Abstract
Apoptosis and autophagy are genetically-regulated, evolutionarily-conserved processes that regulate cell fate. Both apoptosis and autophagy are important in development and normal physiology and in a wide range of diseases. Recent studies show that despite the marked differences between these two processes, their regulation is intimately connected and the same regulators can sometimes control both apoptosis and autophagy. In this review, I discuss some of these findings, which provide possible molecular mechanisms for crosstalk between apoptosis and autophagy and suggest that it may be useful to think of these processes as different facets of the same cell death continuum rather than completely separate processes.
Keywords: Autophagy; Apoptosis; Bcl-2, FADD; Atg 5
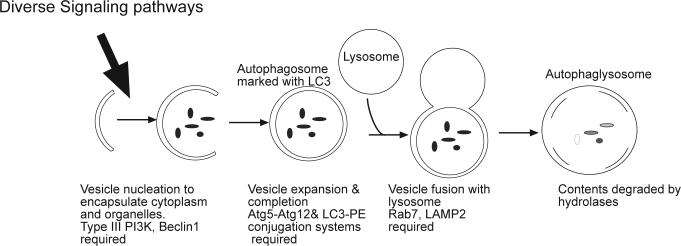
In the early to mid 1990s there was a rapid increase in our understating about the regulation of apoptosis. Fifteen or so years later we are in the midst of a similarly exciting period for understanding autophagy with very rapid growth of publications on the regulation and function of this process (1). Autophagy (the form of autophagy discussed here is macroautophagy, other forms– microautophagy and chaperone-mediated autophagy share the same lysosomal degradation mechanism but differ in the way that material is delivered to the lysosome) is a degradative process involving sequestration of parts of the cytoplasm in double membrane vesicles (autophagic vesicles or autophagosomes) that fuse with lysosomes forming the autophagolysosome. In the autophagolysosome, the cytoplasmic material that was engulfed is hydrolyzed and the resulting amino acids and other macromolecular precursors can be recycled (2-4), see Fig 1 for a schematic of the process. Autophagy is a mechanism by which organelles are removed and is the primary degradation mechanism for long-lived proteins and thus maintains quality control for proteins and organelles. Autophagy is ubiquitous in eukaryotic cells and important in development and in diverse pathophysiological conditions (5)– for example providing protection against neurodegeneration (6, 7), infections (8, 9) and tumor development (10-13). Cells that are deficient in autophagy also demonstrate enhanced chromosomal instability (14, 15), which may be related to the tumorigenesis associated with autophagy deficiency.
Figure 1. Schematic of macroautophagy.
A membrane forms and expands to sequester cytoplasm and organelles in a double-membrane autophagosome. The initial step requires the activity of a Type III PI3kinase and the Beclin1 protein and is regulated by diverse signaling pathways providing both positive and negative signals. Completion and expansion of the autophagosome requires the products of two protein conjugation systems that produce Atg5-Atg12 and Atg8/LC3-PE. The mature autophagosome fuses with a lysosome through a step that requires the Rab7 GTPase and the LAMP2 protein to produce the autophagolysosome in which the contents are degraded.
Autophagy is important in cell death decisions and can protect cells by preventing them from undergoing apoptosis. For example, increased autophagy in nutrient deprived or growth factor-withdrawn cells allows cell survival (16, 17) by inhibiting apoptosis. Autophagy also protects cells from various other apoptotic stimuli (18). It is not clear how autophagy stops cells from undergoing apoptosis; one suggested mechanism is the sequestration of damaged mitochondria (18) thus preventing released cytochrome c from being able to form a functional apoptosome in the cytoplasm. Recent work suggests that autophagy can also protect cells from caspase-independent death that occurs after Mitochondrial Outer Membrane Permeabilization (MOMP) in the presence of caspase inhibitors. Interestingly this study showed that as long as energy can still be generated (in this case by increased glycolysis caused by elevated GAPDH), cells can use autophagy to survive MOMP and the release of cytochrome c and other apoptogenic proteins and recover to continue to grow (19). Thus, increased levels of autophagy can protect cells from apoptosis and this kind of caspase-independent death. Unfortunately, at least from the perspective of a simple story, things are not so straightforward because autophagy can also do the opposite– it can also kill cells.
Autophagic cell death (also known as Type II programmed cell death to distinguish it from apoptosis or Type I programmed cell death) (20-22) has been described as a distinct form of cell death that differs from other death mechanisms such as apoptosis and necrosis. One example of autophagic cell death was caused by increased reactive oxygen species resulting from autophagic degradation of catalase (23). Unlike apoptosis, which relies upon the activation of caspases that cleave hundreds of target proteins (24), autophagic cell death is usually thought of as caspase-independent (21). Indeed, two of the best examples of autophagic cell death where the demise of the cell was shown to require the autophagy machinery took place in cells with profound defects in the apoptosis machinery (25) or in the presence of caspase inhibitors (26). Cells undergoing autophagic cell death or apoptosis look different (27); the characteristic cellular morphology associated with apoptosis is usually due to caspase cleavage of cytoskeletal and other structural proteins (24) so that apoptotic cells show early degradation of the cytoskeleton but preserve organelles until fairly late in the process. In contrast, autophagic cell death is associated with accumulation of large numbers of autophagic vesicles, which degrade organelles early in the process while the cytoskeleton remains intact and functional until late in the process (5). With this background, it is perhaps not surprising that there would be close connections between the regulatory machinery that control autophagy and apoptosis. As discussed below, recent data from a number of groups has started to describe these connections.
