Abstract
Plant stem-cell pools, the source for all organs, are first established during embryogenesis. It has been known for decades that cytokinin and auxin interact to control organ regeneration in cultured tissue1. Auxin plays a critical role for root-stem cell specification in zygotic embryogenesis2,3, but the early embryonic function of cytokinin is obscure4-6. Here, we introduce a synthetic reporter to universally visualise cytokinin output in vivo. Surprisingly, the first embryonic signal is detected in the hypophysis, the founder cell of the root stem-cell system. Its apical daughter cell, precursor of the quiescent centre, maintains phosphorelay activity, whereas the basal daughter cell represses signalling output. Intriguingly, auxin activity levels exhibit the inverse profile. We show that auxin antagonizes cytokinin output in the basal cell-lineage by direct transcriptional activation of ARABIDOPSIS RESPONSE REGULATOR (ARR) genes, ARR7 and ARR15, feedback repressors of cytokinin signalling. Loss of ARR7 and ARR15 function or ectopic cytokinin signalling in the basal cell during early embryogenesis results in a defective root-stem cell system. The results provide a molecular model of transient and antagonistic interaction between auxin and cytokinin critical for specifying the first root-stem cell niche.
Cytokinins are adenine-derived signalling molecules that play many essential roles in postembryonic growth and development. However, the role of cytokinin signalling in early embryogenesis remained unclear4-6. To visualise cytokinin’s signalling output in vivo, we aimed at designing a synthetic reporter that overcomes the limitations of current reporters, typically immediate-early cytokinin target genes. The discrete expression patterns of these markers7,8 indicate that they integrate unknown secondary input that reflects cytokinin-independent regulation.
Cytokinin signalling is mediated by a multistep two-component circuitry through histidine and aspartate phosphorelay9. Nuclear B-type response regulators (RRs) mediate transcriptional activation in response to phosphorelay signalling activity, while A-type RRs repress signalling in a negative-feedback loop. The DNA-binding domains of diverse B-type RR family members are conserved and bind a common DNA-target sequence (A/G)GAT(T/C) in vitro10-12. This motif is significantly enriched in the cis-regulatory region of immediate-early cytokinin target genes13, suggesting its in vivo relevance. To generate a universal cytokinin reporter we tested and optimised synthetic reporter designs using luciferase (LUC) activity in Arabidopsis mesophyll protoplast assays14,15. The resulting synthetic reporter TCS-LUC (Two-Component-output-Sensor) harboured the concatemerised B-type ARR-binding motifs10-12 and a minimal 35S promoter14. Only cytokinins activated TCS-LUC, while other plant hormones, auxin, abscisic acid and gibberellic acid, had no effect (Fig. 1a, b). All three known cytokinin receptors contributed to its cytokinin-dependent induction in vivo, as cells isolated from double cytokinin receptor mutants were compromised in their ability to induce TCS-LUC expression (Fig.1c). The extent of this reduction was correlated with the in planta contribution of the different receptors6. B-type ARR family members promoted strong TCS-LUC induction in a co-transfection assay14 (Fig. 1d). Conversely, co-expression of A-type ARR family members inhibited cytokinin-dependent TCS-LUC activity (Fig. 1e). TCS-LUC displayed concentration-dependent activation by cytokinin from as low as 100 pM up to about 1 μM. Furthermore, TCS mediated significantly higher induction compared to the native ARR6 promoter14 (Fig. 1f). Addition of a viral translational enhancer (Ω)16 amplified the response further (Fig. 1f). Taken together, TCS-LUC could specifically report even low levels of phosphorelay output triggered by any of the three endogenous cytokinin receptors and relayed to any response regulator tested.
Figure 1. Sensitive and specific response of TCS.

