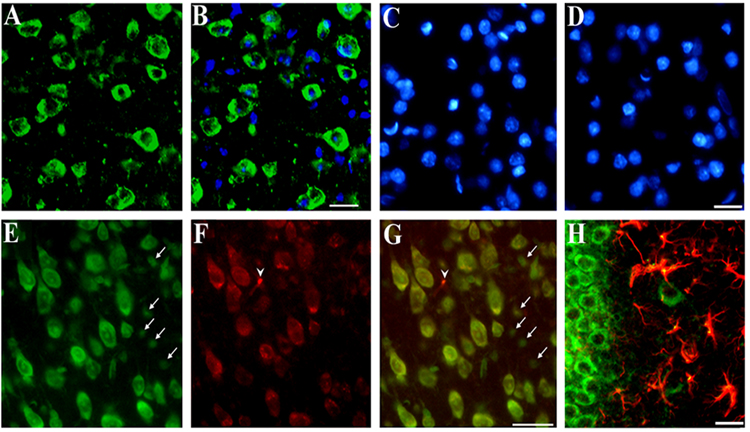
Both neuronal and non-neuronal cells express CXCR4 (Banisadr et al., 2002; Lavi et al., 1997; Lazarini et al., 2000; Westmoreland et al., 2002). We therefore used histological analyses to examine the relationship between low levels of BDNF and cells expressing CXCR4. We first analyzed the cerebral cortex and hippocampus of 6-month-old WT mice for CXCR4 immunoreactivity, using a CXCR4 specific antibody together with Nissl, glial fibrillary acidic protein (GFAP) and CD11 staining to detect neurons, astrocytes and microglia, respectively. CXCR4 positive and negative cells were observed throughout the cerebral cortex (Figs. 2A and B). Specificity of the antibody was confirmed by omitting the primary (Fig. 2C) or secondary antibody (Fig. 2D). In both cases, no CXCR4 immunoreactivity was detected (Figs 2C and D). Most of CXCR4 immunoreactivity was detected in several Nissl positive cells in layers II/III (data not shown) and V (Figs. 2E and G). Some Nissl positive cells were CXCR4 negative (Figs. 2E and G, arrows). Occasionally, we observed CXCR4 immunoreactivity in Nissl negative cells (Figs. 2F and G, arrowhead).
Figure 2. Immunohistochemical analysis of CXCR4.
Serial coronal sections (16 µm) from the frontal cortex or hippocampus were obtained from WT mice. Examples of cortical sections stained with (A) a CXCR4 antibody (green) counterstained with (B) DAPI (blue) to visualize cellular nuclei. Note that some cells are CXCR4 negative. C and D: Examples of sections in which the primary or secondary antibody, respectively, were omitted. Sections were counterstained with DAPI. E and F: Sections stained with Nissl and CXCR4 antibody, respectively (see Experimental Procedures). G: E and F were merged to show colocalization of CXCR4 and neurons (yellow). Note that in E small Nissl positive cells are CXCR4 negative (arrows). In F, one CXCR4 positive cell (arrowhead) is Nissl negative. H: Example of a hippocampal section stained with CXCR4 (green) and GFAP (red) and showing colocalization of CXCR4 and GFAP (yellow). Bars=40 µm.
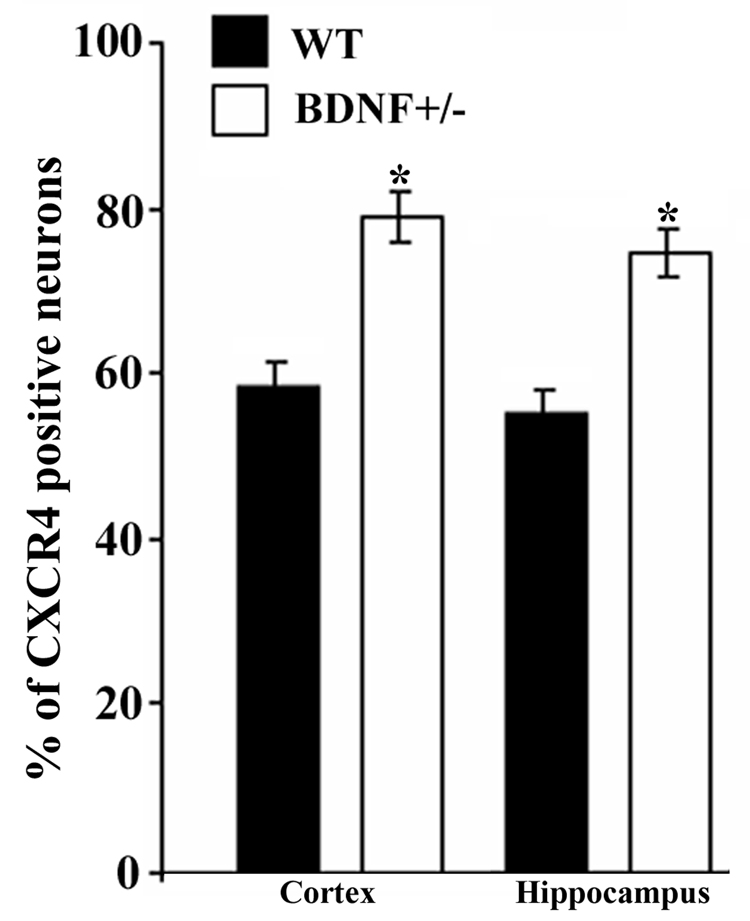
In the hippocampus, CXCR4 immunoreactivity was seen in neurons as well as in astrocytes (Fig. 2H), supporting previous data that in adult rats several cell types exhibit CXCR4 immunoreactivity (Banisadr et al., 2002). Sections from WT and BDNF+/− mice were then used to compare the number of CXCR4 positive cells. The number of CXCR4 positive cells was determined in layer V of the frontal cortex and the dorsolateral portion of the hippocampus corresponding to the CA2 area. Semi-quantitative analysis using Metamorph® revealed that more neurons were positive for CXCR4 in BDNF+/− mice than in WT in both regions (Fig. 3). There was no difference in CXCR4 positive astrocytes between WT and heterozygous mice. Moreover, no CD11 immunoreactivity (microglia) was seen in either WT or BDNF+/− mice (data not shown).
Figure 3. BDNF+/− mice exhibit more CXCR4 immunoreactivity.
Serial coronal sections obtained as described in Fig. 2 and in Experimental Procedures were used to determine the number of CXCR4 positive neurons in the frontal cortex and hippocampus. The number of CXCR4 positive neurons was determined in layer V of the cortex and CA2 region of the hippocampus in an area of 1 mm2 per section using MetaMorph® software. An average of 8000 neurons per animal per area was counted. Data, expressed as % of Nissl positive cells per section, are the mean ± SEM of 10 sections per animal (n=4 each group). *p<0.05 vs WT.