Regulation of Autophagy
Autophagy is controlled by a group of evolutionarily conserved genes (ATG genes) (28, 29) that control a coordinated process leading to the induction and nucleation of autophagic vesicles, their completion, expansion and fusion with lysosomes and breakdown and recycling. Over 30 ATG genes have been identified in yeast and at least 11 (ATG 1, 3, 4, 5, 6, 7, 8, 9, 10,12 and 16) have orthologs in mammals, Atg6 is also known as Beclin1 and Atg8 is commonly called LC3 in mammals. Not surprisingly given the large number of stimuli that modulate autophagy, numerous upstream signaling pathways (growth factor signaling, PI3 kinase/ Akt, MAP kinases, AMP dependent protein kinase, small GTPases, trimeric G proteins, inositol triphosphates, calcium signaling and others) regulate the process. Many of these pathways work through mTOR, which is a potent inhibitor of autophagy. Correspondingly mTOR inhibitors such as rapamycin activate autophagy. However there are also mTOR-independent mechanisms of inducing autophagy. For example, the disaccharide trehalose is an mTOR-independent activator of autophagy (30). The induction of autophagy occurs through a mechanism that involves the Atg1 kinase, which when overexpressed in Drosophila is sufficient to induce autophagy (31). In mammalian cells the Atg1 ortholog ULK1 also controls autophagy (32). Nucleation of the autophagic vesicle involves a Type III PI3 kinase complex that includes the kinase (Vps34) and Beclin1 (Atg6), which interact through a conserved domain in Beclin 1 (33). This complex contains multiple proteins including another putative tumor suppressor called UVRAG (34) and a protein called Ambra1 (35) , which regulate the activity of the kinase and thus control autophagy induction. Recently it was demonstrated that another protein called Bif-1 binds to UVRAG and that this interaction can also regulate the PI3 kinase activity and induce autophagic vesicle formation (36). The formation and expansion of the autophagosome requires two protein conjugation systems that involve many of the identified Atg proteins (37); Atg8/LC3 and Atg12 are similar to ubiquitin and are activated by Atg7, which acts as the E1 enzyme for both Atg8 and Atg12 leading to their conjugation to the lipid phosphatidylethanolamine (PE) and Atg5 respectively in multi-step processes that also involve Atg3, Atg4 and Atg10. The Atg5-Atg12 conjugate is required for the formation of the LC3 conjugate and both the Atg5-Atg12 and LC3-PE conjugates are associated with the autophagosome. The conjugated form of LC3 is called LC3-II to distinguish it from the unconjugated form (LC3-I) and has faster mobility on SDS gels. Atg5 dissociates from the autophagosome but LC3-II remains associated and is eventually degraded in the autophagolysosomes. Consequently, the processing and degradation of LC3 (or GFP-tagged LC3) is often used as a way to test if autophagy is being increased or decreased (38). Atg9 is a transmembrane protein that shuttles between the Golgi and endosomes and may identify the membranes used to form the autophagic vesicle. Fusion of the autophagosome with the lysosome requires the Rab7 GTPase (39, 40) and the lysosome-associated membrane protein-2 (LAMP2) (41). By interfering with specific regulators it is feasible to block autophagy at different stages in the process. For example, to block autophagy at early stages and prevent autophagosome formation, one could inhibit the Type III PI3 kinase complex with 3-methyl adenine or knockdown Beclin 1 or Atg5 with siRNAs, while to inhibit autophagy at later stages one could prevent fusion between the autophagosome and lysosome by knocking down or inhibiting Rab7 or use pharmacological agents such as the lysomotropic agent chloroquine or bafilomycin A1, an inhibitor of the vacuolar type H+ ATPase. It may be important to discriminate between blocking autophagy at early or later steps because differences in outcome for the cell have been noted when this has been done (42). When the later stages of autophagy are blocked, the cell may accumulate large numbers of autophagosomes. Therefore the appearance of large numbers of autophagosomes after treatment with a stimulus can indicate either an increase in autophagy (i.e. more generation of autophagosomes) or a blocking of autophagy at the fusion step with lysosomes. Measurement of the autophagic flux by monitoring the degradation of LC3 or long-lived proteins helps distinguish between these possibilities. Although much progress has been made in the decade since ATG1 was first identified (1), many unresolved questions remain regarding the molecular regulation of autophagy (43).
Regulation of Apoptosis
Morphological changes and death in apoptotic cells are caused by caspases, which cleave about 400 proteins (24). Apoptotic caspases include “initiator caspases” (caspase-2, −8, −9 and −10) that start an apoptotic cascade and “effector caspases” (caspase-3, −6 and −7) that disassemble the cell. Different pathways leading to caspase activation have been characterized (44). Diverse stimuli cause release of mitochondrial proteins to activate the intrinsic apoptosis pathway (45) leading to MOMP and the release of cytochrome c and other apoptogenic proteins (46). MOMP is regulated by the Bcl family of proteins (47)– these proteins contain one or more Bcl homology (BH) domains and fall into three categories; anti-apoptotic BH1234 proteins (e.g. Bcl-2, Bcl-xL, Mcl-1), which have four BH domains, pro-apoptotic BH123 proteins (e.g. Bax, Bak) and a family of BH3-only proteins (e.g. Bid, Bim, Bad, Noxa and others), which sense different kinds of cell stress/damage. The BH3-only proteins work by directly activating the BH123 proteins or neutralizing the BH1234 proteins (there are competing models about which of these two mechanisms applies (48-50)). Released cytochrome c interacts with Apaf-1, pro-caspase-9 and dATP to form the apoptosome (51). This complex activates caspase-9, which then activates effector caspases. At the same time, release of Smac/Diablo (52, 53) inhibits Inhibitor of Apoptosis Proteins (IAPs), which are themselves caspase inhibitors to provide a coordinated method to efficiently activate caspases.