a, 100 nM trans-zeatin (tZ) induces TCS-LUC, not 1 μM auxin (NAA), 100μM abscisic acid (ABA), or 50 μM gibberellic acid (GA). TCS*-LUC negative control. b, TCS-LUC induced by cZ, cis-zeatin; oT, ortho-topolin; TDZ, thidiazuron; iP, N6-(Δ2-isopentenyl)adenine; AD, adenine (negative control), all at 100 nM. c–e, TCS-LUCinduction by tz (c) reduced in double-mutant ahk2–2, ahk3–3, cre1–16 Combinations, (d) stimulated by B-type ARRs, (e) reduced by A-type ARRs. f, Dose responses. g–k, Embryonic TCS∷GFP activity. s, suspensor; hy, hypophysis; lsc, lens-shaped cell; bc, basal cell lineage; qc, quiescent centre. Error bars s.d. (n=3), scale bars 10μM.
To determine the expression pattern in planta, we generated transgenic Arabidopsis plants carrying the green fluorescent protein (GFP) reporter controlled by the TCS synthetic promoter. The activity of TCS∷GFP in the seedling was consistent with cytokinin actions previously documented, for example in cotyledons7 (Supplementary Fig. 1b), shoot meristem7,17 (Supplementary Fig. 1c), root tip7,17,18 and root vasculature19 (Supplementary Fig. 1d), as well as in emerging lateral root base and primordia20 (Supplementary Fig. 1e, f). Seedlings that were subjected to a short-term incubation with the cytokinin-synthesis inhibitor lovastatin21 abolished TCS∷GFP expression (Supplementary Fig. 2b,e) Importantly,TCS∷GFP expression was restored by the co-administration of a cytokinin together with lovastatin (Supplementary Fig. 2c, f). The analyses validated the physiological response of the novel synthetic cytokinin reporter in intact plants.
To uncover new roles of phosphorelay signalling in vivo, we followed TCS∷GFP expression during early embryogenesis. Since cytokinins have long been implicated in shoot regeneration1, we were surprised to detect the first distinct signal in the founder of the root-stem cells, the hypophysis, at the 32-cell stage (Fig. 1h). By the transition stage, the hypophysis has undergone an asymmetrical cell division. Intriguingly, the resulting large basal daughter cell and its descendants repressed TCS∷GFP expression, while the apical lens-shape cell retained its expression (Fig. 1j). By the heart stage, a second phosphorelay output had appeared near the shoot stem-cell primordium (Fig. 1k).
To identify the signalling components involved in embryonic phosphorelay activity, we determined the transcription levels of all the two-component genes at the transition stage. A subset of candidates for each signalling step was transcribed (Fig. 2a). We were especially interested in the expression patterns of the A-type ARRs, as they represent direct targets commonly used as markers of cytokinin signalling. ARR7 mRNA was abundant at the transition stage. ARR7∷GFP transgenic lines displayed first GFP activity at the late globular stage, just after the asymmetric division of the hypophysis. Surprisingly, ARR7∷GFP activity was high in the basal daughter cell but lower in the lens shape cell and its descendants (Figs 2d and 3c), inverse to TCS∷GFP levels (Figs 2c and 3a). mRNA in-situ hybridisation confirmed a similar endogenous ARR7 expression pattern (Fig. 3s). ARR15, the sister gene of ARR7, is expressed in the comparable domain, albeit at much lower levels (Figs 2a, e and 3d). Therefore, ARR7 and ARR15 expression only partially reflected the phosphorelay output (compare Fig. 2d, e with Fig. 2c and Fig. 3c, d, s with Fig. 3a). The results suggested that additional input might control ARR7 and ARR15 expression.
Figure 2. Inverse correlation between cytokinin and auxin signalling.

a, Two-component gene transcription by quantitative real time reverse transcriptase polymerase chain reaction (qRT-PCR) at transition stage. Error bars s.d. (n=3). b, TCS∷GFP in hy at early globular stage. c, Downregulation in bc at late globular stage. d, First ARR7∷GFP expression peaks in bc. e, ARR15∷GFP expression. f, DR5∷GFP activity highest in bc. Zoomed view in middle row. Schematic interpretation in bottom row. NGE, normalized gene expression; HK/AHK, Arabidopsis histidine kinases; HPT/AHP, histidine-phosphotransfer proteins; RR/ARR, response regulators; CKI1, cytokinin independent 1; ETR1, ethylene response 1; ERS1, ethylene response sensor 1.