TrkB positive neurons exhibit CXCR4 immunoreactivity
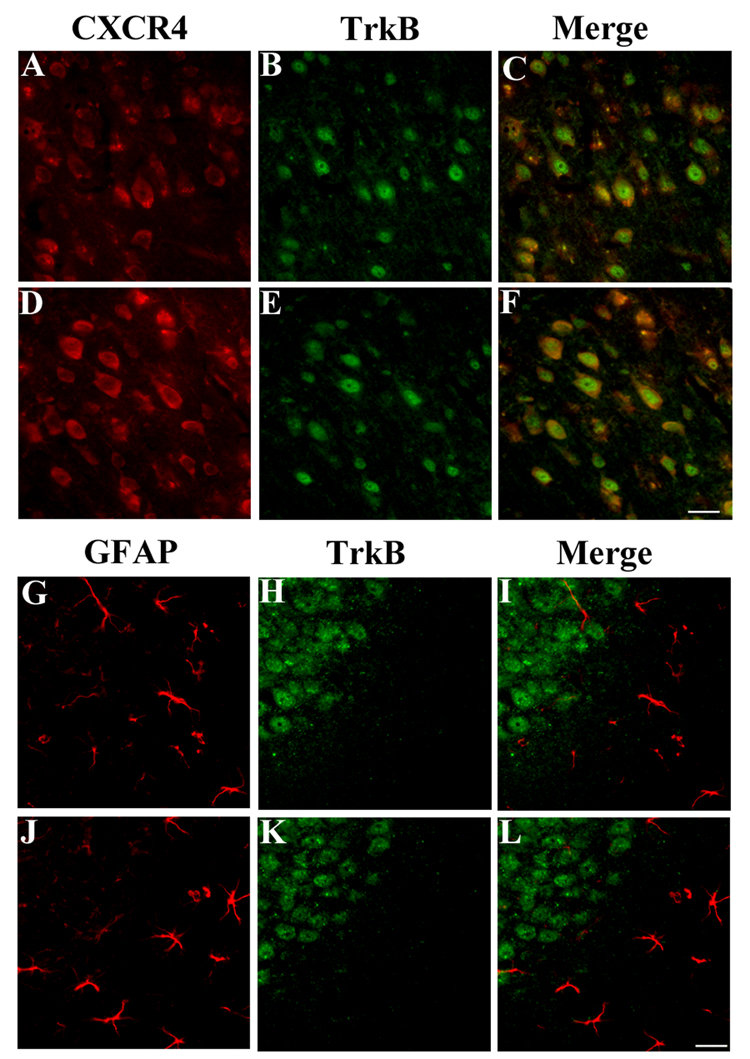
BDNF has been shown to prevent gp120 neurotoxicity by activating the tyrosine kinase receptor TrkB (Mocchetti and Bachis, 2004). Therefore, we used a TrkB specific antibody (TrkB23–36) developed against the extracellular portion of TrkB (Fryer et al., 1996) to provide a correlation between distribution of the TrkB that signals in response to BDNF and CXCR4 expression. This antibody has been shown to stain neurons in the cortex and hippocampus (Fryer et al., 1996). In the cortex (Figs. 4A–C) and hippocampus (data not shown) of WT animals, most of TrkB positive cells exhibited CXCR4 immunoreactivity. To distinguish whether these neurons expressed full-length or truncated forms of TrkB, adjacent sections were stained with an antibody that recognizes the tyrosine kinase intracellular portion of Trk (TrkB-in). All neurons that were stained with TrkB23–36 exhibited the same immunoreactive profile for TrkB-in (not shown), suggesting that these neurons express the full-length TrkB.
Figure 4. CXCR4 co-localizes with TrkB in neurons.
Cortical sections from WT (A–C) and BDNF+/− mice (D–F) were stained with CXCR4 antibody (red) and an antibody that recognizes the extracellular domain of TrkB (αtrkB23–36, green). In C and F, please note that a number of CXCR4 positive cells are also TrkB positive. Sections from the hippocampus of WT (G–I) and BDNF+/− mice (J–L) were stained for GFAP (red, G and J) and TrkB (green, H and K). No astrocytes were TrkB positive. Bar=50 µm.
Neurons are the major source of TrkB immunoreactivity in layer V of the cortex. However, few astrocytes were detected in this layer. To assure the neuronal expression of TrkB, TrkB23–36 was used in conjunction with a GFAP antibody in co-localization studies of hippocampal sections. TrkB immunoreactivity was mostly detected in the dentate gyrus (Fig. 4H), and CA regions of the hippocampus (data not shown) confirming previous data (Fryer et al., 1996). GFAP positive cells were TrkB negative (Fig. 4G–I). Because CXCR4 immnoreactivity was also detected in astrocytes, our data overall suggest that CXCR4 does not always co-localize with TrkB.
Sections from WT and BDNF+/− mice were then used to compare the number of CXCR4 positive cells in TrkB positive/negative neurons. In the cortex, more CXCR4/TrkB positive cells were observed in BDNF+/− animals (Figs. 4D–F) than WT (Figs. 4A–C). However, the total number of TrkB positive neurons was similar in both animal groups (Figs. 4B and E). In the hippocampus, more CXCR4 immunoreactivity was observed in BDNF+/− mice (Fig. 3), but no changes were observed in GFAP (Fig. 4J) or TrkB positive cells (Fig. 4K), suggesting that the observed changes in CXCR4 positive neurons in BDNF+/− mice are not due to alterations of cell number. Overall, these results support previous pharmacological data that TrkB plays a crucial role in the regulation of CXCR4 expression (Mocchetti and Bachis, 2004).