The death receptor or extrinsic pathway involves cell surface receptors (54), which induce apoptosis by forming a multi-protein complex called the Death-Inducing Signaling Complex (DISC) (55). Upon ligand binding, death receptors recruit an adapter protein called FADD (56-58), which brings caspase-8 to the DISC where it is dimerized and catalytically activated (59). Active caspase-8 can then directly activate effector caspases such as caspase-3 or can activate the intrinsic apoptosis pathway by cleaving the BH3-only protein Bid resulting in its translocation to the mitochondria (60). A third apoptosis pathway is activated when the endoplasmic reticulum (ER) is stressed (61). This pathway involves many of the same BH-containing proteins that regulate the mitochondrial pathway. Bax and Bak are also located at the ER membrane (62) where they regulate apoptosis and cause calcium release from the ER through mechanisms that are countered by ER-localized Bcl-2 and Bcl-xL. These signals usually eventually activate the mitochondrial pathway too (63). Apoptosis as a result of mitochondrial permeabilization can also be triggered following lysosomal permeabilization (64) and accumulating evidence indicates that there may be direct linkages between lysosomal functions and both apoptosis and autophagy (65). For example, it was recently reported that a transmembrane protein called TMEM166 that is associated with lysosomes and the ER as been reported to regulate apoptosis and autophagy (66).
Molecular Connections between Autophagy and Apoptosis
Since autophagy can block apoptosis and both autophagy and apoptosis can kill cells, one might expect that their regulation would be coordinated. It is perhaps a little less expected that the same proteins would regulate both processes. However, recent data show that this is often the case. Some connections occur upstream of the autophagic and apoptotic machinery itself where signaling pathways regulate both processes. For example, p53, which is a potent inducer of apoptosis, can also induce autophagy through increased expression of a direct p53 target gene called DRAM (67). Similarly activation of the PI3 kinase/ Akt pathway, which is a well known way to inhibit apoptosis, also inhibits autophagy (68). Thus important signaling pathways apparently simultaneously increase or decrease both autophagy and apoptosis. In addition, proteins that are themselves central components of the apoptosis or autophagy machinery (Bcl family proteins, FADD, and at least one of the Atg proteins) regulate both processes directly.
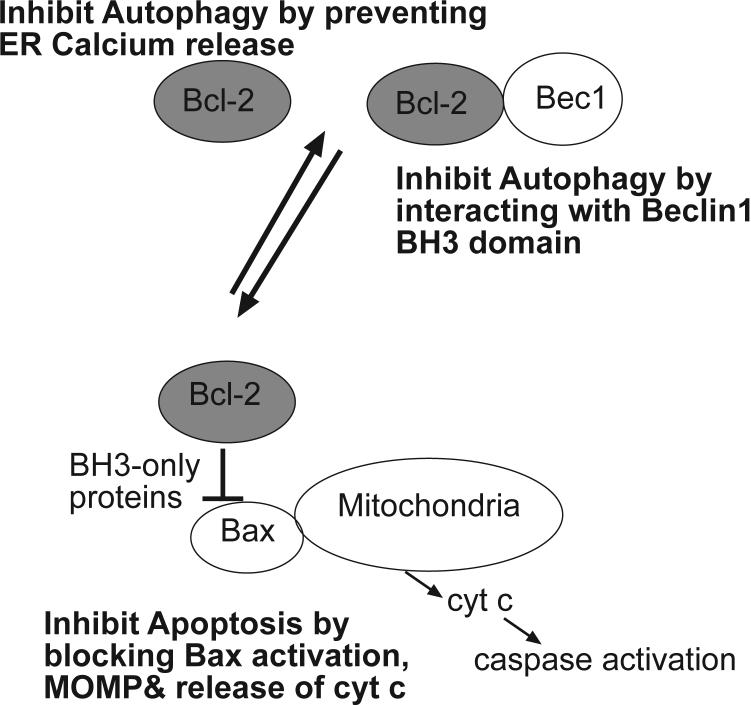
As discussed above, Beclin 1/Atg 6 is part of a Type III PI3 kinase complex that is required for the formation of the autophagic vesicle and interference with Beclin 1 can prevent autophagy induction. Beclin 1 was identified as a Bcl-2 interacting protein (10). Thus a key apoptosis regulator also interacts physically with an autophagy regulator. Beclin1 also interacts with the other major anti-apoptotic Bcl family protein (Bcl-xL) and this interaction has been shown to regulate autophagy so that not only does Bcl-2/ Bcl-xL inhibit apoptosis by binding to and interfering with the action of the pro-apoptotic proteins Bax and Bak, it also inhibits autophagy by binding with Beclin 1 and this interaction is important in the regulation of starvation-induced autophagy (69). There may however be differences in the regulation of these pathways depending on subcellular localization – Bcl-2 is found at the mitochondria and the ER however the autophagy inhibition function of Bcl-2 occurs only at the ER and mitochondrial targeted Bcl-2, which is a potent inhibitor of many apoptotic stimuli (70), cannot inhibit autophagy (69). Recent work from several groups has focused on further analysis of the Beclin 1-Bcl-2/Bcl-xL interaction. Structural and biochemical studies demonstrate that the interaction is via a BH3 domain in Beclin 1(71-74)– i.e. Beclin 1 is a BH3-only protein. Furthermore, elegant experiments from Kroemer and colleagues show that disruption of the interaction between the Beclin 1 BH3 domain and Bcl-2 leads to increased autophagy (71). Thus, in this case a BH3 domain interaction with Bcl-2 serves not to regulate apoptosis but instead to control autophagy. It is also possible that the Beclin 1 BH3 domain may function like other BH3 domains and regulate apoptosis; however clear evidence for this has not been shown.