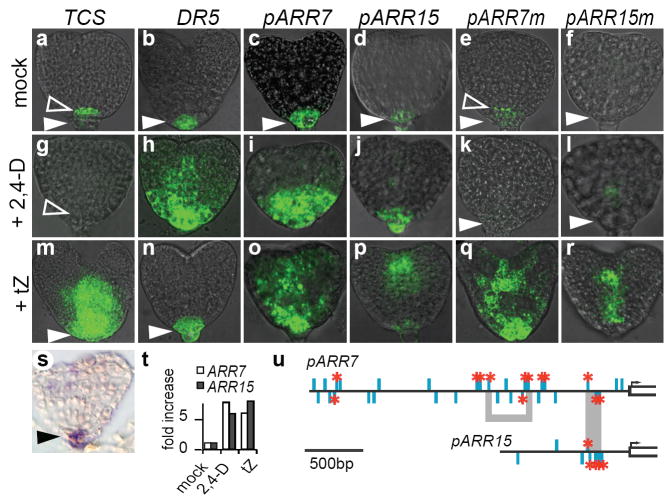
Figure 3. ARR7 and ARR15 mediate auxin control of cytokinin signalling.
a–r, Over-night treatment with water, 50 μM 2,4-D or tZ. pARR7m (e, k, q) pARR15m (f, l, r) explained in (u). Filled arrowheads denote bc, open arrowheads qc. s, In situ hybridisation to ARR7 mRNA. t, ARR7 and ARR15 mRNA induction assayed by qRT-PCR after 4-hour treatment of embryos with 1 μM tZ or 2,4-D. u, ARR7 and ARR15 promoters. TGTC on sense and antisense strands as blue bars. Red asterisks: TGTC -> TGgC. Grey shade connects conserved regions; see text for detail.
Interestingly, auxin signalling output visualised by the synthetic reporter DR5∷GFP2 was highest in the hypophysis-derived basal cell (Fig. 2f), similar to ARR7 and ARR15 expression. This raised the possibility that auxin could induce transcription of the cytokinin repressors ARR7 and ARR15, that in turn prevented phosphorelay output in the basal cell lineage (Fig. 4m). To test the effect of ectopic auxin signalling, TCS∷GFP, DR5∷GFP, ARR7∷GFP and ARR15∷GFP transgenic embryos were treated with the synthetic auxin analogue 2,4-D2 (Fig. 3g–l). Incubation with 2,4-D caused expansion of the DR5∷GFP expression domain (Fig. 3h)2. The levels of ARR7∷GFP and ARR15∷GFP expression also increased (Fig. 3i, j, t) and their domain widened (Fig. 3i, j), while TCS∷GFP activity was abolished (arrowhead, Fig. 3g). Exogenous cytokinin caused a broad expansion of TCS∷GFP expression (Fig. 3m), but left DR5∷GFP expression unaffected (Fig. 3n). Both ARR7∷GFP and ARR15∷GFP expression was increased (Fig. 3o, p, t), reflecting their documented status as direct cytokinin target genes17. These experiments revealed that, besides cytokinin, auxin regulated ARR7 and ARR15 transcription, and supported a model where high endogenous auxin activity suppressed cytokinin output via stimulation of ARR7 and ARR15 transcription (Fig. 4m).
Figure 4. Function of differential phosphorelay output for root-stem cell establishment.

a–l, Embryos are arr15, RPS5a∷AlcA AlcR∷ARR7-i. a–f, Control embryos, no ethanol. g–l, Embryos after ARR7-i induction in the arr15 mutant background. Induction for 10 h (g), 36 h (h) and 60 h (i–l). d–f, j–l, In situ hybridisations. Artificial colours (c) denote stem-cell identity, with qc in pink. Cells shaded in grey (i) have unclear identity. Filled arrowheads point to bc, open arrowheads to qc (d–f, j–l), or missing qc (i–l). m, Model for auxin-dependent phosphorelay downregulation in bc. Scale bars 10 μm.