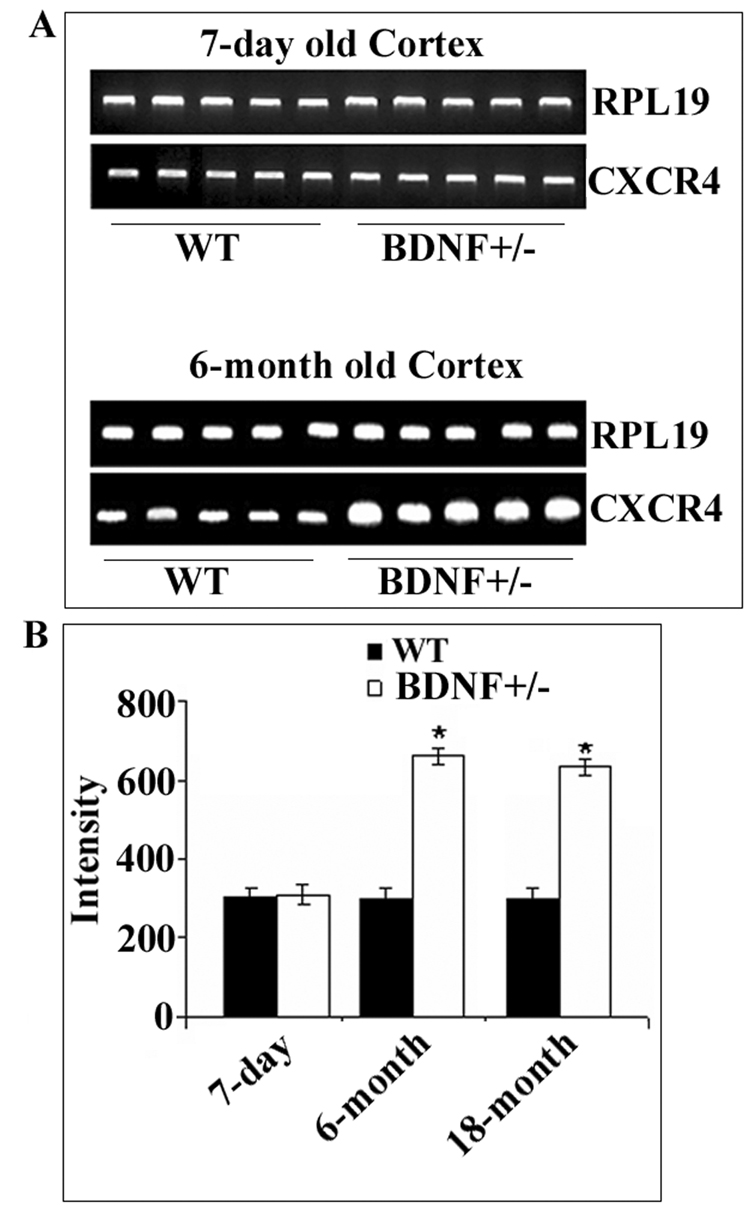
CXCR4 mRNA up-regulation is age-dependent
BDNF+/− mice exhibit an age-dependent reduction of fibers and function of central serotonergic neurons (Lyons et al., 1999). To determine whether BDNF regulation of CXCR4 expression is age-dependent, cortical CXCR4 mRNA levels were measured by PCR (Fig. 5A) in 7-day-old pups, 6- and 18-month-old mice. In WT, there was no significant difference in CXCR4 mRNA levels between 7- day and 6- and 18-month-old mice (Fig. 5), suggesting that maximal expression of CXCR4 occurs already a few days postnatal. Interestingly, while 18-month-old BDNF+/− mice exhibited a ~two-fold increase in CXCR4 mRNA levels (Fig. 5B), 7-day-old WT and BDNF+/− displayed the same amount of CXCR4 mRNA levels (Fig. 5). Thus, BDNF appears to regulate CXCR4 expression only in adulthood.
Figure 5. Age-dependent increase of CXCR4 in BDNF+/− mice.
Cortical CXCR4 mRNA levels were measured in 7-day-old pups, 6 and 18-month-old BDNF+/− mice and WT littermates. RNA extraction and RT-PCR was carried out as described in Experimental Procedures. A. Representative gels showing CXCR4 and RPL19 cDNAs from the cortex of 7-day old and 6-month old mice. B. Semi-quantitative analysis of CXCR4 mRNA was carried out by Quantity One 1-D analysis. Data, expressed as Intensity of cDNA bands, are the mean ± SEM of five independent samples. *p<.005 vs WT.
BDNF reduces membrane associated CXCR4
CXCR4 is a G protein-coupled receptor (GPCR) that undergoes ligand induced internalization (Tarasova et al., 1998). In neurons, internalization of gp120 and retrograde axonal transport are necessary for the toxic activity of gp120 (Bachis et al., 2003; Bachis et al., 2006). Therefore, we examined whether BDNF alters membrane-associated CXCR4. Internalization of CXCR4 by BDNF was determined by cell surface biotinylation. Because this assay requires several micrograms of proteins that cannot be easily obtained from primary neuronal cultures, we used human SH-SY5Y neuroblastoma cells. These cells are sensitive to the toxic action of T tropic gp120 (Bardi et al., 2006). However, SH-SY5Y lacks endogenous expression of trkB (Kaplan et al., 1993). Thus, we used SH-SY5Y cells over-expressing TrkB (clone TB8) under a tetracycline-regulated promoter (Kim et al., 1999). These cells, in addition to be BDNF responsive, differentiate into a neuronal phenotype (Kim et al., 1999). Cells were exposed to medium control or containing BDNF (10 ng/ml) for various times (5, 30 min and 3 hr). Cells were then incubated with membrane impermeant NHS-LC-Biotin for 30 min at 4°C, and biotinylated cell surface proteins were precipitated with immobilized streptavidin and subjected to CXCR4 immunoblotting. Immobilized streptavidin precipitated a complex of CXCR4 immunoreactivity of approximately 42 kDa (Fig. 6A), which is consistent with the molecular weight of CXCR4. In cells exposed to BDNF, CXCR4 immunoreactivity decreased within 5 min of exposure (Fig. 6B), suggesting increased internalization.
Figure 6. BDNF increases internalization of CXCR4.
SY5YTB8 cells (Kim et al., 1999) were incubated with medium control (0.1% BSA) or medium containing BDNF (10 ng/ml) for 5 or 30 min or 3 hr. Cells were then washed and treated with 2 mM NHS-LC-biotin for 30 min at 4°C to biotinylate cell surface proteins. Biotinylated proteins were precipitated with immobilized streptavidin. Precipitates were then electrophoresed on SDS-polyacrylamide gels and subjected to CXCR4 immunoblotting. A. Example of immunoblot. B. Immunoreactive bands were scanned and quantified by Quantity One 1-D Analysis Software. Data, expressed as intensity of immunoreactive bands, are the mean ± SEM of three independent experiments. *p<0.05 vs control.
To determine whether BDNF affects also the expression of CXCR4, SY5TB8 cells were exposed to BDNF (10 ng/ml) for 4 and 8 hr. CXCR4 mRNA levels were then measured by RT-PCR using primers designed to amplify human CXCR4 mRNA (Table 1). The human housekeeping gene cyclophilin was used as an internal control to standardize CXCR4 mRNA levels. BDNF promoted a decrease in CXCR4 mRNA at both 4 and 8 hr (Fig. 7). This data, together with previous results showing reduced CXCR4 protein levels in cerebellar granule cells exposed to BDNF (Bachis et al., 2003), indicates that this trophic factor modulates CXCR4 expression and trafficking.