Another mechanism by which Bcl-2 at the ER can control autophagy has been described (75). In this case Bcl-2 inhibits autophagy not because it interacts with Beclin 1 but instead because it can block calcium release from the ER. The calcium activates Ca2+/ calmodulin-dependent kinase kinase-ß and AMP-activated protein kinase, which leads to inhibition of mTOR to activate autophagy. Therefore two quite separate mechanisms may allow Bcl-2 (and Bcl-xL) to inhibit autophagy (Fig. 2) instead of apoptosis; it is not known if both these mechanisms apply at the same time or if one is more important than the other in different contexts. Added complexity comes from the fact that under other circumstances – in cells that have lost both Bax and Bak and are therefore essentially completely resistant to the intrinsic apoptosis pathway– Bcl-2 (and Bcl-xL) appears to have the opposite effect because increasing their expression stimulates autophagic cell death while Bcl-xL knockdown reduces autophagy (25). Thus while the connections between Bcl family of apoptosis regulators and autophagy appear to be complicated it is clear that this family of proteins are intimately involved in regulating the two processes and that it may be where the proteins are– i.e. at the ER or elsewhere– that determines whether they regulate apoptosis or autophagy.
Figure 2. Regulation of apoptosis and autophagy by Bcl-2 and Bcl-xL.
Bcl-2 (and Bcl-xL) inhibits apoptosis by blocking activation of Bax (and Bak) to prevent MOMP and subsequent release of cytochrome c and other apoptogenic proteins from the mitochondria. Alternatively, Bcl-2 at the endoplasmic reticulum can inhibit autophagy by interacting with the Beclin 1 BH3 domain and by preventing calcium release from the ER to block the activation of a signaling pathway involving CAMKK-ß, AMPK which represses mTOR to activate autophagy.
Key components of the other well-characterized apoptosis pathway, the extrinsic death receptor pathway, can also control autophagy. Binding of the adaptor protein FADD to the ligand-bound death receptor (or other adaptors) is a required step in the formation of the DISCs that mediate death receptor signaling (55) with FADD functioning as a platform upon which caspase-8 dimerization and activation occurs. FADD consists of two protein interaction domains, a death domain and a death effector domain. Unexpectedly, in normal epithelial cells the death domain of FADD is able to induce a novel cell death mechanism that involves high levels of autophagy (76). Since the FADD death domain has no catalytic activity it presumably induces autophagy by physically interacting with another protein. We don't know what that protein is as yet (although, as discussed below, it can interact with Atg5). Interestingly, and in keeping with a theme that will be discussed further below, the autophagy response is more easily observed when apoptosis is blocked, suggesting that autophagy and apoptosis are induced simultaneously by FADD death domain in the normal epithelial cells. This mechanism of FADD-regulated autophagy also applies when FADD-dependent signaling is induced by TRAIL, a cytokine that activates the DR4 and DR5 death receptors. A FADD-independent mechanism of inducing autophagy and autophagic cell death from the DR5 receptor was also recently proposed in response to an activating antibody (77). Thus, there may be more than one way to activate autophagy from death receptors.
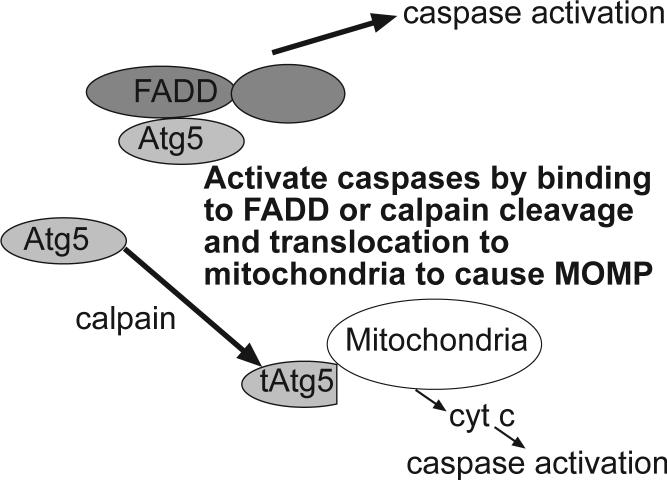
The above studies indicate that components of the core apoptosis machinery regulating the intrinsic and extrinsic pathways can also control autophagy. Other recent studies show the converse – i.e. that autophagy regulators can control apoptosis. In experiments examining interferon- and Atg5-induced autophagic cell death, it was shown that FADD can interact with Atg5 (78). This study suggested that the interaction resulted in cell death (but not the autophagic vesicle formation) that required FADD and involved caspases. The implication of these studies is that Atg5 can regulate components of the extrinsic apoptosis pathway. Another mechanism by which Atg5 can regulate apoptosis has also been described (79). The key step in this mechanism is the cleavage of Atg5 by calpain to create a truncated form of the protein that translocates to the mitochondria to cause cytochrome c release and activation of the intrinsic apoptosis pathway that can be blocked by Bcl-2. The general significance of this mechanism is suggested by data showing that Atg5 knockdown protects tumor cells towards a variety of apoptosis stimuli while Atg5 expression sensitized to these apoptotic stimuli. Again the situation may be rather complex and calpain activity may both increase and decrease autophagy because while calpain cleavage of Atg5 produces a protein that cannot activate autophagy (79), other studies show that calpain activity is required for autophagy induction by rapamycin and amino acid starvation (80). Further discussion of interactions between apoptosis and autophagy can also be found in a recent review article by Maiuri et.al. (81).