To explore the possibility that auxin signalling directly induced transcription of ARR7 and ARR15, we analysed their cis-regulatory regions for motifs that might mediate auxin input. Auxin response elements (AuxRE) have been defined as TGTCTC, however, careful in vitro analysis suggested that only the first four nucleotides are essential for auxin response factor (ARF) binding22. Therefore, we screened the promoters of ARR7 and ARR15 for the TGTC motif. We found 32 occurrences for ARR7 and 8 for ARR15 (Fig. 3u). Previously characterised functional AuxRE have been categorized as “simple” (defined by repetitive motifs) or “composite” where the motif is flanked by a cofactor-binding site23. These criteria guided us in focusing on putative functional TGTC hits. Point mutations shown to abolish ARF binding22 were introduced specifically in TGTC motifs occurring at least twice in a 30bp window or flanked by sequence conserved within or between the ARR7 and ARR15 promoters (Fig. 3u). The resulting mutated reporters ARR7m∷GFP and ARR15m∷GFP showed strongly reduced expression in the auxin signalling domain (filled arrowhead Fig. 3e, f). Furthermore, ectopic auxin signalling was unable to stimulate their expression (arrowheads Fig. 3k, l). In contrast, they retained responsiveness to cytokinin (Fig. 3q, r). Intriguingly, uncoupled from auxin input, ARR7m∷GFP showed an expression pattern similar to that contributed by the cytokinin reporter TCS∷GFP (compare Fig 3e with Fig. 3a). Consequently, exogenous auxin application caused repression of ARR7m∷GFP (Fig. 3k, l), presumably due to higher endogenous ARR7 and ARR15 expression (Fig. 3i, j, t) that prevents cytokinin response (Fig. 3g). The results suggested that auxin signalling directly induces transcription of ARR7 and ARR15 via conserved TGTC elements. The sensitivity of the ARR7 and ARR15 promoters to auxin seemed to be confined to early embryogenesis as expression of ARR7∷GFP and ARR15∷GFP in the root tip was undetectable by the upturned-U stage (Supplementary Fig. 3), whereas localised auxin signalling persisted.
What is the function of ARR7 and ARR15, expressed early in embryogenesis under the control of auxin? No embryo defect was observed in the arr7 or arr15 single mutants (ref. 8,Supplementary Fig. 6a–f, Fig. 4a–f, and data not shown). The arr7 arr15 double mutants were reported to cause female gamethophytic lethality8, precluding analysis of embryonic function. Therefore, we generated conditional double loss-of-function arr7 arr15 embryos by expressing an ethanol-inducible24 RNA interference construct against ARR7 (ARR7-i) (Supplementary Fig. 4) in arr15 background with or without the TCS∷GFP reporter (Fig. 4). Control experiments were performed with single arr15 mutant embryos carrying un-induced ARR7-i (Fig. 4a–f) and ethanol-induced ARR7-i mutant embryos (Supplementary Fig. 6a–f). Ten hours after ARR7-i transgene induction in arr15 embryos, ectopic phosphorelay output, revealed by TCS∷GFP expression, was observed in the basal cell lineage (Fig. 4g). After 36 hours, in addition to ectopic cytokinin signalling, cell shapes and number became irregular (Fig. 4h). Later, after 60 hours, the morphology of the root stem-cell system was severely distorted (Fig. 4i–l), and the attribution of stem-cell identity based on shape and position was ambiguous in the double mutant (Fig. 4i). Furthermore, the expression of key transcription factors required for root stem cell specification and function, SCARECROW (SCR)25, PLETHORA1 (PLT1)26 and WUSCHEL-RELATED-HOMEOBOX5 (WOX5)27, was abolished or severely reduced (Fig. 4j–l). Eventually, embryo development arrested (not shown). The single mutant control embryos (Fig. 4a–f, Supplementary Fig. 6a–f), did not show any of these phenotypes. These results suggested that loss of both ARR7 and ARR15 causes ectopic cytokinin signalling in the basal cell lineage (Fig. 4g), which interferes with the stereotypical cell division pattern (Fig. 4h) and prevents the establishment of normal embryonic pattern, in particular the root stem cell system, as judged by morphology (Fig. 4i–l) and expression of key marker genes (Fig. 4j–l).