Figure 7. BDNF decreases CXCR4 mRNA levels.
SY5YTB8 cells were exposed to medium control (0.1% BSA) or containing BDNF (10 ng/ml) for 4 and 8 hr and CXCR4 mRNA levels were measured by RT-PCR using primers designed to amplify human CXCR4 (see Table 1). Bands corresponding to CXCR4 cDNA were quantified using Quantity One 1-D Analysis Software. Data, expressed as intensity of bands, are the mean ± SEM of three independent experiments done (n=9). *p<0.05 vs control.
CCR5 and CXCR3 mRNA levels are up-regulated in BDNF +/− mice
HIV-1 can generally use CCR5 as co-receptors for cell entry (Signoret et al., 2005). In addition, CXCR3 appears to mediate the toxic action of CXCR4 ligands (Vergote et al., 2006). Therefore, it is important to establish whether BDNF alters the expression profile of these chemokine receptors. Total RNA was extracted from the cerebral cortex, hippocampus and striatum of 6-month-old mice and analyzed by RT-PCR (Fig. 8A). BDNF+/− mice displayed higher CXCR3 and CCR5 mRNA levels than WT in all regions examined (Fig. 8B). However, the hippocampus of BDNF+/− mice exhibited a robust 7-fold increase in CCR5 mRNA levels (Fig. 8C). Thus, chemokine receptor expression by BDNF follows a region-specific pattern of regulation.
Figure 8. Modulation of chemokine receptors by BDNF is area-specific.
Total RNA was extracted from the indicated brain areas of 6-month-old BDNF+/− mice or WT littermates. RT-PCR was performed using primers designed to amplify mouse CCR5 and CXCR3. A. Representative gels showing CCR5, CXCR3 and RPL19 cDNAs from the indicated brain areas. B. Semi-quantitative analysis of chemokine receptor cDNAs was carried out by Quantity One 1-D analysis. Data, expressed as intensity of cDNA bands, are the mean ± SEM of five independent samples. *p<0.05; **p<0.001 vs WT.
3. Discussion
A number of HIV positive individuals develop HAD, which is characterized pathologically by neuronal loss and dysfunction (Everall et al., 2005). HIV infection is mediated by the co-receptors CXCR4 and CCR5. Thus, in this report, we investigated whether chemokine receptor expression can be regulated in the brain. We used BDNF+/− mice because BDNF exhibits neuroprotection in vitro against two strains of gp120 that binds to CXCR4 and CCR5 (Bachis and Mocchetti, 2005). Moreover, BDNF+/− mice are more sensitive to gp120 toxicity (Nosheny et al., 2004). In these animals, we found an increase of CXCR4, CCR5 and CXCR3 mRNAs in several brain areas and an accumulation of CXCR4 immunoreactivity in neurons. Previous data have shown that BDNF down-regulates CXCR4 levels in vitro (Bachis et al., 2003) and in vivo (Nosheny et al., 2007). Moreover, we show that in SY5YTB8 neuroblastoma cells, BDNF accelerates CXCR4 trafficking and reduces CXCR4 mRNA levels. These data, taken together, indicate that BDNF modulates expression and availability of chemokine receptors both physiologically and pharmacologically. Based on these findings, we propose that down-regulation of CXCR4 is a mechanism that may account for the neuroprotective property of BDNF against gp120.
CXCR4 immunoreactivity has been detected in neurons as well as non-neuronal cells, including microglia and astrocytes in numerous brain areas of different species (Banisadr et al., 2002; Klein et al., 1999; Lazarini et al., 2003; van der Meer et al., 2000). In this report, we confirm that neurons and astrocytes express CXCR4. However, BDNF appears to affect CXCR4 immunoreactivity only in neurons. Indeed, in BDNF+/− mice we observed an increase in CXCR4 positive neurons in both the cortex and hippocampus without a concomitant increase in CXCR4 immunoreactivity in astrocytes. This result is not surprising because neurons are known to express catalytically active TrkB (Fryer et al., 1996), the receptor that mediates the neurotrophic activity of BDNF, including its neuroprotective effect against gp120 (Mocchetti and Bachis, 2004). Indeed, in our study, astrocytes were not labeled by a TrkB specific antibody (Fryer et al., 1996), while a number of neurons in layer V of the cortex, dentate gyrus and CA regions of the hippocampus were both CXCR4 and TrkB positive. Thus, BDNF may not regulate CXCR4 expression in non-neuronal cells because of the absence of TrkB. These data support previous pharmacological results showing that TrkB is the main neurotrophin receptor that, by decreasing CXCR4 levels, modulates the neuroprotective property of BDNF (Mocchetti and Bachis, 2004).
BDNF+/− mice exhibited an increase in CXCR4 mRNA content in the cerebral cortex, hippocampus and striatum. We were surprised to find that the content of CXCR4 mRNA in the hypothalamus and cerebellum of BDNF+/− mice was similar to that of WT. The reason of this anatomically-restricted effect is unclear at present. Both the cerebellum and hypothalamus express TrkB (Givalois et al., 2004; Ohira and Hayashi, 2003). Nevertheless, in these areas, CXCR4 may not be co-localized with TrkB. Our data have shown that astrocytes can be CXCR4 positive but TrkB negative. Moreover, glial cells express the truncated isoform of TrkB (Fryer et al., 1996), which does not appear to mediate most of the neurotrophic effect of BDNF; rather this receptor prevents BDNF signaling (Dorsey et al., 2006). Only detailed anatomical studies will reveal why BDNF has a restricted action on CXCR4 in these areas. On the other hand, both cerebellum and hypothalamus exhibit the highest levels of CXCR4 mRNA among all regions examined, suggesting that the expression of this receptor may be influenced by other stimuli. Defining the molecular mechanisms of this regulation may help a better understanding of the role of CXCR4 in adult brain.