Are autophagic and apoptotic cell death different facets of the same cell death mechanism?
We tend to think of autophagic and apoptotic cell death as being very different mechanisms (21) and much effort in the literature is devoted to defining whether your favorite cell death is occurring through one or the other mechanism, by, for example, asking if autophagy is promoting or preventing death. This makes sense based on the clear morphological differences that can be seen between cells that are thought to be dying by apoptosis versus autophagic cell death and could also have practical importance. For example, since the mechanism of tumor cell killing by an anti-cancer agent determines the way that resistance to the therapy can be selected for, a drug that induces apoptosis and one that induces autophagic cell death might provide quite different selective pressures on the tumor cell and might therefore be more or less effective against a particular tumor (22, 82). On the other hand, the close connections between the apoptosis machinery and the autophagic machinery that are being discovered would be expected to lead to a situation where there may be simultaneous activation of the processes. This, in turn suggests that cell death may actually involve both processes. Direct evidence for this idea was obtained from the studies showing that FADD death domain could induce both apoptosis and autophagy as it kills normal epithelial cells (76) and the finding that Atg5-induced death, which involved the generation of large numbers of autophagosomes required FADD binding and involved caspases (78). However this idea extends beyond FADD because similar connections have been found in other circumstances too.
Neufeld and colleagues performed experiments to overexpress Atg1 in Drosophila and found that this was sufficient to increase autophagy, to inhibit cell growth and to induce cell death (31). However, while the death required that the autophagy signal from Atg1 be propagated – mutation of a downstream autophagy regulator, Atg8 blocked the Atg1-induced death– the actual mechanism of cell death appeared to be through apoptosis and was associated with increased caspase activity and blocked by a caspase inhibitor. Similar effects also occur in a physiological cell death response that occurs during Drosophila metamorphosis. Steroid regulated death during Drosophila metamorphosis occurs by a process that looks like autophagic cell death (83). However, regulators of the apoptotic machinery such as grim, reaper and dronk control the process (84), and caspase inhibition prevents the death (85). Thus although the morphological studies of these dying cells would lead us to conclude that they are dying by autophagic Type II cell death, this death requires regulators that are usually thought of as controlling apoptotic cell death. Similar mechanisms apply in mammalian cells too; nutrient deprivation in cells that lack LAMP2 and thus cannot fuse autophagosomes with lysosomes causes a cell death response with morphological characteristics of autophagic cell death. However, the actual killing of the cells involves a later activation of apoptosis with caspase activation occurring subsequent to MOMP (41). These examples suggest that the idea that there are two clearly different programmed cell death mechanisms (Type I and Type II, apoptosis and autophagic cell death) is oversimplified. Instead, perhaps what we are really seeing are different facets of the same integrated cell death mechanism– i.e. there aren't really two different programmed cell death mechanisms (with necrosis as a less regulated but distinct third mechanism), instead there is one integrated cell death mechanism that involves both apoptotic and autophagic components working together and regulating each other in a cell death continuum that also involves necrotic cell death (Fig. 4). In this scenario, there is no clear cut distinction between different death mechanisms and what we see when we study dying cells depends on which facet of the mechanism is most prominent at the time we happen to be looking. Moreover, by manipulating the continuum it may be feasible to alter both the amount of cell death and the characteristics (i.e. the relative amount of apoptosis, necrosis, autophagy etc,) that occurs as the cells die because if everything is connected as in Fig. 4, alteration of one mechanism should also affect the others.
Figure 4. Cell death continuum.
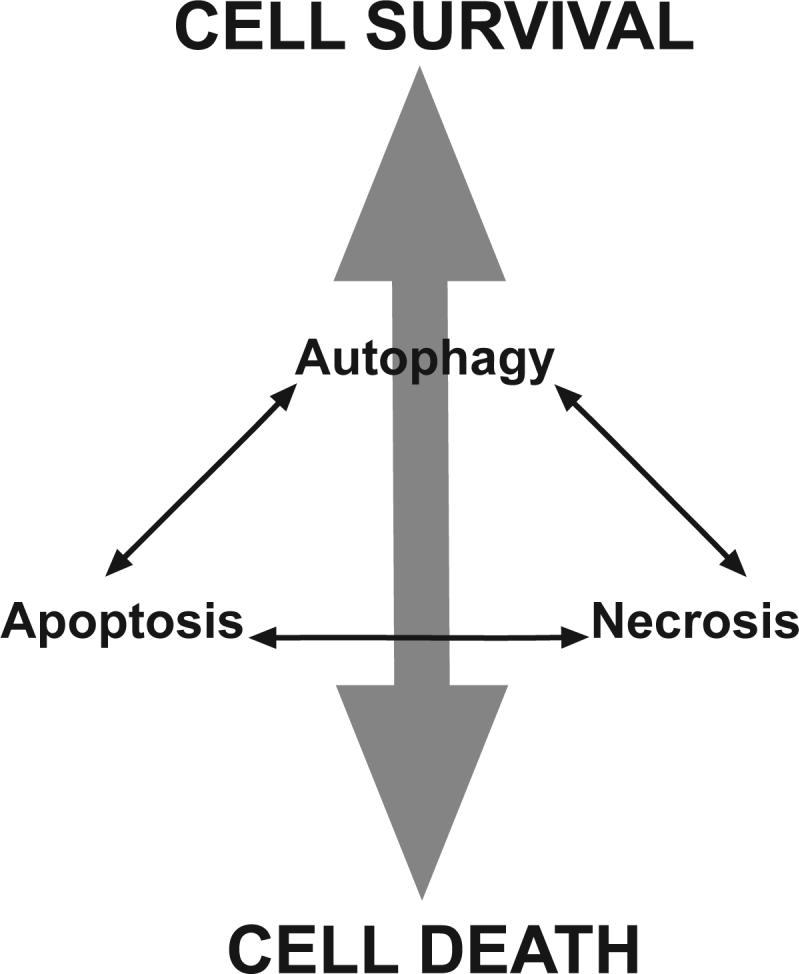
Mechanistic interactions between regulators of apoptosis autophagy and necrosis control whether a cell lives or dies and determine the particular facets of the death mechanism with characteristics of apoptosis, necrosis etc. that occur in the dying cells. Perturbation of these interactions may alter the amount and characteristics of cell death.