To test whether directly activating cytokinin signalling in the basal cell lineage also affects stem cell development, we used the DR5 promoter to direct the expression of a constitutively active variant of the B-type ARR10, most abundantly expressed in early embryos (Fig. 2a), in auxin-signalling cells. Mutation of the aspartate residue at position 69 to glutamate (D69E) mimics the phosphorylated, active state of ARR1028. Indeed, early embryonic expression of ARR10D69E in auxin-maximum cells resulted in a phenotype comparable to loss of ARR7 and ARR15 function (Supplementary Fig. 6g). Finally, we addressed the requirement of phosphorelay signalling in early embryo development. It has been reported that mutations in three cytokinin receptors had no obvious effect on embryonic pattern formation5-7. Residual, or phosphorelay activity independent of known cytokinin receptors might still occur in these conditions. We chose to dominantly interfere with transcriptional activation executed by B-type ARR proteins and converted the abundant (Fig. 2a) positive regulator ARR10 into a potent dominant-acting transcriptional repressor by adding an EAR repression domain29 (Supplementary Fig. 5). Induced ubiquitous expression of ARR10-EAR in early globular embryos led to strong pattern defects (Supplementary Fig. 6h). As the lens-shaped cell is prominently marked by TCS∷GFP expression during early embryogenesis (Fig 2c), the result suggested its phosphorelay activity is also important for stem cell specification. Interestingly, manipulations of cytokinin signalling initiated later, at embryonic heart stage, had no obvious effect on root stem cell organization (Supplementary Fig. 7). Thus, differential phosphorelay output seems transiently required for successful development of the hypophysis-derived daughter cells into an operational root stem-cell system (Fig. 4m).
By combining a new visualisation tool and inducible genetic manipulations, we have uncovered a locally and temporally defined antagonistic interaction between auxin and cytokinin that controls the establishment of the first root stem cell niche. In the prevailing view, A-type RRs such as ARR7 and ARR15 act in the negative feedback loop to cytokinin signalling. As a result, A-type RR levels are in balance with signalling levels, and their expression domains are centred on the pathway output. In contrast, cytokinin activity will be reduced or eliminated where other signals induce A-type RR. Thus, gaining control of feedback regulators represents a simple yet effective mechanism to define the output domain of other pathways, and enables dynamic and quantitative interactions among signalling pathways to promote the complex plant developmental programs.
METHODS SUMMARY
Plasmid constructs
TCS contains 6 direct repeats of aaAATCTacaaAATCTttttGGATT-ttgtGGATTttctagc (core B-type ARR pentamers10-12 in capital letters), negative control TCS* has 6 repeats of aaAATgTacaaAATgTttttGcATTttg-tGcATTttctagc. TCS was cloned in front of a minimal 35S promoter with a TATA box30, followed by the LUC, ΩLUC or ΩGFP30 coding regions. The integrated reporter construct with the Ω translational enhancer16 was designated TCS∷GFP. The constructs, RPS5a-AlcR/AlcA-ARR7-i and DR5rev-AlcR/AlcA-ARR10D69E, were cloned based on a binary vector received from E. Lam (personal communication). For the ARR7-i construct, the first exon and intron of the ARR7 gene were cloned in sense orientation followed by the first exon in antisense orientation.