BDNF down-regulation of CXCR4 expression is believed to underlie its neuroprotective property against gp120 (Bachis et al., 2003; Nosheny et al., 2007). A similar cellular mechanism for neuroprotection has been proposed for another growth factor, basic fibroblast growth factor (FGF2), a member of the heparin binding growth factor that has neuroprotective properties similar to BDNF. In fact, FGF2 limits gp120 and SDF toxicity in SH-SY5Y neuroblastoma cells by reducing CXCR4 levels (Sanders et al., 2000). Most of the neuroprotective activities of BDNF and FGF2 occur through the activation of high affinity tyrosine kinase receptors TrkB and FGF receptor 1, respectively. Both receptors share a similar signal transduction mechanism. BDNF or FGF2 binding to TrkB or FGF receptor leads to activation of a number of target proteins including phosphatidylinositol-3 kinase, phospholipase c-γ, and extracellular signal regulated kinase (Miller and Kaplan, 2003). Thus, it is not surprising to observe a similar and perhaps overlapping neuroprotective profile between these two trophic factors. Indeed, blocking signaling of either TrkB by K252a (Mocchetti and Bachis, 2004) or FGF receptor by 5’-methylthioadenosine (Sanders et al., 2000) abolishes the neuroprotective effect of BDNF and FGF2, respectively. The fact that both trophic factors inhibit gp120 and SDF toxicity by reducing CXCR4 levels adds to the notion that modulation of CXCR4 is a viable strategy to limit HIV-mediated apoptosis.
In this report, we utilized BDNF+/− mice to analyze the effect of BDNF on CCR5 and CXCR3 mRNA levels. CCR5 is another co-receptor used by HIV for cell entry (Signoret et al., 2005). CXCR3 mediates the toxic action of CXCR4 ligands (Vergote et al., 2006). Both CXCR3 and CCR5 mRNA levels were increased in BDNF+/− mice in the cerebral cortex, hippocampus and striatum. Moreover, we observed an age-dependent increase in CXCR4 mRNA levels in these animals. Intriguingly, while the net increase in CXCR3 and CXCR4 mRNAs was similar in all regions examined, CCR5 mRNA levels in the hippocampus exhibited the highest increase. The physiological significance of this effect is unclear. In the adult hippocampus, CCR5 expression is mainly localized in neurons of the CA1 region and dentate gyrus (Torres-Munoz et al., 2004; Westmoreland et al., 2002) and within neural progenitor cells (Tran et al., 2007). Multipotent neural progenitor cells are still present in the adult hippocampus and are capable of giving rise to functional neurons and glial cells (Lledo et al., 2006). BDNF has a crucial role in hippocampal neurogenesis (Schmidt and Duman, 2007). Therefore, it may be worth speculating that a balance between BDNF and chemokine receptors may contribute to modulate adult neurogenesis.
Molecular and cellular mechanisms of neuroprotection against HAD are crucial for the development of new adjunct therapies. In this and other reports (Bachis et al., 2003; Nosheny et al., 2007), we have provided evidence that BDNF regulates the expression of chemokine receptors implicated in HIV infection. Therefore, one may speculate that BDNF, by reducing membrane associated CXCR4, may help diminish the neurotoxic effect of T-tropic virus. These findings raise hope for a pharmacological intervention in HAD in adjunction to the current antiviral therapy.
4. Experimental Procedures
Animals
7-day, 6-month and 18-month-old BDNF+/− mice and WT littermates were generated in the C57BL/6 genetic background as previously described (Lyons et al., 1999) and were back-crossed for 10–12 generations to a C57BL/6 genetic background, a strategy resulting in great reduction of the genetic heterogeneity present in the original 129 Sv-C57BL/6 mixed background. Mice were group-housed under standard conditions, three to five per cage with food and water available ad libitum and were maintained on a 12-hr light/dark cycle. Both male and female mice were used for the analyses.
Cell cultures
SH-SY5Y neuroblastoma cells over-expressing TrkB (clone TB8) were prepared and grown as previously described (Kim et al., 1999). Cells were plated onto precoated 100 mm dishes (Corning Inc., Corning, NY) and grown in RPMI 1640 (Cambrex Bio Science, Walkersville, MD) containing 10% fetal bovine serum, 2 mmol/L glutamine and 0.5 µg/ml puromycin.
Semi-Quantitative RT-PCR
Total RNA was isolated from brain areas or SY5YTB8 cells using absolute RNA PCR isolation kit (Strategene, La Jolla, CA) according to the manufacturer’s protocol. Total RNA (5µg) was treated with RNAse-free DNAse and then reverse-transcribed using SuperScript and oligo(dT)-primer (Invitrogen, Grand Island, NY) according to the manufacturer’s protocol in 20 µl of reaction mixture. The resulting cDNA was amplified by PCR using sense and antisense primers described in Table 1.
PCR reactions were performed in a Genius thermal Cycler using GoTag green master mix (Promega Corp. Madison, WI). The thermal cycling parameters were as follows: 1 min 30 sec at 94°C followed by 25 cycles (housekeeping gene RPL19 and cyclophilin) and 30 cycles (CXCR3, CXCR4 and CCR5) of 30 sec at 94°C, 1 min 30 sec at 59°C, 1 min at 72°C and final incubation for 5 min at 72°C. Different dilutions of cDNA samples were used for different genes to provide a linear range of PCR reactions. The products were separated on a 2% agarose gel in Tris/acetate/EDTA buffer containing ethidium bromide. Relative abundance of mRNAs for chemokine receptors was estimated based on the intensity of cDNA bands using Quantity One 1-D Analysis Software (BioRad, Lab., Inc. Hercules, CA).
Immunohistochemistry
Mice were anesthetized with 2-2-2-tribromoethanol (125 mg/kg, Sigma) and perfused transcardially with 4% paraformaldehyde. The brain was removed and post-fixed in 4% paraformaldehyde, then transferred into buffered graded sucrose (10%, 20% and 30%) and serial cross sections (16 µm) throughout the somatosensory cortex were prepared. For CXCR4 and Nissl, sections were incubated for 24 hr at 4°C with a CXCR4 monoclonal antibody (5 µg/ml, NIH AIDS Research Reference Reagent Program, Rockville, MD) followed by fluorescein-conjugated secondary antibody (Alexafluor 546, cat #A-11030, 1:500 dil, Molecular Probes, Eugene, OR) for 1 hr at room temperature to visualize CXCR4. Sections were then incubated with NeuroTrace™ green fluorescent Nissl Stain (cat # N21489, 1:100 dil, Molecular Probes). For CXCR4 and TrkB, sections were incubated for 24 hr at 4°C with CXCR4 antibody as above, together with a polyclonal TrkB antibody (αTrkB23–36 or TrkB-in, 1:200 dilution, a gift from Dr. D.R. Kaplan, Hospital for Sick Children, University of Toronto, Toronto, Ontario, Canada). The characterization of these antibodies has been described elsewhere (Fryer et al., 1996). Alexafluor 546 (Molecular probe) and FITC (1:100, cat# F7367, Sigma St. Louis, MO) were used as secondary antibodies. For CXCR4 and GFAP, sections were incubated with the CXCR4 antibody described above for 24 hr at 4°C together with polyclonal GFAP antibody (4 µg/ml, Advanced Immunochemical, Long Beach, CA) followed by corresponding secondary antibodies FITC or Cy3, (cat # 706-165-148, 1:100 dil, Jackson ImmunoReserach Lab., West Grove, PA). For TrkB and microglia, sections were incubated with the TrkB antibody as above together with a monoclonal CD11b antibody (10 µg/ml, Serotec, Raleight, NC) followed by corresponding secondary antibodies FITC and Alexafluor 546. All primary and secondary antibodies were diluted in blocking reagent (0.01% triton, 2% BSA in 1× PBS). Reaction was visualized with a Zeiss fluorescence microscope Axiophot2. Sections were examined using a 20X objective. Total intensity of fluorescent signal was determined by using MetaMorph® software (Universal Imaging Corporation™, Downingtown, PA) as previously described (Bachis et al., 2003; Nosheny et al., 2004). A total of 10 sections (1 section every 80 µm) per animal was used (N=4 mice per group).