These ideas could have practical implications as we try to manipulate these pathways to better treat disease. For example, when treating cancer we often use drugs that cause severe damage to the cell. Most cancer cells still have functional apoptosis machinery (86) and many anti-cancer agents activate caspases and induce apoptosis. So, when cancer cells are treated, the induction of autophagy and the interactions between autophagy and apoptosis, e.g. mediated by Bcl-2's ability to regulate both processes, could have profound effects on the outcome for the tumor cell. Autophagy may reduce the apoptosis sufficiently to keep the cell under the threshold of caspase activity that is needed to cause death. If the inhibition of apoptosis by autophagy is able to keep the tumor cells alive long enough to repair the damage, metabolize the drug etc., the long term effect might be that the cells regain their ability to grow and re-populate the tumor. In a situation like this, it would be useful to combine the drug with an autophagy inhibitor because this will increase the overall efficiency of the anti-cancer treatment i.e. increase the number of tumor cells killed for a given dose of drug. Recent studies suggest that this may indeed be useful and lead to increased tumor regression in animal models (87).
However, what might happen when the cellular damage is so severe that it cannot be repaired or there is sustained interference with essential cellular processes such as protein synthesis such that no matter what happens, the tumor cell cannot survive in the long term; under these circumstances, does the relative amount of autophagy and apoptosis matter? In this case, the role of autophagy may be less to do with the absolute number of tumor cells that are eventually killed but instead could control where on the continuum of cell death mechanisms the death occurs. For example, when autophagy blocks apoptosis, the cells may die by a non-apoptotic mechanism when autophagy occurs but may activate caspases and die by apoptosis when autophagy is inhibited. In tumor cells that have profound defects in apoptosis and are induced to undergo increased autophagy through hypoxia and nutrient deprivation, inhibition of autophagy has also been shown to cause tumor cells to die with characteristics of necrosis including release of HMGB1 (88). In the real world of cancer treatment, these differences in the ways that cells die could have significant practical implications; release of HMGB1 from necrotic cells (it is not released from apoptotic cells) is a pro-inflammatory signal (89). Therefore, one might find increased inflammation in tumors where the apoptosis-autophagy-necrosis death continuum is perturbed. Indeed in the hypoxic region of a tumor that has both apoptosis and autophagy inhibited, increased necrosis with infiltration of macrophages and production of cytokines and chemokines has been observed in mouse models (88). In addition some forms of apoptosis lead to a more or less immunogenic cell death (90, 91) that can affect the efficiency of tumor regression through a “bystander effect” whereby the immune system attacks the tumor too. However, sustained inflammation e.g. after necrotic cell death with release of HMGB1 may also promote tumor growth (92). Manipulation of these processes to increase the anti-tumor immune response while limiting the inflammatory signals that promote tumor growth could perhaps allow one to optimize the anti-tumor response (93). Manipulation of particular regulators (Atg proteins, Bcl family members and the like) to skew the arrows connecting the various states in the schematic shown in Fig. 4 and thus control the amount of apoptosis, autophagy and necrosis that takes place in the tumor may provide a way to do this. Of course, similar concepts can be applied to other pathological conditions that involve perturbation of cell death. To achieve this goal, we need to better understand the mechanisms that were discussed here and uncover the other interactions between these supposedly separate processes that are surely out there.
Figure 3. Regulation of apoptosis by Atg 5.
The autophagy regulator Atg 5 induces caspase activation by interacting with the adaptor protein FADD, a component of the extrinsic apoptosis pathway. Alternatively, cleavage of Atg 5 by calpain causes the truncated Atg 5 protein to translocate to the mitochondria to induce MOMP resulting in cytochrome c release and caspase activation.
Acknowledgements
Research in my laboratory is supported by grants from the NCI. I thank David Virshup for helpful comments on the manuscript.