Plant material and treatment
Plants were Columbia background and grown at 12 h light/23 °C and 12 h dark/20 °C cycle. In vitro embryo culture was performed as described in ref. 2. Ethanol (0.5-1.0%) with no effect on normal embryogenesis was used to induce transgene expression. The arr15 (WISCDSLOX334D02) mutant harbours a T-DNA insertion in the first exon of the ARR15 gene, after bp 73 from the translation start (details in Supplementary Fig. 8). The corresponding seed stock CS851593 was obtained from the Arabidopsis Biological Resource Center (ABRC, USA). Arabidopsis protoplasts were isolated and transfected as described in refs. 14,15.
Quantitative RT-PCR
RNA was extracted and amplified from ten pooled embryos, using PicoPure RNA isolation and RiboAMP RNA amplification kits (Arcturus, Mountain View, CA, USA). For expression in wild-type transition-stage embryos, three biological replicates were processed. qRT-PR was performed as described in ref. 8. Amplification of EIF-4A and TUB4 served as standards. Primer sequences are provided in Supplementary Information (Table 1).
In situ hybridisation
In situ hybridisation was performed as in ref. 8.
Supplementary Material
Supplementary Information accompanies the paper on www.nature.com/nature.
Acknowledgments
We thank T. Kakimoto and C. Ueguchi for providing ahk mutant seeds, J. Friml for the DR5-GFP plasmid and DR5∷GFP seeds, E. Lam for providing the AlcA/AlcR vector, A. Jazwinska for help with mRNA in-situ hybridisations, and S. Riku for help with plant growth and protoplast experiments. We are grateful to C. Ping and S. Howell, to Y. Guo, Y. Tan and G. Selvaraj, T. Mizuno, J. Zuo; D. Jackson, and to J. To for sharing unpublished results. This work was supported by a Fellowship for Prospective Researchers by the Swiss National Science Foundation (SNF), a Long Term Fellowship of the International Human Frontier Science Program (HFSP) organization to B.M., and grants from the National Science Foundation and National Institutes of Health to J.S.
Footnotes
The authors declare no competing interests.
References
- 1.Skoog F, Miller CO. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol. 1957;54:118–130. [PubMed] [Google Scholar]
- 2.Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 3.Weijers D, Jürgens G. Auxin and embryo axis formation: the ends in sight? Curr Opin Plant Biol. 2005;8:32–37. doi: 10.1016/j.pbi.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Riefler M, Novak O, Strnad M, Schmülling T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell. 2006;18:40–54. doi: 10.1105/tpc.105.037796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell. 2004;16:1365–1377. doi: 10.1105/tpc.021477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higuchi M, et al. In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci U S A. 2004;101:8821–8826. doi: 10.1073/pnas.0402887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.To JP, Haberer G, Ferreira FJ, Deruere J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell. 2004;16:658–671. doi: 10.1105/tpc.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature. 2005;438:1172–1175. doi: 10.1038/nature04270. [DOI] [PubMed] [Google Scholar]
- 9.Müller B, Sheen J. Advances in cytokinin signaling. Science. 2007;318:68–69. doi: 10.1126/science.1145461. [DOI] [PubMed] [Google Scholar]
- 10.Sakai H, Aoyama T, Oka A. Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 2000;24:703–711. doi: 10.1046/j.1365-313x.2000.00909.x. [DOI] [PubMed] [Google Scholar]
- 11.Hosoda K, Imamura A, Katoh E, Hatta T, Tachiki M, Yamada H, Mizuno T, Yamazaki T. Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell. 2002;14:2015–2029. doi: 10.1105/tpc.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imamura A, Kiba T, Tajima Y, Yamashino T, Mizuno T. In vivo and in vitro characterization of the ARR11 response regulator implicated in the His-to-Asp phosphorelay signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 2003;44:122–131. doi: 10.1093/pcp/pcg014. [DOI] [PubMed] [Google Scholar]