Cell surface biotinylation and streptavidin precipitation
Cell surface proteins were isolated using Pinpoint Cell Surface Protein Isolation Kit (Pierce, Rockford, IL) according to the manufacturer’s protocol. In brief, the day of the experiment, SY5YTB8 (Kim et al., 1999) were exposed to human recombinant BDNF (Amgen Inc., Thousand Oaks, CA), for various time points at 37°C. Medium was then replaced by PBS and cells were incubated with sulfo-NHS-LC-biotin (2 mM; Pierce, Rockford, IL) in PBS for 30 min at 4°C. Cells were then harvested and centrifuged at 500×g for 3 min and the pellet was washed once by adding 5 ml of TBS. Cells were lysed using a lysis buffer containing protease inhibitor cocktail (P 8340 Sigma, Saint Louis, MO), for 30 min at 4°C. After removal of insoluble proteins, supernatants were incubated for 1 hr at room temperature in a column containing 500 µl of immobilized neutravidin gel slurry. The biotin–streptavidin–agarose complexes were collected after centrifugation and washed three times in wash buffer. The complexes were then incubated with SDS-PAGE sample buffer, 50 mM DTT for 1 hr at room temperature. Biotinylated surface protein was finally isolated by 2 min centrifugation at 1000× g. Proteins were loaded into a 4–12% SDS-PAGE gel, and blots were probed with a CXCR4 antibody (1:1000, Santa Cruz Biotec., Inc., Santa Cruz, CA).
Data analysis
Statistical analysis was done by ANOVA followed by Student’s t test or ANOVA and Scheffe’s test for multiple comparisons (SigmaStat, SPSS Science, San Raphael, CA).
Acknowledgements
Supported by HHS grant NS 040670 and by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Special thank to NIH AIDS Research Reference Reagent Program for the CXCR4 antibody, Amgen for the gift of BDNF and Dr. D. R. Kaplan for the TrkB antibody.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The abbreviations used are: BDNF, brain-derived neurotrophic factor; CNS, central nervous system; FGF2, basic fibroblast growth factor; gp120, glycoprotein 120; GPCR: G protein-coupled receptor; HIV, Human Immunodeficiency Virus-1; HAD, HIV-associated dementia; PCR, polymerase chain reaction; RPL19, ribosomal protein 19; SDF, stromal-cell derived factor α; WT, wild type.
References
- Bachis A, Major EO, Mocchetti I. Brain-derived neurotrophic factor inhibits human immunodeficiency virus-1/gp120-mediated cerebellar granule cell death by preventing gp120 internalization. J Neurosci. 2003;23:5715–5722. doi: 10.1523/JNEUROSCI.23-13-05715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachis A, Mocchetti I. Brain-derived neurotrophic factor is neuroprotective against human immunodeficiency virus-1 envelope proteins. Ann NY Acad Sci. 2005;1053:247–257. doi: 10.1196/annals.1344.022. [DOI] [PubMed] [Google Scholar]
- Bachis A, Aden SA, Nosheny RL, Andrews PM, Mocchetti I. Axonal transport of Human Immunodeficiency Virus Type 1 envelope glycoprotein 120 is found in association with neuronal apoptosis. J. Neurosci. 2006;26:6771–6780. doi: 10.1523/JNEUROSCI.1054-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagri A, Gurney T, He X, Zou YR, Littman DR, Tessier-Lavigne M, Pleasure SJ. The chemokine SDF1 regulates migration of dentate granule cells. Development. 2002;129:4249–4260. doi: 10.1242/dev.129.18.4249. [DOI] [PubMed] [Google Scholar]
- Banisadr G, Fontanges P, Haour F, Kitabgi P, Rostene W, Parsadaniantz SM. Neuroanatomical distribution of CXCR4 in adult rat brain and its localization in cholinergic and dopaminergic neurons. Eur J Neurosci. 2002;16:1661–1671. doi: 10.1046/j.1460-9568.2002.02237.x. [DOI] [PubMed] [Google Scholar]
- Bansal AK, Mactutus CF, Nath A, Maragos W, Hauser KF, Booze RM. Neurotoxicity of HIV-1 proteins gp120 and Tat in the rat striatum. Brain Res. 2000;879:42–49. doi: 10.1016/s0006-8993(00)02725-6. [DOI] [PubMed] [Google Scholar]
- Bardi G, Sengupta R, Khan MZ, Patel JP, Meucci O. Human immunodeficiency virus gp120-induced apoptosis of human neuroblastoma cells in the absence of CXCR4 internalization. J Neurovirol. 2006;12:211–218. doi: 10.1080/13550280600848373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson JF, Long D, Doranz BJ, Rucker J, Jirik FR, Doms RW. A seven-transmembrane domain receptor involved in fusion and entry of T- cell-tropic human immunodeficiency virus type 1 strains. J. Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biard-Piechaczyk M, Robert-Hebmann V, Richard V, Roland J, Hipskind RA, Devaux C. Caspase-dependent apoptosis of cells expressing the chemokine receptor CXCR4 is induced by cell membrane-associated human immunodeficiency virus type 1 envelope glycoprotein (gp120) Virology. 2000;268:329–344. doi: 10.1006/viro.1999.0151. [DOI] [PubMed] [Google Scholar]
- Chao MV. Neurotrophins and their receptors: A convergence point for many signalling pathways. Nat Rev Neurosci. 2003;4:299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- Dorsey SG, Renn CL, Carim-Todd L, Barrick CA, Bambrick L, Krueger BK, Ward CW, Tessarollo L. In vivo restoration of physiological levels of truncated TrkB.T1 receptor rescues neuronal cell death in a trisomic mouse model. Neuron. 2006;51:21–28. doi: 10.1016/j.neuron.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Everall IP, Hansen LA, Masliah E. The shifting patterns of HIV encephalitis neuropathology. Neurotox Res. 2005;8:51–61. doi: 10.1007/BF03033819. [DOI] [PubMed] [Google Scholar]
- Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;10:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- Fryer RH, Kaplan DR, Feinstein SC, Radeke MJ, Grayson DR, Kromer LF. Developmental and mature expression of full-length and truncated TrkB receptors in the rat forebrain. J Comp Neurol. 1996;374:21–40. doi: 10.1002/(SICI)1096-9861(19961007)374:1<21::AID-CNE2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Givalois L, Arancibia S, Alonso G, Tapia-Arancibia L. Expression of Brain-Derived Neurotrophic Factor and its receptors in the median eminence cells with sensitivity to stress. Endocrinol. 2004;145:4737–4747. doi: 10.1210/en.2004-0616. [DOI] [PubMed] [Google Scholar]
- Gray F, Adle-Biassette H, Chretien F, Lorin de la Grandmaison G, Force G, Keohane C. Neuropathology and neurodegeneration in human immunodeficiency virus infection. Pathogenesis of HIV-induced lesions of the brain, correlations with HIV-associated disorders and modifications according to treatments. Clin Neuropathol. 2001;20:146–155. [PubMed] [Google Scholar]
- He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay CR, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- Hesselgesser J, Taub D, Baskar P, Greenberg M, Hoxie J, Kolson DL, Horuk R. Neuronal apoptosis induced by HIV-1 gp120 and the chemokine SDF-1 alpha is mediated by the chemokine receptor CXCR4. Curr Biol. 1998;8:595–598. doi: 10.1016/s0960-9822(98)70230-1. [DOI] [PubMed] [Google Scholar]
- James HJ, Sharer LR, Zhang Q, Wang HG, Epstein LG, Reed JC, Gelbard HA. Expression of caspase-3 in brains from paediatric patients with HIV-1 encephalitis. Neuropathol Appl Neurobiol. 1999;25:380–386. doi: 10.1046/j.1365-2990.1999.00195.x. [DOI] [PubMed] [Google Scholar]
- Kaplan DR, Matsumoto K, Lucarelli E, Thiele CJ. Induction of TrkB by retinoic acid mediates biologic responsiveness to BDNF and differentiation of human neuroblastoma cells. Neuron. 1993;11:321–331. doi: 10.1016/0896-6273(93)90187-v. [DOI] [PubMed] [Google Scholar]
- Kaul M, Lipton SA. Chemokines and activated macrophages in HIV gp120-induced neuronal apoptosis. Proc Natl Acad Sci U S A. 1999;96:8212–8216. doi: 10.1073/pnas.96.14.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CJ, Matsuo T, Lee K-H, Thiele CJ. Up-regulation of Insulin-Like Growth Factor-II expression is a feature of TrkA but not TrkB activation in SH-SY5Y neuroblastoma cells. Am J Pathol. 1999;155:1661–1670. doi: 10.1016/S0002-9440(10)65481-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RS, Williams KC, Alvarez-Hernandez X, Westmoreland S, Force T, Lackner AA, Luster AD. Chemokine receptor expression and signaling in macaque and human fetal neurons and astrocytes: implications for the neuropathogenesis of AIDS. J Immunol. 1999;163:1636–1646. [PubMed] [Google Scholar]
- Kruman II, Nath A, Mattson MP. HIV-1 protein Tat induces apoptosis of hippocampal neurons by a mechanism involving caspase activation, calcium overload, and oxidative stress. Exp Neurol. 1998;154:276–288. doi: 10.1006/exnr.1998.6958. [DOI] [PubMed] [Google Scholar]
- Lavi E, Strizki JM, Ulrich AM, Zhang W, Fu L, Wang Q, O'Connor M, Hoxie JA, Gonzalez-Scarano F. CXCR-4 (Fusin), a co-receptor for the type 1 human immunodeficiency virus (HIV-1), is expressed in the human brain in a variety of cell types, including microglia and neurons. Am J Pathol. 1997;151:1035–1042. [PMC free article] [PubMed] [Google Scholar]
- Lazarini F, Casanova P, Tham TN, De Clercq E, Arenzana-Seisdedos F, Baleux F, Dubois-Dalcq M. Differential signalling of the chemokine receptor CXCR4 by stromal cell-derived factor 1 and the HIV glycoprotein in rat neurons and astrocytes. Eur J Neurosci. 2000;12:117–125. doi: 10.1046/j.1460-9568.2000.00894.x. [DOI] [PubMed] [Google Scholar]
- Lazarini F, Tham TN, Casanova P, Arenzana-Seisdedos F, Dubois-Dalcq M. Role of the alpha-chemokine stromal cell-derived factor (SDF-1) in the developing and mature central nervous system. Glia. 2003;42:139–148. doi: 10.1002/glia.10139. [DOI] [PubMed] [Google Scholar]
- Lledo PM, Alonso M, Grubb MS. Adult neurogenesis and functional plasticity in neuronal circuits. Nat Rev Neurosci. 2006;7:179–193. doi: 10.1038/nrn1867. [DOI] [PubMed] [Google Scholar]
- Lu M, Grove EA, Miller RJ. Abnormal development of the hippocampal dentate gyrus in mice lacking the CXCR4 chemokine receptor. Proc Natl Acad Sci U S A. 2002;99:7090–7095. doi: 10.1073/pnas.092013799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons WE, Mamounas LA, Ricaurte GA, Coppola V, Reid SW, Bora SH, Wihler C, Koliatsos VE, Tessarollo L. Brain-derived neurotrophic factor-deficient mice develop aggressiveness and hyperphagia in conjunction with brain serotonergic abnormalities. Proc Natl Acad Sci U S A. 1999;96:15239–15244. doi: 10.1073/pnas.96.26.15239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Jones D, Borghesani PR, Segal RA, Nagasawa T, Kishimoto T, Bronson RT, Springer TA. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad Sci U S A. 1998;95:9448–9453. doi: 10.1073/pnas.95.16.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masliah E, DeTeresa RM, Mallory ME, Hansen LA. Changes in pathological findings at autopsy in AIDS cases for the last 15 years. AIDS. 2000;14:69–74. doi: 10.1097/00002030-200001070-00008. [DOI] [PubMed] [Google Scholar]