References
- 1.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007 doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 2.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 3.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 5.Cuervo AM. Autophagy: in sickness and in health. Trends Cell Biol. 2004;14:70–77. doi: 10.1016/j.tcb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 7.Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 8.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 9.Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat Rev Microbiol. 2004;2:301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 11.Liang XH, Yu J, Brown K, Levine B. Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res. 2001;61:3443–3449. [PubMed] [Google Scholar]
- 12.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathew R, Kongara S, Beaudoin B, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007 doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karantza-Wadsworth V, Patel S, Kravchuk O, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boya P, Gonzalez-Polo RA, Casares N, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lum JJ, Bauer DE, Kong M, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 18.Ravikumar B, Berger Z, Vacher C, O'Kane CJ, Rubinsztein DC. Rapamycin pre-treatment protects against apoptosis. Hum Mol Genet. 2006;15:1209–1216. doi: 10.1093/hmg/ddl036. [DOI] [PubMed] [Google Scholar]
- 19.Colell A, Ricci JE, Tait S, et al. GAPDH and Autophagy Preserve Survival after Apoptotic Cytochrome c Release in the Absence of Caspase Activation. Cell. 2007;129:983–997. doi: 10.1016/j.cell.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 20.Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–2906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- 21.Tsujimoto Y, Shimizu S. Another way to die: autophagic programmed cell death. Cell Death Differ. 2005;12(Suppl 2):1528–1534. doi: 10.1038/sj.cdd.4401777. [DOI] [PubMed] [Google Scholar]
- 22.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu L, Wan F, Dutta S, et al. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci U S A. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luthi AU, Martin SJ. The CASBAH: a searchable database of caspase substrates. Cell Death Differ. 2007;14:641–650. doi: 10.1038/sj.cdd.4402103. [DOI] [PubMed] [Google Scholar]
- 25.Shimizu S, Kanaseki T, Mizushima N, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 26.Yu L, Alva A, Su H, et al. Regulation of an ATG7-beclin 1 Program of Autophagic Cell Death by Caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 27.Galluzzi L, Maiuri MC, Vitale I, et al. Cell death modalities: classification and pathophysiological implications. Cell Death Differ. 2007;14:1237–1243. doi: 10.1038/sj.cdd.4402148. [DOI] [PubMed] [Google Scholar]
- 28.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meijer AJ, Codogno P. Regulation and role of autophagy in mammalian cells. Int J Biochem Cell Biol. 2004;36:2445–2462. doi: 10.1016/j.biocel.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 31.Scott RC, Juhasz G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multi-domain modulator of autophagy. J Biol Chem. 2007 doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- 33.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 34.Liang C, Feng P, Ku B, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–698. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 35.Maria Fimia G, Stoykova A, Romagnoli A, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi Y, Coppola D, Matsushita N, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohsumi Y, Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol. 2004;15:231–236. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 39.Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 40.Jager S, Bucci C, Tanida I, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 41.Gonzalez-Polo RA, Boya P, Pauleau AL, et al. The apoptosis/autophagy paradox: autophagic vacuolization before apoptotic death. J Cell Sci. 2005;118:3091–3102. doi: 10.1242/jcs.02447. [DOI] [PubMed] [Google Scholar]
- 42.Kanzawa T, Germano IM, Komata T, Ito H, Kondo Y, Kondo S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004;11:448–457. doi: 10.1038/sj.cdd.4401359. [DOI] [PubMed] [Google Scholar]
- 43.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams JM. Ways of dying: multiple pathways to apoptosis. Genes Dev. 2003;17:2481–2495. doi: 10.1101/gad.1126903. [DOI] [PubMed] [Google Scholar]
- 45.Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- 46.Green DR, Kroemer G. The pathophysiology of mitochondrial cell death. Science. 2004;305:626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 47.Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 48.Youle RJ. Cell biology. Cellular demolition and the rules of engagement. Science. 2007;315:776–777. doi: 10.1126/science.1138870. [DOI] [PubMed] [Google Scholar]
- 49.Hacker G, Weber A. BH3-only proteins trigger cytochrome c release, but how? Arch Biochem Biophys. 2007;462:150–155. doi: 10.1016/j.abb.2006.12.022. [DOI] [PubMed] [Google Scholar]
- 50.Green DR. At the gates of death. Cancer Cell. 2006;9:328–330. doi: 10.1016/j.ccr.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 51.Li P, Nijhawan D, Budihardjo I, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 52.Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 53.Verhagen AM, Ekert PG, Pakusch M, et al. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102:43–53. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- 54.Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 55.Thorburn A. Death Receptor-induced cell killing. Cellular Signalling. 2004;16:139–144. doi: 10.1016/j.cellsig.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 56.Chinnaiyan AM, O'Rourke K, Tewari M, Dixit VM. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 57.Thomas LR, Henson A, Reed JC, Salsbury FR, Thorburn A. Direct binding of FADD to the TRAIL receptor DR5 is regulated by the death effector domain of FADD. J Biol Chem. 2004;279:32780–32785. doi: 10.1074/jbc.M401680200. [DOI] [PubMed] [Google Scholar]