- 13.Rashotte AM, Carson SD, To JP, Kieber JJ. Expression profiling of cytokinin action in Arabidopsis. Plant Physiol. 2003;132:1998–2011. doi: 10.1104/pp.103.021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang I, Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413:383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- 15.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 16.Gallie DR. The 5’-leader of tobacco mosaic virus promotes translation through enhanced recruitment of eIF4F. Nucleic Acids Res. 2002;30:3401–3411. doi: 10.1093/nar/gkf457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’Agostino IB, Deruere J, Kieber JJ. Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 2000;124:1706–1717. doi: 10.1104/pp.124.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aloni R, Langhans M, Aloni E, Ullrich CI. Role of cytokinin in the regulation of root gravitropism. Planta. 2004;220:177–182. doi: 10.1007/s00425-004-1381-8. [DOI] [PubMed] [Google Scholar]
- 19.Mähönen AP, et al. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science. 2006;311:94–98. doi: 10.1126/science.1118875. [DOI] [PubMed] [Google Scholar]
- 20.Lohar DP, Schaff JE, Laskey JG, Kieber JJ, Bilyeu KD, Bird DM. Cytokinins play opposite roles in lateral root formation, and nematode and Rhizobial symbioses. Plant J. 2004;38:203–214. doi: 10.1111/j.1365-313X.2004.02038.x. [DOI] [PubMed] [Google Scholar]
- 21.Orchard CB, et al. Tobacco BY-2 cells expressing fission yeast cdc25 bypass a G2/M block on the cell cycle. Plant J. 2005;44:290–299. doi: 10.1111/j.1365-313X.2005.02524.x. [DOI] [PubMed] [Google Scholar]
- 22.Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19:309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- 23.Guilfoyle T, Hagen G, Ulmasov T, Murfett J. How does auxin turn on genes? Plant Physiol. 1998;118:341–347. doi: 10.1104/pp.118.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roslan HA, et al. Characterization of the ethanol-inducible alc geneexpression system in Arabidopsis thaliana. Plant J. 2001;28:225–235. doi: 10.1046/j.1365-313x.2001.01146.x. [DOI] [PubMed] [Google Scholar]
- 25.Sabatini S, Heidstra R, Wildwater M, Scheres B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 2003;17:354–358. doi: 10.1101/gad.252503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aida M, et al. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119:109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 27.Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- 28.Hass C, et al. The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J. 2004;23:3290–3302. doi: 10.1038/sj.emboj.7600337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003;34:733–739. doi: 10.1046/j.1365-313x.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- 30.Ottenschläger I, Wolff P, Wolverton C, Bhalerao RP, Sandberg G, Ishikawa H, Evans M, Palme K. Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc Natl Acad Sci U S A. 2003;100:2987–2991. doi: 10.1073/pnas.0437936100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weijers D, Franke-van Dijk M, Vencken RJ, Quint A, Hooykaas P, Offringa R. An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development. 2001;128:4289–4299. doi: 10.1242/dev.128.21.4289. [DOI] [PubMed] [Google Scholar]
- 32.Xiang C, Han P, Lutziger I, Wang K, Oliver DJ. A mini binary vector series for plant transformation. Plant Mol Biol. 1999;40:711–717. doi: 10.1023/a:1006201910593. [DOI] [PubMed] [Google Scholar]
- 33.Sauer M, Friml J. In vitro culture of Arabidopsis embryos within their ovules. Plant J. 2004;40:835–843. doi: 10.1111/j.1365-313X.2004.02248.x. [DOI] [PubMed] [Google Scholar]
- 34.Laureys F, Dewitte W, Witters E, Van Montagu M, Inze D, Van Onckelen H. Zeatin is indispensable for the G2-M transition in tobacco BY-2 cells. FEBS Lett. 1998;426:29–32. doi: 10.1016/s0014-5793(98)00297-x. [DOI] [PubMed] [Google Scholar]
- 35.Laule O, Furholz A, Chang HS, Zhu T, Wang X, Heifetz PB, Gruissem W, Lange M. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2003;100:6866–6871. doi: 10.1073/pnas.1031755100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information accompanies the paper on www.nature.com/nature.