- McArthur JC. HIV dementia: an evolving disease. J Neuroimmunol. 2004;157:3–10. doi: 10.1016/j.jneuroim.2004.08.042. [DOI] [PubMed] [Google Scholar]
- Meucci O, Fatatis A, Simen AA, Bushell TJ, Gray PW, Miller RJ. Chemokines regulate hippocampal neuronal signaling and gp120 neurotoxicity. Proc Natl Acad Sci U S A. 1998;95:14500–14505. doi: 10.1073/pnas.95.24.14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller FD, Kaplan DR. Signaling mechanisms underlying dendrite formation. Curr Opin Neurobiol. 2003;13:391–398. doi: 10.1016/s0959-4388(03)00072-2. [DOI] [PubMed] [Google Scholar]
- Miller RJ, Tran PB. Chemokinetics. Neuron. 2005;47:621. doi: 10.1016/j.neuron.2005.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocchetti I, Bachis A. Brain-derived neurotrophic factor activation of TrkB protects neurons from HIV-1/gp120-induced cell death. Crit Rev Neurobiol. 2004;16:51–57. doi: 10.1615/critrevneurobiol.v16.i12.50. [DOI] [PubMed] [Google Scholar]
- Moore JP, Kitchen SG, Pugach P, Zack JA. The CCR5 and CXCR4 coreceptors--central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res Hum Retroviruses. 2004;20:111–126. doi: 10.1089/088922204322749567. [DOI] [PubMed] [Google Scholar]
- Nosheny RL, Bachis A, Acquas E, Mocchetti I. Human immunodeficiency virus type 1 glycoprotein gp120 reduces the levels of brain-derived neurotrophic factor in vivo: potential implication for neuronal cell death. Eur J Neurosci. 2004;20:2857–2864. doi: 10.1111/j.1460-9568.2004.03764.x. [DOI] [PubMed] [Google Scholar]
- Nosheny RL, Amhed F, Yakovlev AG, Meyer EM, Ren K, Tessarollo L, Mocchetti I. Brain-derived neurotrophic factor prevents the nigrostriatal degeneration induced by human immunodeficiency virus-1 glycoprotein 120 in vivo. Eur. J. Neurosci. 2007;25:2275–2284. doi: 10.1111/j.1460-9568.2007.05506.x. [DOI] [PubMed] [Google Scholar]
- Ohira K, Hayashi M. Expression of TrkB subtypes in the adult monkey cerebellar cortex. Journal of Chemical Neuroanatomy. 2003;25:175–183. doi: 10.1016/s0891-0618(02)00096-0. [DOI] [PubMed] [Google Scholar]
- Sanders VJ, Everall IP, Johnson RW, Masliah E. Fibroblast growth factor modulates HIV coreceptor CXCR4 expression by neural cells. HNRC Group. J Neurosci Res. 2000;59:671–679. doi: 10.1002/(SICI)1097-4547(20000301)59:5<671::AID-JNR10>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Schmidt HD, Duman RS. The role of neurotrophic factors in adult hippocampal neurogenesis, antidepressant treatments and animal models of depressive-like behavior. Behav Pharmacol. 2007;18:391–418. doi: 10.1097/FBP.0b013e3282ee2aa8. [DOI] [PubMed] [Google Scholar]
- Signoret N, Hewlett L, Wavre S, Pelchen-Matthews A, Oppermann M, Marsh M. Agonist-induced Endocytosis of CC Chemokine Receptor 5 Is Clathrin Dependent. Mol Biol Cell. 2005;16:902–917. doi: 10.1091/mbc.E04-08-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumm RK, Zhou C, Ara T, Lazarini F, Dubois-Dalcq M, Nagasawa T, Hollt V, Schulz S. CXCR4 regulates interneuron migration in the developing neocortex. J. Neurosci. 2003;23:5123–5130. doi: 10.1523/JNEUROSCI.23-12-05123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasova NI, Stauber RH, Michejda CJ. Spontaneous and ligand-induced trafficking of CXC-chemokine receptor 4. J Biol Chem. 1998;273:15883–15886. doi: 10.1074/jbc.273.26.15883. [DOI] [PubMed] [Google Scholar]
- Toggas SM, Masliah E, Rockenstein EM, Rall GF, Abraham CR, Mucke L. Central nervous system damage produced by expression of the HIV-1 coat protein gp120 in transgenic mice. Nature. 1994;367:188–193. doi: 10.1038/367188a0. [DOI] [PubMed] [Google Scholar]
- Torres-Munoz JE, Van Waveren C, Keegan MG, Bookman RJ, Petito CK. Gene expression profiles in microdissected neurons from human hippocampal subregions. Mol Brain Res. 2004;127:105–114. doi: 10.1016/j.molbrainres.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Tran PB, Banisadr G, Ren D, Chenn A, Miller RJ. Chemokine receptor expression by neural progenitor cells in neurogenic regions of mouse brain. J. Comp. Neurol. 2007;500:1007–1034. doi: 10.1002/cne.21229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer P, Ulrich AM, Gonzalez-Scarano F, Lavi E. Immunohistochemical analysis of CCR2, CCR3, CCR5, and CXCR4 in the human brain: potential mechanisms for HIV dementia. Exp Mol Pathol. 2000;69:192–201. doi: 10.1006/exmp.2000.2336. [DOI] [PubMed] [Google Scholar]
- Vergote D, Butler GS, Ooms M, Cox JH, Silva C, Hollenberg MD, Jhamandas JH, Overall CM, Power C. Proteolytic processing of SDF-1{alpha} reveals a change in receptor specificity mediating HIV-associated neurodegeneration. Proc. Natl. Acad. Sci. 2006;103:19182–19187. doi: 10.1073/pnas.0604678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland SV, Alvarez X, deBakker C, Aye P, Wilson ML, Williams KC, Lackner AA. Developmental expression patterns of CCR5 and CXCR4 in the rhesus macaque brain. J Neuroimmunol. 2002;122:146–158. doi: 10.1016/s0165-5728(01)00457-x. [DOI] [PubMed] [Google Scholar]
- Zheng J, Thylin MR, Ghorpade A, Xiong H, Persidsky Y, Cotter R, Niemann D, Che M, Zeng YC, Gelbard HA, Shepard RB, Swartz JM, Gendelman HE. Intracellular CXCR4 signaling, neuronal apoptosis and neuropathogenic mechanisms of HIV-1-associated dementia. J Neuroimmunol. 1999;98:185–200. doi: 10.1016/s0165-5728(99)00049-1. [DOI] [PubMed] [Google Scholar]