- 58.Thomas LR, Johnson RL, Reed JC, Thorburn A. The C-terminal tails of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and Fas receptors have opposing functions in Fas associated death domain (FADD) recruitment and can regulate agonist-specific mechanisms of receptor activation. J Biol Chem. 2004;279:52479–52486. doi: 10.1074/jbc.M409578200. [DOI] [PubMed] [Google Scholar]
- 59.Boatright KM, Renatus M, Scott FL, et al. A unified model for apical caspase activation. Mol Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 60.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 61.Boyce M, Yuan J. Cellular response to endoplasmic reticulum stress: a matter of life or death. Cell Death Differ. 2006;13:363–373. doi: 10.1038/sj.cdd.4401817. [DOI] [PubMed] [Google Scholar]
- 62.Zong WX, Li C, Hatzivassiliou G, et al. Bax and Bak can localize to the endoplasmic reticulum to initiate apoptosis. J Cell Biol. 2003;162:59–69. doi: 10.1083/jcb.200302084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boya P, Cohen I, Zamzami N, Vieira HL, Kroemer G. Endoplasmic reticulum stress-induced cell death requires mitochondrial membrane permeabilization. Cell Death Differ. 2002;9:465–467. doi: 10.1038/sj.cdd.4401006. [DOI] [PubMed] [Google Scholar]
- 64.Boya P, Gonzalez-Polo RA, Poncet D, et al. Mitochondrial membrane permeabilization is a critical step of lysosome-initiated apoptosis induced by hydroxychloroquine. Oncogene. 2003;22:3927–3936. doi: 10.1038/sj.onc.1206622. [DOI] [PubMed] [Google Scholar]
- 65.Kroemer G, Jaattela M. Lysosomes and autophagy in cell death control. Nat Rev Cancer. 2005 doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- 66.Wang L, Yu C, Lu Y, et al. TMEM166, a novel transmembrane protein, regulates cell autophagy and apoptosis. Apoptosis. 2007;12:1489–1502. doi: 10.1007/s10495-007-0073-9. [DOI] [PubMed] [Google Scholar]
- 67.Crighton D, Wilkinson S, O'Prey J, et al. DRAM, a p53-Induced Modulator of Autophagy, Is Critical for Apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 68.Arico S, Petiot A, Bauvy C, et al. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem. 2001;276:35243–35246. doi: 10.1074/jbc.C100319200. [DOI] [PubMed] [Google Scholar]
- 69.Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 70.Zhu W, Cowie A, Wasfy GW, Penn LZ, Leber B, Andrews DW. Bcl-2 mutants with restricted subcellular location reveal spatially distinct pathways for apoptosis in different cell types. Embo J. 1996;15:4130–4141. [PMC free article] [PubMed] [Google Scholar]
- 71.Maiuri MC, Le Toumelin G, Criollo A, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. Embo J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282:13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 73.Erlich S, Mizrachy L, Segev O, et al. Differential Interactions Between Beclin 1 and Bcl-2 Family Members. Autophagy. 2007:3. doi: 10.4161/auto.4713. [DOI] [PubMed] [Google Scholar]
- 74.Feng W, Huang S, Wu H, Zhang M. Molecular Basis of Bcl-xL's Target Recognition Versatility Revealed by the Structure of Bcl-xL in Complex with the BH3 Domain of Beclin-1. J Mol Biol. 2007;372:223–235. doi: 10.1016/j.jmb.2007.06.069. [DOI] [PubMed] [Google Scholar]
- 75.Hoyer-Hansen M, Bastholm L, Szyniarowski P, et al. Control of Macroautophagy by Calcium, Calmodulin-Dependent Kinase Kinase-beta, and Bcl-2. Mol Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 76.Thorburn J, Moore F, Rao A, et al. Selective Inactivation of a FADD-dependent Apoptosis and Autophagy Pathway in Immortal Epithelial Cells. Mol Biol Cell. 2005;16:1189–1199. doi: 10.1091/mbc.E04-10-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Park KJ, Lee SH, Kim TI, et al. A human scFv antibody against TRAIL receptor 2 induces autophagic cell death in both TRAIL-sensitive and TRAIL-resistant cancer cells. Cancer Res. 2007;67:7327–7334. doi: 10.1158/0008-5472.CAN-06-4766. [DOI] [PubMed] [Google Scholar]
- 78.Pyo JO, Jang MH, Kwon YK, et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280:20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- 79.Yousefi S, Perozzo R, Schmid I, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 80.Demarchi F, Bertoli C, Copetti T, et al. Calpain is required for macroautophagy in mammalian cells. J Cell Biol. 2006;175:595–605. doi: 10.1083/jcb.200601024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 82.Hippert MM, O'Toole PS, Thorburn A. Autophagy and cancer: good bad or both? Cancer Research. 2006;66:9349–9351. doi: 10.1158/0008-5472.CAN-06-1597. [DOI] [PubMed] [Google Scholar]
- 83.Baehrecke EH. Autophagic programmed cell death in Drosophila. Cell Death Differ. 2003;10:940–945. doi: 10.1038/sj.cdd.4401280. [DOI] [PubMed] [Google Scholar]
- 84.Lee CY, Cooksey BA, Baehrecke EH. Steroid regulation of midgut cell death during Drosophila development. Dev Biol. 2002;250:101–111. doi: 10.1006/dbio.2002.0784. [DOI] [PubMed] [Google Scholar]
- 85.Martin DN, Baehrecke EH. Caspases function in autophagic programmed cell death in Drosophila. Development. 2004;131:275–284. doi: 10.1242/dev.00933. [DOI] [PubMed] [Google Scholar]
- 86.Green DR, Evan GI. A matter of life and death. Cancer Cell. 2002;1:19–30. doi: 10.1016/s1535-6108(02)00024-7. [DOI] [PubMed] [Google Scholar]
- 87.Amaravadi RK, Yu D, Lum JJ, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Degenhardt K, Mathew R, Beaudoin B, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 90.Casares N, Pequignot MO, Tesniere A, et al. Caspase-dependent immunogenicity of doxorubicin-induced tumor cell death. J Exp Med. 2005;202:1691–1701. doi: 10.1084/jem.20050915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Obeid M, Tesniere A, Ghiringhelli F, et al. Calreticulin exposure dictates the immunogenicity of cancer cell death. Nat Med. 2007;13:54–61. doi: 10.1038/nm1523. [DOI] [PubMed] [Google Scholar]
- 92.Vakkila J, Lotze MT. Inflammation and necrosis promote tumour growth. Nat Rev Immunol. 2004;4:641–648. doi: 10.1038/nri1415. [DOI] [PubMed] [Google Scholar]
- 93.Lake RA, van der Most RG. A better way for a cancer cell to die. N Engl J Med. 2006;354:2503–2504. doi: 10.1056/NEJMcibr061443. [DOI] [PubMed] [Google Scholar]